PixelBNN: Augmenting the PixelCNN with Batch Normalization and the Presentation of a Fast Architecture for Retinal Vessel Segmentation
Abstract
:1. Introduction
2. Material and Methods
2.1. Datasets
2.1.1. DRIVE
2.1.2. STARE
2.1.3. CHASE_DB1
2.2. Preprocessing
2.2.1. Continuous Pixel Space
2.2.2. Image Enhancement
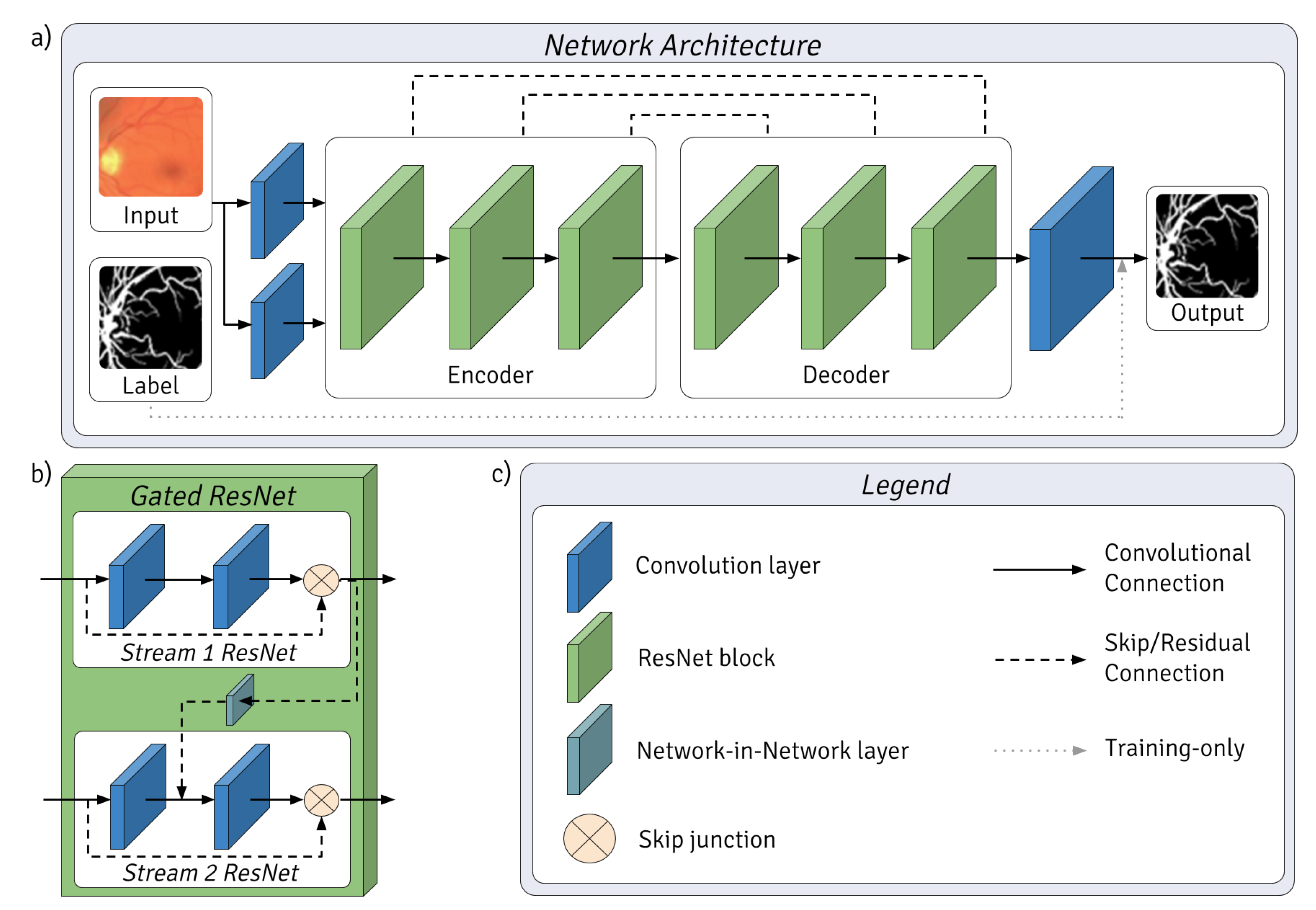
2.3. Network Architecture
- The use of two parallel input streams resembles bipolar cells in the retina, each stream possessing different yet potentially overlapping feature spaces initialized by different convolutional kernels.
- The layer structure was based on that of the lateral geniculate nucleus, visual cortices (V1, V2) and medial temporal Gyrus, whereby each is represented by an encoder–decoder pair of gated ResNet blocks.
- Final classification was executed by a convolutional layer which concatenates the outputs of the final gated ResNet block, as the inferotemporal cortex is believed to do.
