Preliminary Results of Clover and Grass Coverage and Total Dry Matter Estimation in Clover-Grass Crops Using Image Analysis
Abstract
1. Introduction
1.1. Related Work
2. Materials and Methods
2.1. Testbed and Data Acquisition
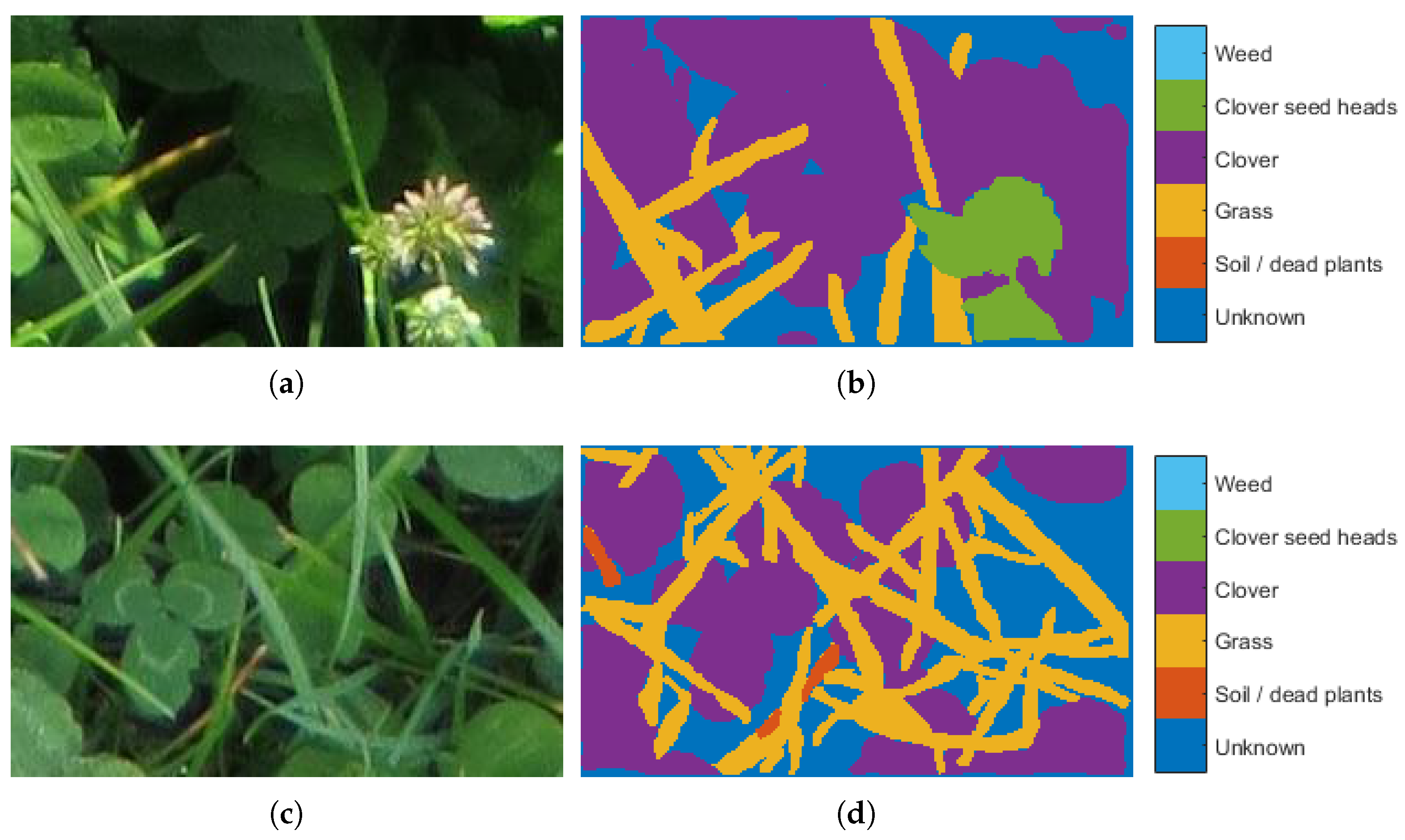
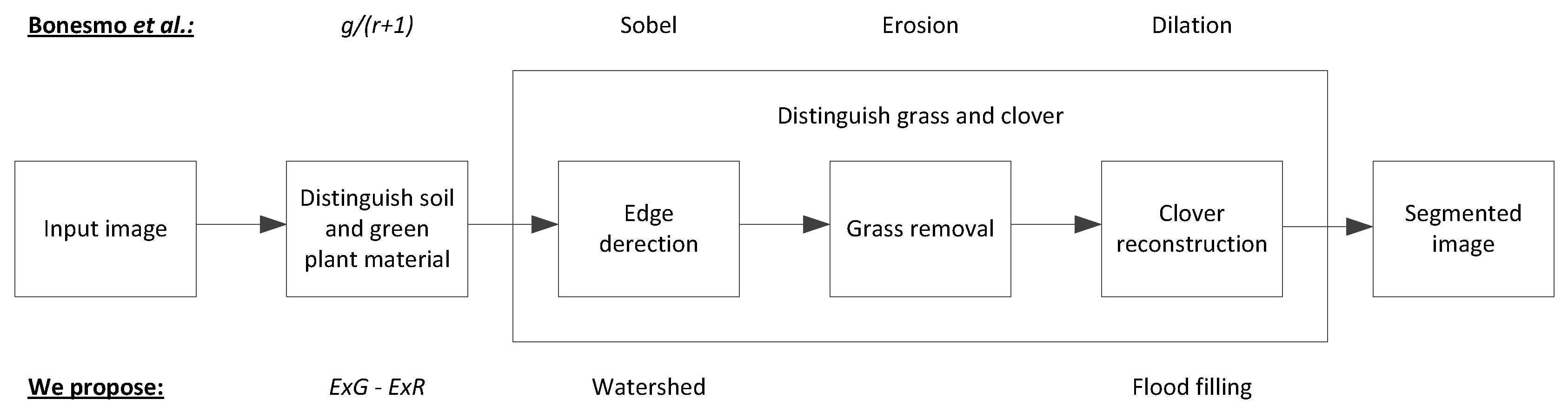
2.2. Clover-Grass Segmentation
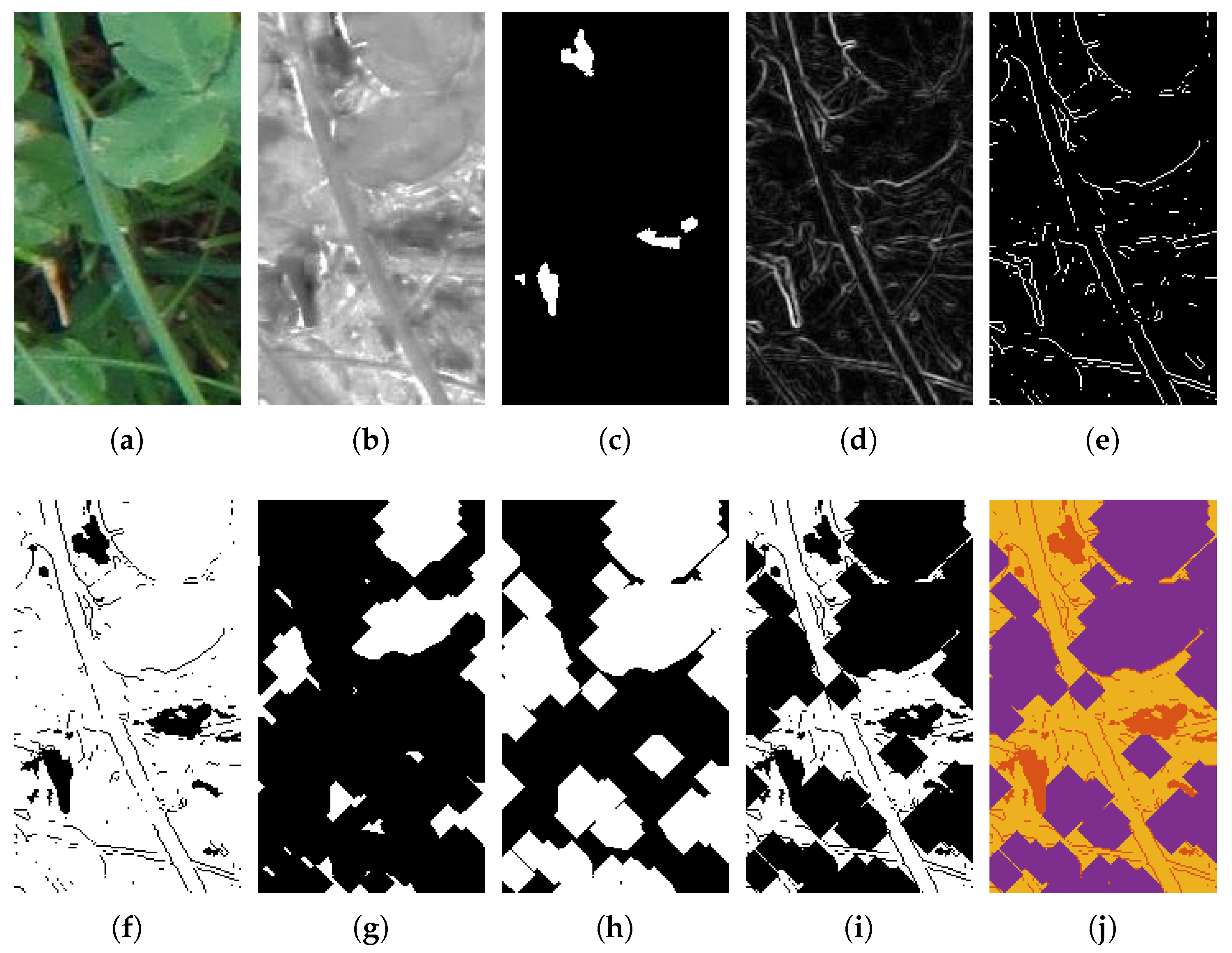
2.2.1. Distinguishing Soil and Green Plant Material
2.2.2. Edge Detection
2.2.3. Clover Reconstruction
2.2.4. Illumination Classification
2.2.5. Training Clover-Grass Segmentation
2.3. Dry Matter Estimation
3. Results
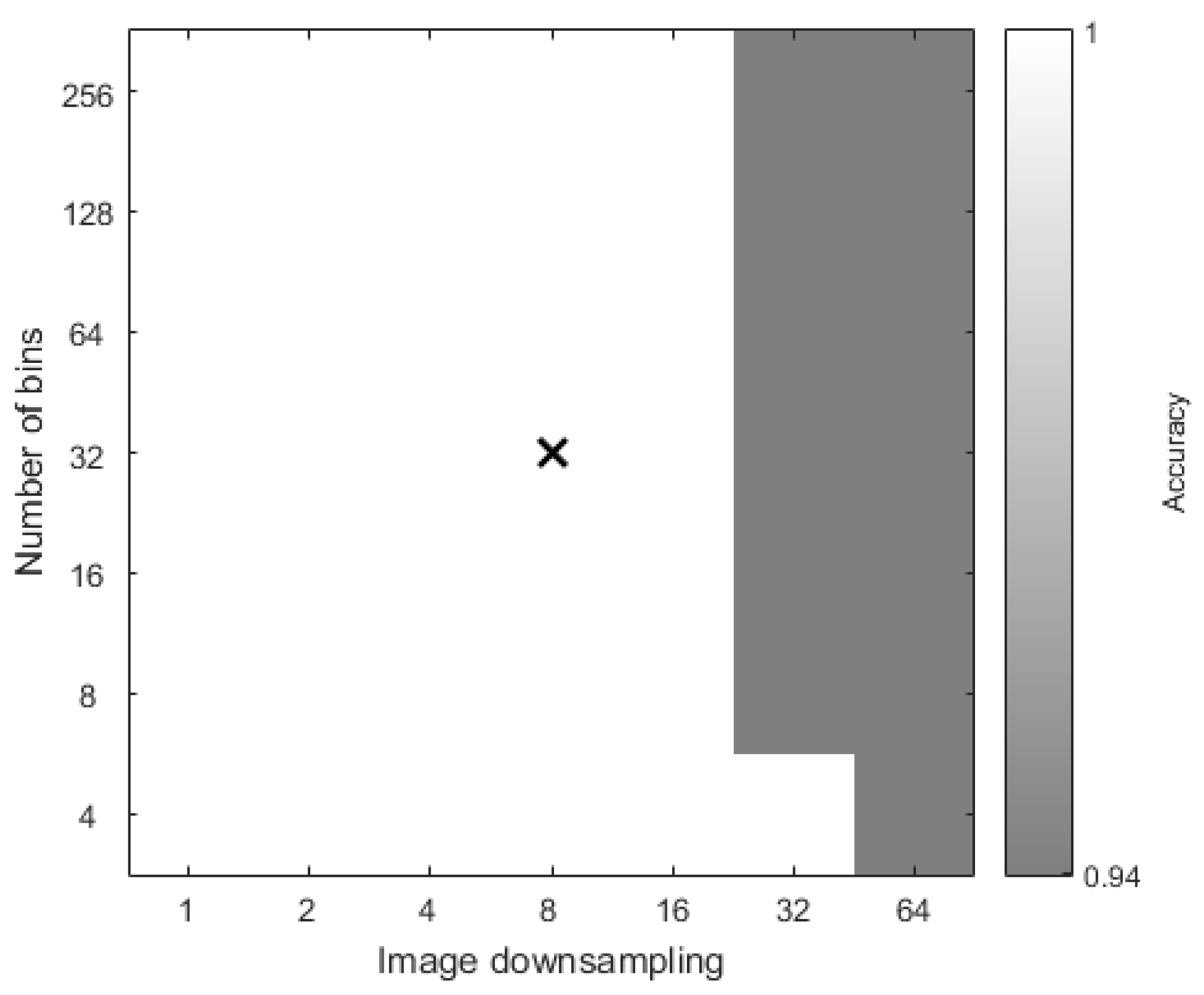
3.1. Illumination Classification
3.2. Image Segmentation
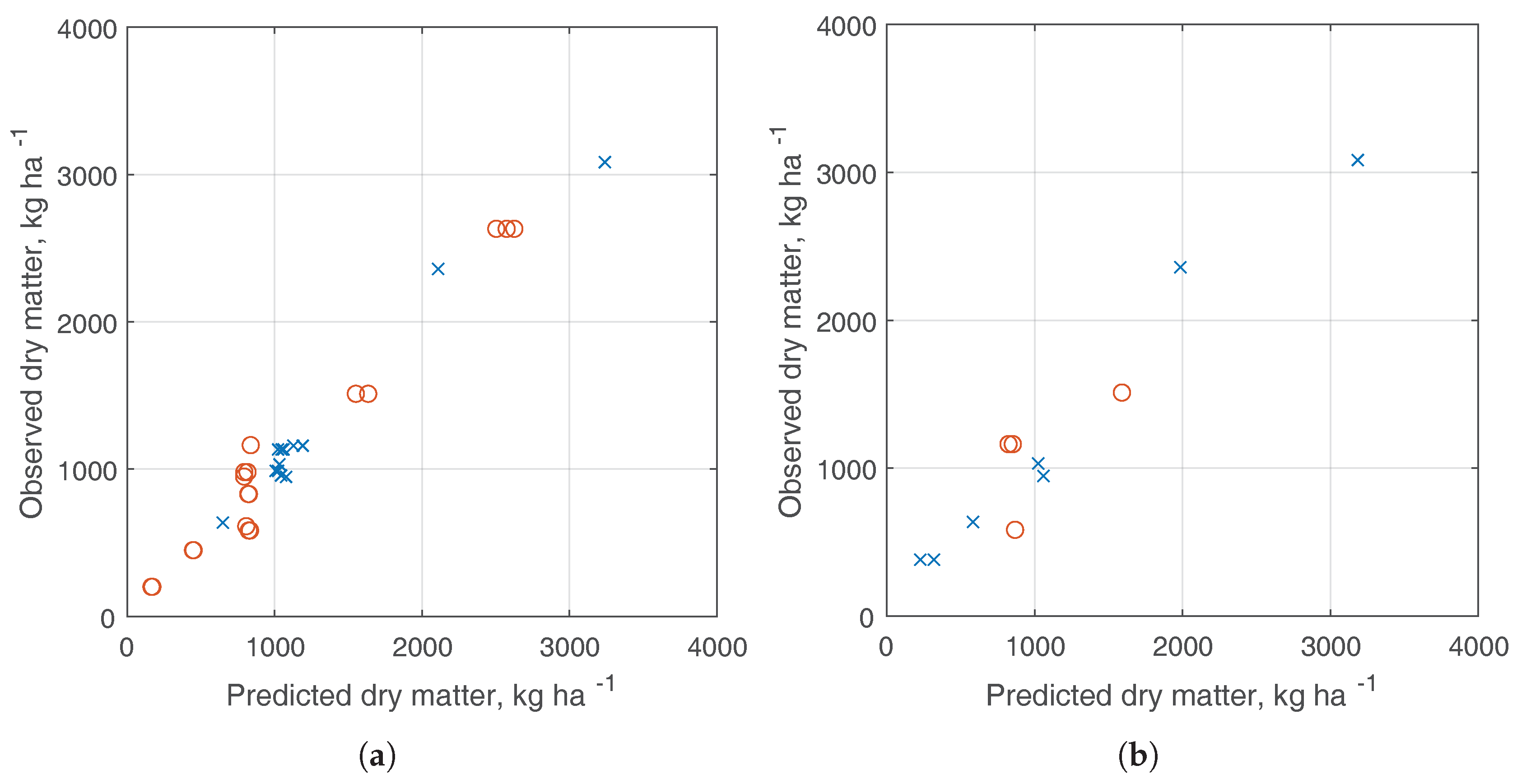
3.3. Dry Matter Estimation
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| DM | Dry Matter |
| Excess Green | |
| Excess Red | |
| Excess Green – Excess Red | |
| ExGR from Chromaticity | |
| ExGR from Normalized color values | |
| fwIoU | Frequency-weighted Intersection over Union |
| K | Potassium |
| MAE | Mean Absolute Error |