2.4. Platform
2.5. Experiment Design
2.6. Performance Indicators
2.7. Training Details
3. Results
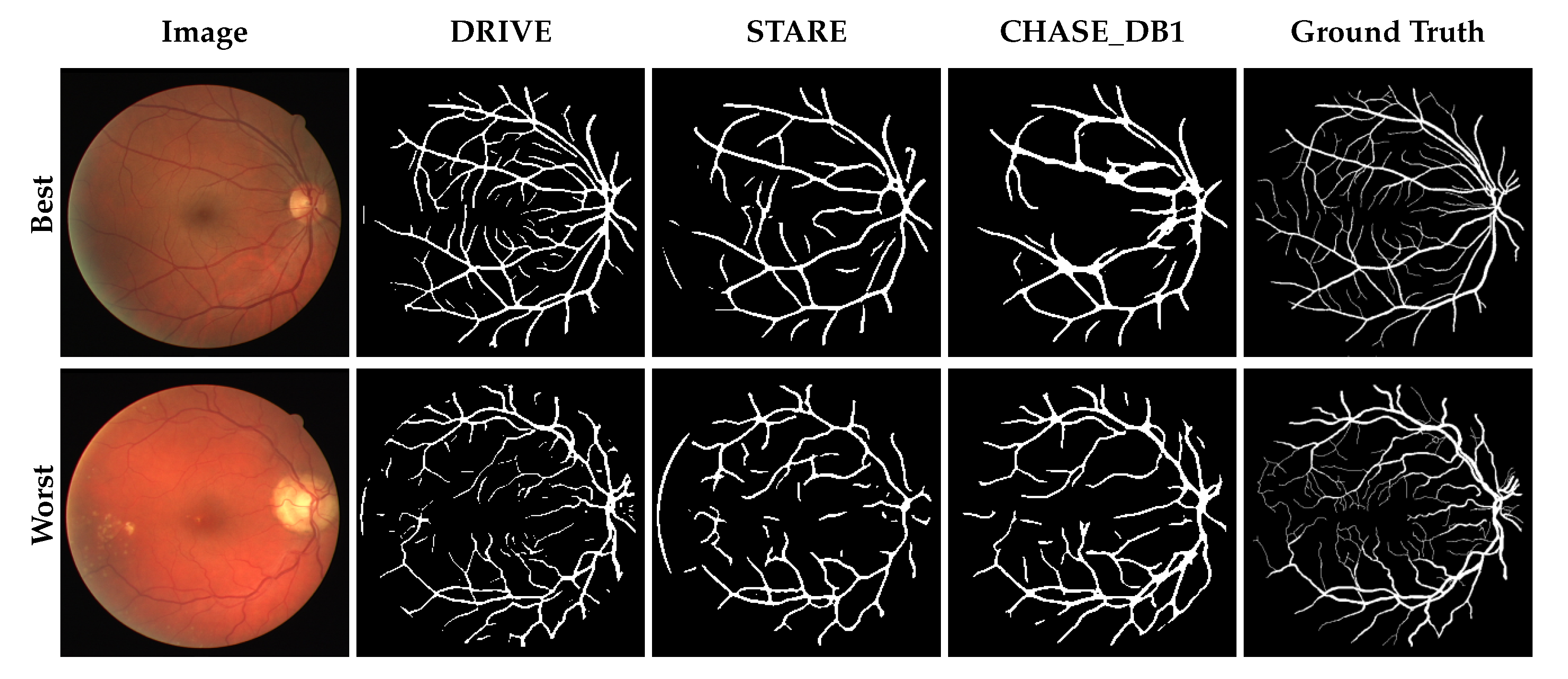
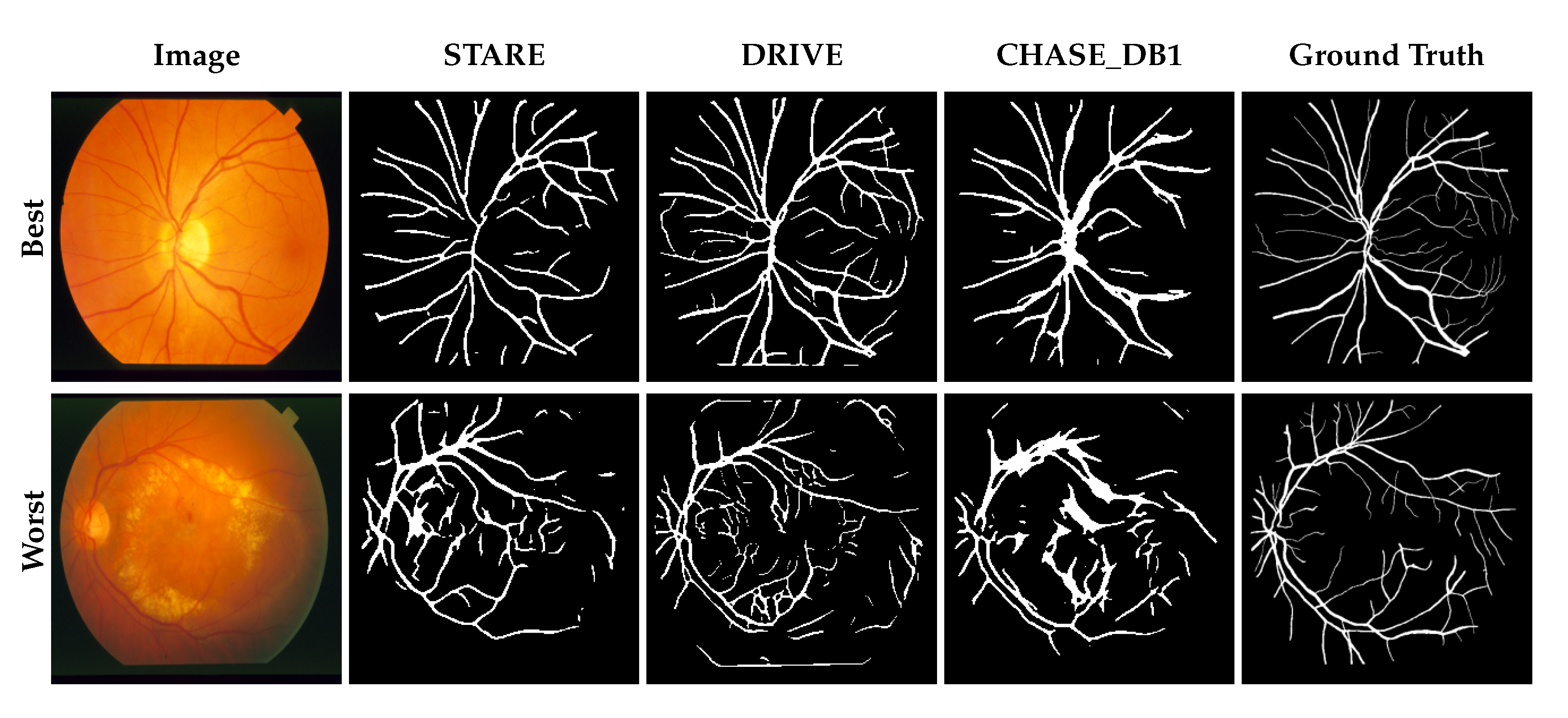
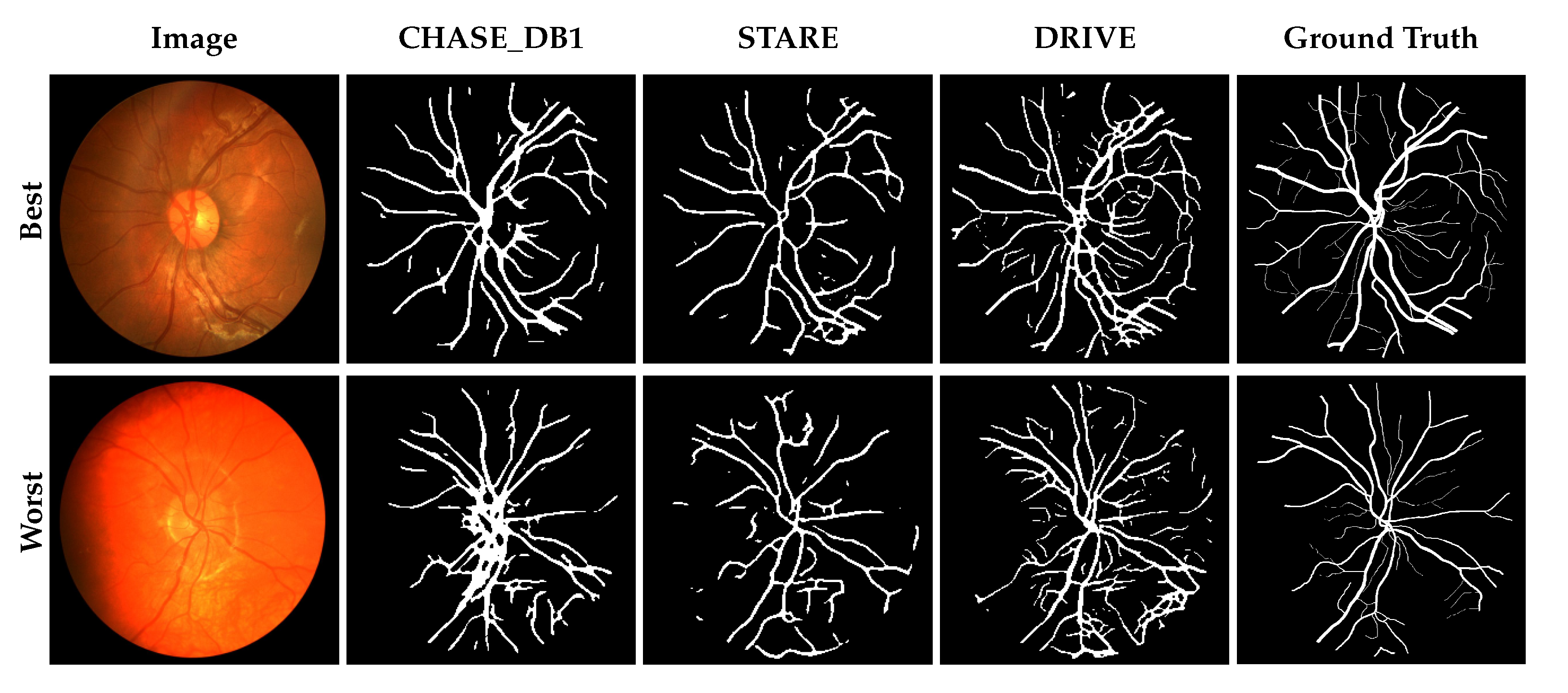
3.1. Performance Comparison
3.2. Computation Time
4. Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Hoover, A.; Kouznetsova, V.; Goldbaum, M. Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response. IEEE Trans. Med. Imaging 2000, 19, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Jelinek, H.; Cree, M.J. Automated Image Detection of Retinal Pathology; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Ricci, E.; Perfetti, R. Retinal Blood Vessel Segmentation Using Line Operators and Support Vector Classification. IEEE Trans. Med. Imaging 2007, 26, 1357–1365. [Google Scholar] [CrossRef] [PubMed]
- Cree, M.J. The Waikato Microaneurysm Detector; Technical Report; The University of Waikato: Hamilton, New Zealand, 2008. [Google Scholar]
- Fraz, M.; Welikala, R.; Rudnicka, A.; Owen, C.; Strachan, D.; Barman, S. QUARTZ: Quantitative Analysis of Retinal Vessel Topology and size—An automated system for quantification of retinal vessels morphology. Expert Syst. Appl. 2015, 42, 7221–7234. [Google Scholar] [CrossRef]
- Abramoff, M.D.; Garvin, M.K.; Sonka, M. Retinal Imaging and Image Analysis. IEEE Rev. Biomed. Eng. 2010, 3, 169–208. [Google Scholar] [CrossRef] [PubMed]
- Fraz, M.M.; Remagnino, P.; Hoppe, A.; Uyyanonvara, B.; Rudnicka, A.R.; Owen, C.G.; Barman, S.A. An Ensemble Classification-Based Approach Applied to Retinal Blood Vessel Segmentation. IEEE Trans. Biomed. Eng. 2012, 59, 2538–2548. [Google Scholar] [CrossRef] [PubMed]
- Azzopardi, G.; Strisciuglio, N.; Vento, M.; Petkov, N. Trainable COSFIRE filters for vessel delineation with application to retinal images. Med Image Anal. 2015, 19, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Fraz, M.; Remagnino, P.; Hoppe, A.; Uyyanonvara, B.; Rudnicka, A.; Owen, C.; Barman, S. Blood vessel segmentation methodologies in retinal images—A survey. Comput. Methods Programs Biomed. 2012, 108, 407–433. [Google Scholar] [CrossRef] [PubMed]
- Soares, J.V.; Leandro, J.J.; Cesar, R.M.; Jelinek, H.F.; Cree, M.J. Retinal vessel segmentation using the 2-D Gabor wavelet and supervised classification. IEEE Trans. Med. Imaging 2006, 25, 1214–1222. [Google Scholar] [CrossRef]
- Almazroa, A.; Burman, R.; Raahemifar, K.; Lakshminarayanan, V. Optic Disc and Optic Cup Segmentation Methodologies for Glaucoma Image Detection: A Survey. J. Ophthalmol. 2015, 2015, 180972. [Google Scholar] [CrossRef]
- Lupascu, C.A.; Tegolo, D.; Trucco, E. FABC: Retinal Vessel Segmentation Using AdaBoost. IEEE Trans. Inf. Technol. Biomed. 2010, 14, 1267–1274. [Google Scholar] [CrossRef]
- Bengio, Y.; Courville, A.; Vincent, P. Representation Learning: A Review and New Perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1798–1828. [Google Scholar] [CrossRef]
- Hinton, G.E.; Osindero, S.; Teh, Y.W. A Fast Learning Algorithm for Deep Belief Nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef] [PubMed]
- Van den Oord, A.; Kalchbrenner, N.; Kavukcuoglu, K. Pixel Recurrent Neural Networks. In Proceedings of the 33rd International Conference on Machine Learning, New York, NY, USA, 19–24 June 2016; Balcan, M.F., Weinberger, K.Q., Eds.; PMLR: New York, NY, USA, 2016; Volume 48, pp. 1747–1756. [Google Scholar]
- Kalchbrenner, N.; van den Oord, A.; Simonyan, K.; Danihelka, I.; Vinyals, O.; Graves, A.; Kavukcuoglu, K. Video Pixel Networks. arXiv, 2016; arXiv:1610.00527. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Chen, H.; Qi, X.; Cheng, J.Z.; Heng, P.A. Deep Contextual Networks for Neuronal Structure Segmentation. In Proceedings of the Thirtieth AAAI Conference on Artificial Intelligence (AAAI’16), Phoenix, AZ, USA, 12–17 February 2016; pp. 1167–1173. [Google Scholar]
- Drozdzal, M.; Vorontsov, E.; Chartrand, G.; Kadoury, S.; Pal, C. The Importance of Skip Connections in Biomedical Image Segmentation. In Deep Learning and Data Labeling for Medical Applications: Proceedings of the First International Workshop, LABELS 2016, and Second International Workshop, DLMIA 2016, Held in Conjunction with MICCAI 2016, Athens, Greece, 21 October 2016; Carneiro, G., Mateus, D., Peter, L., Bradley, A., Tavares, J.M.R.S., Belagiannis, V., Papa, J.P., Nascimento, J.C., Loog, M., Lu, Z., et al., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 179–187. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. In Proceedings of the ICLR 2016 Workshop, San Juan, Puerto Rico, 2–4 May 2016. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Abràmoff, M.D.; Lou, Y.; Erginay, A.; Clarida, W.; Amelon, R.; Folk, J.C.; Niemeijer, M. Improved Automated Detection of Diabetic Retinopathy on a Publicly Available Dataset Through Integration of Deep LearningDeep Learning Detection of Diabetic Retinopathy. Investig. Ophthalmol. Vis. Sci. 2016, 57, 5200. [Google Scholar] [CrossRef] [PubMed]
- Leopold, H.A.; Zelek, J.S.; Lakshminarayanan, V. Deep Learning for Retinal Analysis. In Signal Processing and Machine Learning for Biomedical Big Data; Book Chapter 17; Sejdić, E., Falk, T.H., Eds.; CRC Press: Boca Raton, FL, USA, 2018; pp. 329–367. [Google Scholar]
- Van den Oord, A.; Kalchbrenner, N.; Espeholt, L.; kavukcuoglu, k.; Vinyals, O.; Graves, A. Conditional Image Generation with PixelCNN Decoders. In Advances in Neural Information Processing Systems 29; Lee, D.D., Sugiyama, M., Luxburg, U.V., Guyon, I., Garnett, R., Eds.; Curran Associates, Inc.: Vancouver, BC, Canada, 2016; pp. 4790–4798. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv, 2014; arXiv:1412.6980. [Google Scholar]
- Leopold, H.; Orchard, J.; Lakshminarayanan, V.; Zelek, J. A deep learning network for segmenting retinal vessel morphology. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; p. 3144. [Google Scholar]
- Salimans, T.; Karpathy, A.; Chen, X.; Kingma, D.P.; Bulatov, Y. PixelCNN++: A PixelCNN Implementation with Discretized Logistic Mixture Likelihood and Other Modifications. In Proceedings of the ICLR 2017, Toulon, France, 24–26 April 2017. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 6–11 July 2015; Bach, F., Blei, D., Eds.; PMLR: Lille, France, 2015; Volume 37, pp. 448–456. [Google Scholar]
- Staal, J.; Abràmoff, M.D.; Niemeijer, M.; Viergever, M.A.; van Ginneken, B. Ridge-based vessel segmentation in color images of the retina. IEEE Trans. Med. Imaging 2004, 23, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.D.; Philip, S.; Goatman, K.A.; Olson, J.A.; Sharp, P.F. Automated Assessment of Diabetic Retinal Image Quality Based on Clarity and Field Definition. Investig. Ophthalmol. Vis. Sci. 2006, 47, 1120. [Google Scholar] [CrossRef] [PubMed]