| MARE | Mean Absolute Relative Error |
| N | Nitrogen |
| NRMSE | Normalized Root Mean Square Error |
| RGB | Red Green Blue |
| RMSE | Root Mean Square Error |
References
- Søegaard, K. Nitrogen fertilization of grass/clover swards under cutting or grazing by dairy cows. Acta Agric. Scand. Sect. B 2009, 59, 139–150. [Google Scholar]
- Kuoppala, K. Influence of Harvesting Strategy on Nutrient Supply and Production of Dairy Cows Consuming Diets Based on Grass and Red Clover Silage. Ph.D. Thesis, Department of Agricultural Sciences, University Helsinki, Helsinki, Finland, 2010. [Google Scholar]
- Barrett, P.; Laidlaw, A.; Mayne, C. GrazeGro: A European herbage growth model to predict pasture production in perennial ryegrass swards for decision support. Eur. J. Agron. 2005, 23, 37–56. [Google Scholar] [CrossRef]
- Biewer, S.; Erasmi, S.; Fricke, T.; Wachendorf, M. Prediction of yield and the contribution of legumes in legume-grass mixtures using field spectrometry. Precis. Agric. 2009, 10, 128–144. [Google Scholar] [CrossRef]
- Friedl, M.A.; Schimel, D.S.; Michaelsen, J.; Davis, F.W.; Walker, H. Estimating grassland biomass and leaf area index using ground and satellite data. Int. J. Remote Sens. 1994, 15, 1401–1420. [Google Scholar] [CrossRef]
- Bonesmo, H.; Kaspersen, K.; Kjersti Bakken, A. Evaluating an image analysis system for mapping white clover pastures. Acta Agric. Scand. Sect. B 2004, 54, 76–82. [Google Scholar] [CrossRef]
- Himstedt, M.; Fricke, T.; Wachendorf, M. The benefit of color information in digital image analysis for the estimation of legume contribution in legume-grass mixtures. Crop Sci. 2012, 52, 943–950. [Google Scholar] [CrossRef]
- McRoberts, K.C.; Benson, B.M.; Mudrak, E.L.; Parsons, D.; Cherney, D.J. Application of local binary patterns in digital images to estimate botanical composition in mixed alfalfa–grass fields. Comput. Electron. Agric. 2016, 123, 95–103. [Google Scholar] [CrossRef]
- Schapendonk, A.; Stol, W.; van Kraalingen, D.; Bouman, B. LINGRA, a sink/source model to simulate grassland productivity in Europe. Eur. J. Agron. 1998, 9, 87–100. [Google Scholar] [CrossRef]