- Kingma, D.P.; Salimans, T.; Jozefowicz, R.; Chen, X.; Sutskever, I.; Welling, M. Improved Variational Inference with Inverse Autoregressive Flow. In Advances in Neural Information Processing Systems 29; Lee, D.D., Sugiyama, M., Luxburg, U.V., Guyon, I., Garnett, R., Eds.; Curran Associates, Inc.: Vancouver, BC, Canada, 2016; pp. 4743–4751. [Google Scholar]
- Szeliski, R. Computer Vision: Algorithms and Applications, 1st ed.; Springer: New York, NY, USA, 2010. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention, Part III, Proceedings of the MICCAI 2015, 18th International Conference, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Shang, W.; Sohn, K.; Almeida, D.; Lee, H. Understanding and Improving Convolutional Neural Networks via Concatenated Rectified Linear Units. In Proceedings of the 33rd International Conference on Machine Learning, New York, NY, USA, 19–24 June 2016; Balcan, M.F., Weinberger, K.Q., Eds.; PMLR: New York, NY, USA; Volume 48, pp. 2217–2225. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. arXiv, 2016; arXiv:1603.04467. [Google Scholar]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Orlando, J.I.; Prokofyeva, E.; Blaschko, M.B. A Discriminatively Trained Fully Connected Conditional Random Field Model for Blood Vessel Segmentation in Fundus Images. IEEE Trans. Biomed. Eng. 2017, 64, 16–27. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Garcia, E.A. Learning from Imbalanced Data. IEEE Trans. Knowl. Data Eng. 2009, 21, 1263–1284. [Google Scholar]
- Glorot, X.; Bordes, A.; Bengio, Y. Deep Sparse Rectifier Neural Networks. In Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics, Fort Lauderdale, FL, USA, 11–13 April 2011; Gordon, G., Dunson, D., Dudík, M., Eds.; PMLR: Fort Lauderdale, FL, USA, 2011; Volume 15, pp. 315–323. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Lam, B.S.Y.; Gao, Y.; Liew, A.W.C. General Retinal Vessel Segmentation Using Regularization-Based Multiconcavity Modeling. IEEE Trans. Med. Imaging 2010, 29, 1369–1381. [Google Scholar] [CrossRef] [PubMed]
- Kovács, G.; Hajdu, A. A self-calibrating approach for the segmentation of retinal vessels by template matching and contour reconstruction. Med. Image Anal. 2016, 29, 24–46. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dashtbozorg, B.; Bekkers, E.; Pluim, J.P.W.; Duits, R.; ter Haar Romeny, B.M. Robust Retinal Vessel Segmentation via Locally Adaptive Derivative Frames in Orientation Scores. IEEE Trans. Med. Imaging 2016, 35, 2631–2644. [Google Scholar] [CrossRef] [PubMed]
- Roychowdhury, S.; Koozekanani, D.D.; Parhi, K.K. Iterative Vessel Segmentation of Fundus Images. IEEE Trans. Biomed. Eng. 2015, 62, 1738–1749. [Google Scholar] [CrossRef] [PubMed]
- Comparative Study of Retinal Vessel Segmentation Methods on a New Publicly Available Database; International Society for Optics and Photonics: San Diego, CA, USA, 2004; Volume 5370.
- Marín, D.; Aquino, A.; Gegúndez-Arias, M.E.; Bravo, J.M. A new supervised method for blood vessel segmentation in retinal images by using gray-level and moment invariants-based features. IEEE Trans. Med. Imaging 2011, 30, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Fraz, M.M.; Remagnino, P.; Hoppe, A.; Uyyanonvara, B.; Owen, C.G.; Rudnicka, A.R.; Barman, S.A. Retinal Vessel Extraction Using First-Order Derivative of Gaussian and Morphological Processing. In Advances in Visual Computing, Proceedings of the 7th International Symposium (ISVC 2011), Las Vegas, NV, USA, 26–28 September 2011; Bebis, G., Boyle, R., Parvin, B., Koracin, D., Wang, S., Kyungnam, K., Benes, B., Moreland, K., Borst, C., DiVerdi, S., et al., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 410–420. [Google Scholar]
- Fraz, M.M.; Basit, A.; Barman, S.A. Application of Morphological Bit Planes in Retinal Blood Vessel Extraction. J. Digit. Imaging 2013, 26, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Vega, R.; Sanchez-Ante, G.; Falcon-Morales, L.E.; Sossa, H.; Guevara, E. Retinal vessel extraction using Lattice Neural Networks with dendritic processing. Comput. Biol. Med. 2015, 58, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Feng, B.; Xie, L.; Liang, P.; Zhang, H.; Wang, T. A Cross-Modality Learning Approach for Vessel Segmentation in Retinal Images. IEEE Trans. Med. Imaging 2016, 35, 109–118. [Google Scholar] [CrossRef]
- Liskowski, P.; Krawiec, K. Segmenting Retinal Blood Vessels With Deep Neural Networks. IEEE Trans. Med. Imaging 2016, 35, 2369–2380. [Google Scholar] [CrossRef]
- Leopold, H.A.; Orchard, J.; Zelek, J.; Lakshminarayanan, V. Segmentation and feature extraction of retinal vascular morphology. In Proceedings SPIE Medical Imaging; International Society for Optics and Photonics: San Diego, CA, USA, 2017; Volume 10133. [Google Scholar]
- Leopold, H.A.; Orchard, J.; Zelek, J.; Lakshminarayanan, V. Use of Gabor filters and deep networks in the segmentation of retinal vessel morphology. In Proceedings SPIE Medical Imaging; International Society for Optics and Photonics: San Diego, CA, USA, 2017; Volume 10068. [Google Scholar]
- Mo, J.; Zhang, L. Multi-level deep supervised networks for retinal vessel segmentation. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 2181–2193. [Google Scholar] [CrossRef]
| KPI | Description | Value |
|---|---|---|
| True Positive Rate (TPR) | Probability of detection | |
| False Positive Rate (FPR) | Probability of false detection | |
| Accuracy (Acc) | The frequency a pixel is properly classified | |
| Sensitivity aka Recall (SN) | The proportion of true positive results detected by the classifier | or |
| Precision (Pr) | Proportion of positive samples properly classified | |
| Specificity (SP) | The proportion of negative samples properly classified | or |
| Kappa () | Agreement between two observers | |
| Probability of Agreement ( ) | Probability each observer selects a category k for N items | |
| G-mean (G) | Balance measure of SN and SP | |
| F1 Score (F1) | Harmonic mean of precision and recall | or |
| Matthews correlation coefficient (MCC) | Measure from −1 to 1 of agreement between manual and predicted binary segmentations | |
| N = TP + FP + TN + FN S = TP + FN × N P = TP + FP × N |
| Datasets | DRIVE | STARE | CHASE_DB1 |
|---|---|---|---|
| Image Dimensions | 565 × 584 | 700 × 605 | 1280 × 960 |
| Colour Channels | RGB | RGB | RGB |
| Total Images | 40 | 20 | 28 |
| Source Grouping | 20 train and 20 test | - | 14 Patients (2 images in each) |
| Method Summary | |||
| Train—Test Schedule | One-off on 20 train, test on the other 20 | 4-fold cross-validation over 20 images | four-fold cross-validation over 14 patients |
| Information Loss | 5.0348 | 6.4621 | 18.7500 |
| Methods | SN | SP | Pr | Acc | AUC | kappa | G | MCC | F1 |
|---|---|---|---|---|---|---|---|---|---|
| Human (2nd Observer) | 0.7760 | 0.9730 | 0.8066 | 0.9472 | - | 0.7581 | 0.8689 | 0.7601 | 0.7881 |
| Unsupervised Methods | |||||||||
| Lam et al. [42] | - | - | - | 0.9472 | 0.9614 | - | - | - | - |
| Azzopardi et al. [8] | 0.7655 | 0.9704 | - | 0.9442 | 0.9614 | - | 0.8619 | 0.7475 | - |
| Kovács and Hajdu [43] | 0.7270 | 0.9877 | - | 0.9494 | - | - | 0.8474 | - | - |
| Zhang et al. [44] | 0.7743 | 0.9725 | - | 0.9476 | 0.9636 | - | 0.8678 | - | - |
| Roychowdhury et al. [45] | 0.7395± 0.062 | 0.9782± 0.0073 | - | 0.9494± 0.005 | 0.9672 | - | 0.8505 | - | - |
| Niemeijer et al. [46] | 0.6793± 0.0699 | 0.9801± 0.0085 | - | 0.9416± 0.0065 | 9294± 0.0152 | 0.7145 | 0.8160 | - | - |
| Supervised Methods | |||||||||
| Soares et al. [10] | 0.7332 | 0.9782 | - | 0.9461± 0.0058 | 0.9614 | 0.7285 | 0.8469 | - | - |
| Ricci and Perfetti [3] | - | - | - | 0.9595 | 0.9633 | - | - | - | - |
| Marin et al. [47] | 0.7067 | 0.9801 | - | 0.9452 | 0.9588 | - | 0.8322 | - | - |
| Lupascu et al. [12] | - | - | - | 0.9597± 0.0054 | 0.9561 | 0.7200 | 0.8151 | - | - |
| Fraz et al. [48] | 0.7152 | 0.9768 | 0.8205 | 0.9430 | - | - | 0.8358 | 0.7333 | 0.7642 |
| Fraz et al. [7] | 0.7406 | 0.9807 | - | 0.9480 | 0.9747 | - | 0.8522 | - | - |
| Fraz et al. [49] | 0.7302 | 0.9742 | 0.8112 | 0.9422 | - | - | 0.8434 | 0.7359 | 0.7686 |
| Vega et al. [50] | 0.7444 | 0.9600 | - | 0.9412 | - | - | 0.8454 | 0.6617 | 0.6884 |
| Li et al. [51] | 0.7569 | 0.9816 | - | 0.9527 | 0.9738 | - | 0.8620 | - | - |
| Liskowski et al. [52] | 0.7811 | 0.9807 | - | 0.9535 | 0.9790 | 0.7910 | 0.8752 | - | - |
| Leopold et al. [53] | 0.6823 | 0.9801 | - | 0.9419 | 0.9707 | - | 0.8178 | - | - |
| Leopold et al. [54] | 0.7800 | 0.9727 | - | 0.9478 | 0.9689 | - | 0.8710 | - | - |
| Orlando et al. [38] | 0.7897 | 0.9684 | 0.7854 | - | - | - | 0.8741 | 0.7556 | 0.7857 |
| Mo et al. [55] | 0.7779± 0.0849 | 0.9780± 0.0091 | - | 0.9521± 0.0057 | 0.9782± 0.0059 | 0.7759± 0.0329 | 0.8722± 0.0278 | - | - |
| PixelBNN | 0.6963± 0.0489 | 0.9573± 0.0089 | 0.7770± 0.0458 | 0.9106± 0.0121 | 0.8268± 0.0247 | 0.6795± 0.0414 | 0.8159± 0.0286 | 0.6820± 0.0399 | 0.7328± 0.0335 |
| Methods | SN | SP | Pr | Acc | AUC | kappa | G | MCC | F1 |
|---|---|---|---|---|---|---|---|---|---|
| Human (2nd Observer) | 0.8951 | 0.9387 | 0.6424 | 0.9353 | - | 0.7046 | 0.9166 | 0.7225 | 0.7401 |
| Unsupervised Methods | |||||||||
| Lam et al. [42] | - | - | - | 0.9567 | 0.9739 | - | - | - | - |
| Azzopardi et al. [8] | 0.7716 | 0.9701 | - | 0.9497 | 0.9563 | - | 0.8652 | 0.7335 | - |
| Kovács and Hajdu [43] | 0.7665 | 0.9879 | - | - | 0.9711 | - | 0.8702 | - | - |
| Zhang et al. [44] | 0.7791 | 0.9758 | - | 0.9554 | 0.9748 | - | 0.8719 | - | - |
| Roychowdhury et al. [45] | 0.7317± 0.053 | 0.9842± 0.0069 | - | 0.9560± 0.0095 | 0.9673 | - | 0.8486± 0.0178 | - | - |
| Supervised Methods | |||||||||
| Soares et al. [10] | 0.7207 | 0.9747 | - | 0.9479 | 0.9671 | - | 0.8381 | - | - |
| Ricci et al. [3] | - | - | - | 0.9584 | 0.9602 | - | - | - | - |
| Marin et al. [47] | 0.6944 | 0.9819 | - | 0.9526 | 0.9769 | - | 0.8257 | - | - |
| Fraz et al. [48] | 0.7409 | 0.9665 | 0.7363 | 0.9437 | - | - | 0.8462 | 0.7003 | 0.7386 |
| Fraz et al. [7] | 0.7548 | 0.9763 | - | 0.9534 | 0.9768 | - | 0.8584 | - | - |
| Fraz et al. [49] | 0.7318 | 0.9660 | 0.7294 | 0.9423 | - | - | 0.8408 | 0.6908 | 0.7306 |
| Vega et al. [50] | 0.7019 | 0.9671 | - | 0.9483 | - | - | 0.8239 | 0.5927 | 0.6082 |
| Li et al. [51] | 0.7726 | 0.9844 | - | 0.9628 | 0.9879 | - | 0.8721 | - | - |
| Liskowski et al. [52] | 0.8554± 0.0286 | 0.9862± 0.0018 | - | 0.9729± 0.0027 | 0.9928± 0.0014 | 0.8507± 0.0155 | 0.9185± 0.0072 | - | - |
| Mo et al. [55] | 0.8147± 0.0387 | 0.9844± 0.0034 | - | 0.9674± 0.0058 | 0.9885± 0.0035 | 0.8163± 0.0310 | 0.8955± 0.0115 | - | - |
| Orlando et al. [38] | 0.7680 | 0.9738 | 0.7740 | - | - | - | 0.8628 | 0.7417 | 0.7644 |
| PixelBNN | 0.6433± 0.0593 | 0.9472± 0.0212 | 0.6637± 0.1135 | 0.9045± 0.0207 | 0.7952± 0.0315 | 0.5918± 0.0721 | 0.7797± 0.0371 | 0.5960± 0.0719 | 0.6465± 0.0621 |
| Methods | SN | SP | Pr | Acc | AUC | kappa | G | MCC | F1 |
|---|---|---|---|---|---|---|---|---|---|
| Human (2nd Observer) | 0.7425 | 0.9793 | 0.8090 | 0.9560 | - | 0.7529 | 0.8527 | 0.7475 | 0.7686 |
| Unsupervised Methods | |||||||||
| Azzopardi et al. [8] | 0.7585 | 0.9587 | - | 0.9387 | 0.9487 | - | 0.8527 | 0.6802 | - |
| Zhang et al. [44] | 0.7626 | 0.9661 | - | 0.9452 | 0.9606 | - | 0.8583 | - | - |
| Roychowdhury et al. [45] | 0.7615± 0.0516 | 0.9575± 0.003 | - | 0.9467± 0.0076 | 0.9623 | - | 0.8539± 0.0124 | - | - |
| Supervised Methods | |||||||||
| Fraz et al. [7] | 0.7224 | 0.9711 | - | 0.9469 | 0.9712 | - | 0.8376 | - | - |
| Li et al. [51] | 0.7507 | 0.9793 | - | 0.9581 | 0.9716 | - | 0.8574 | - | - |
| Liskowski et al. [52] | 0.7816± 0.0178 | 0.9836± 0.0022 | - | 0.9628± 0.0020 | 0.9823± 0.0016 | 0.7908± 0.0111 | 0.8768± 0.0063 | - | - |
| Mo et al. [55] | 0.7661 ± 0.0533 | 0.9816± 0.0076 | - | 0.9599± 0.0050 | 0.9812± 0.0040 | 0.8672± 0.0201 | 0.7689± 0.0263 | - | - |
| Orlando et al. [38] | 0.7277 | 0.9712 | 0.7438 | - | - | - | 0.8403 | 0.7046 | 0.7332 |
| PixelBNN | 0.8618± 0.0232 | 0.8961± 0.0150 | 0.3951± 0.0603 | 0.8936± 0.0138 | 0.878959± 0.0138 | 0.4889± 0.0609 | 0.8787± 0.0140 | 0.5376± 0.0491 | 0.5391± 0.0587 |
| Methods | SN | SP | Pr | Acc | AUC | kappa | G | MCC | F1 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Test images from: DRIVE | |||||||||||
| Model trained on: STARE | Soares et al. [10] | - | - | - | 0.9397 | - | - | - | - | - | |
| Ricci et al. [3] | - | - | - | 0.9266 | - | - | - | - | - | ||
| Marin et al. [47] | - | - | - | 0.9448 | - | - | - | - | - | ||
| Fraz et al. [7] | 0.7242 | 0.9792 | - | 0.9456 | 0.9697 | - | 0.8421 | - | - | ||
| Li et al. [51] | 0.7273 | 0.9810 | - | 0.9486 | 0.9677 | - | 0.8447 | - | - | ||
| Liskowski et al. [52] | - | - | - | 0.9416 | 0.9605 | - | - | - | - | ||
| Mo et al. [55] | 0.7412 | 0.9799 | - | 0.9492 | 0.9653 | - | 0.8522 | - | - | ||
| PixelBNN | 0.5110± 0.0362 | 0.9533± 0.0094 | 0.7087± 0.0554 | 0.8748± 0.0126 | 0.7322± 0.0199 | 0.5193± 0.0404 | 0.6974± 0.0258 | 0.5309± 0.0422 | 0.5907± 0.0348 | ||
| Model trained on: CHASE_DB1 | Li et al. [51] | 0.7307 | 0.9811 | - | 0.9484 | 0.9605 | - | 0.8467 | - | - | |
| Mo et al. [55] | 0.7315 | 0.9778 | - | 0.9460 | 0.9650 | - | 0.8457 | - | - | ||
| PixelBNN | 0.6222± 0.0441 | 0.9355± 0.0085 | 0.6785± 0.0383 | 0.8796± 0.0090 | 0.7788± 0.0204 | 0.5742± 0.0282 | 0.7622± 0.0254 | 0.5768± 0.0279 | 0.6463± 0.0237 | ||
| Test images from: STARE | |||||||||||
| Model trained on: DRIVE | Soares et al. [10] | - | - | - | 0.9327 | - | - | - | - | - | |
| Ricci et al. [3] | - | - | - | 0.9464 | - | - | - | - | - | ||
| Marin et al. [47] | - | - | - | 0.9528 | - | - | - | - | - | ||
| Fraz et al. [7] | 0.7010 | 0.9770 | - | 0.9493 | 0.9660 | - | 0.8276 | - | - | ||
| Li et al. [51] | 0.7027 | 0.9828 | - | 0.9545 | 0.9671 | - | 0.8310 | - | - | ||
| Liskowski et al. [52] | - | - | - | 0.9505 | 0.9595 | - | - | - | - | ||
| Mo et al. [55] | 0.7009 | 0.9843 | - | 0.9570 | 0.9751 | - | 0.8306 | - | - | ||
| PixelBNN | 0.7842± 0.0552 | 0.9265± 0.0196 | 0.6262± 0.1143 | 0.9070± 0.0181 | 0.8553± 0.0323 | 0.6383± 0.0942 | 0.8519± 0.0343 | 0.6465± 0.0873 | 0.6916± 0.0868 | ||
| Model trained on: CHASE_DB1 | Li et al. [51] | 0.6944 | 0.9831 | - | 0.9536 | 0.9620 | - | 0.8262 | - | - | |
| Mo et al. [55] | 0.7387 | 0.9787 | - | 0.9549 | 0.9781 | - | 0.8503 | - | - | ||
| PixelBNN | 0.6973± 0.0372 | 0.9062± 0.0189 | 0.5447± 0.0957 | 0.8771± 0.0157 | 0.8017± 0.0226 | 0.5353± 0.0718 | 0.7941± 0.0245 | 0.5441± 0.0649 | 0.6057± 0.0674 | ||
| Test images from: CHASE_DB1 | |||||||||||
| Model trained on: DRIVE | Li et al. [51] | 0.7118 | 0.9791 | - | 0.9429 | 0.9628 | - | 0.8348 | - | - | |
| Mo et al. [55] | 0.7003 | 0.9750 | - | 0.9478 | 0.9671 | - | 0.8263 | - | - | ||
| PixelBNN | 0.9038± 0.0196 | 0.8891± 0.0089 | 0.3886± 0.0504 | 0.8901± 0.0088 | 0.8964± 0.0116 | 0.4906± 0.0516 | 0.8963± 0.0116 | 0.5480± 0.0413 | 0.5416± 0.0513 | ||
| Model trained on: STARE | Fraz et al. [7] | 0.7103 | 0.9665 | - | 0.9415 | 0.9565 | - | 0.8286 | - | - | |
| Li et al. [51] | 0.7240 | 0.9768 | - | 0.9417 | 0.9553 | - | 0.8410 | - | - | ||
| Mo et al. [55] | 0.7032 | 0.9794 | - | 0.9515 | 0.9690 | - | 0.8299 | - | - | ||
| PixelBNN | 0.7525± 0.0233 | 0.9302± 0.0066 | 0.4619± 0.0570 | 0.9173± 0.0059 | 0.8413± 0.0132 | 0.5266± 0.0482 | 0.8365± 0.0143 | 0.5475± 0.0412 | 0.5688± 0.0475 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leopold, H.A.; Orchard, J.; Zelek, J.S.; Lakshminarayanan, V. PixelBNN: Augmenting the PixelCNN with Batch Normalization and the Presentation of a Fast Architecture for Retinal Vessel Segmentation. J. Imaging 2019, 5, 26. https://doi.org/10.3390/jimaging5020026
Leopold HA, Orchard J, Zelek JS, Lakshminarayanan V. PixelBNN: Augmenting the PixelCNN with Batch Normalization and the Presentation of a Fast Architecture for Retinal Vessel Segmentation. Journal of Imaging. 2019; 5(2):26. https://doi.org/10.3390/jimaging5020026
Chicago/Turabian StyleLeopold, Henry A., Jeff Orchard, John S. Zelek, and Vasudevan Lakshminarayanan. 2019. "PixelBNN: Augmenting the PixelCNN with Batch Normalization and the Presentation of a Fast Architecture for Retinal Vessel Segmentation" Journal of Imaging 5, no. 2: 26. https://doi.org/10.3390/jimaging5020026
APA StyleLeopold, H. A., Orchard, J., Zelek, J. S., & Lakshminarayanan, V. (2019). PixelBNN: Augmenting the PixelCNN with Batch Normalization and the Presentation of a Fast Architecture for Retinal Vessel Segmentation. Journal of Imaging, 5(2), 26. https://doi.org/10.3390/jimaging5020026