- Jing, Q.; Bélanger, G.; Baron, V.; Bonesmo, H.; Virkajärvi, P.; Young, D. Regrowth simulation of the perennial grass timothy. Ecol. Model. 2012, 232, 64–77. [Google Scholar] [CrossRef]
- Bonesmo, H.; Baron, V.S.; Young, D.; Bélanger, G.; Jing, Q. Adapting the CATIMO grass model to meadow bromegrass grown in western Canada. Can. J. Plant Sci. 2014, 94, 61–71. [Google Scholar] [CrossRef]
- Adam, E.; Mutanga, O.; Rugege, D. Multispectral and hyperspectral remote sensing for identification and mapping of wetland vegetation: A review. Wetl. Ecol. Manag. 2010, 18, 281–296. [Google Scholar] [CrossRef]
- Xie, Y.; Sha, Z.; Yu, M. Remote sensing imagery in vegetation mapping: A review. J. Plant Ecol. 2008, 1, 9–23. [Google Scholar] [CrossRef]
- Rembold, F.; Atzberger, C.; Savin, I.; Rojas, O. Using Low Resolution Satellite Imagery for Yield Prediction and Yield Anomaly Detection. Remote Sens. 2013, 5, 1704–1733. [Google Scholar] [CrossRef]
- Joshi, N.; Baumann, M.; Ehammer, A.; Fensholt, R.; Grogan, K.; Hostert, P.; Jepsen, M.R.; Kuemmerle, T.; Meyfroidt, P.; Mitchard, E.T.A.; et al. A Review of the Application of Optical and Radar Remote Sensing Data Fusion to Land Use Mapping and Monitoring. Remote Sens. 2016, 8, 70. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Cohen, W.B.; Parker, G.G.; Harding, D.J. Lidar Remote Sensing for Ecosystem StudiesLidar, an emerging remote sensing technology that directly measures the three-dimensional distribution of plant canopies, can accurately estimate vegetation structural attributes and should be of particular interest to forest, landscape, and global ecologists. Bioscience 2002, 52, 19–30. [Google Scholar]
- Ju, J.; Roy, D.P. The availability of cloud-free Landsat ETM+ data over the conterminous United States and globally. Remote Sens. Environ. 2008, 112, 1196–1211. [Google Scholar] [CrossRef]
- Åstrand, B.; Baerveldt, A.J. An agricultural mobile robot with vision-based perception for mechanical weed control. Auton. Robot. 2002, 13, 21–35. [Google Scholar] [CrossRef]
- Giselsson, T.M.; Midtiby, H.S.; Jørgensen, R.N. Seedling discrimination with shape features derived from a distance transform. Sensors 2013, 13, 5585–5602. [Google Scholar] [CrossRef] [PubMed]
- Dyrmann, M.; Christiansen, P. Automated Classification of Seedlings Using Computer Vision; Aarhus University: Aarhus, Denmark, 2014. [Google Scholar]
- McCarthy, C.L.; Hancock, N.H.; Raine, S.R. Applied machine vision of plants: A review with implications for field deployment in automated farming operations. Intell. Serv. Robot. 2010, 3, 209–217. [Google Scholar] [CrossRef]
- Gebhardt, S.; Schellberg, J.; Lock, R.; Kühbauch, W. Identification of broad-leaved dock (Rumex obtusifolius L.) on grassland by means of digital image processing. Precis. Agric. 2006, 7, 165–178. [Google Scholar] [CrossRef]
- Gebhardt, S.; Kühbauch, W. A new algorithm for automatic Rumex obtusifolius detection in digital images using colour and texture features and the influence of image resolution. Precis. Agric. 2007, 8, 1–13. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Proceedings of Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer; pp. 234–241. [Google Scholar]
- Mortensen, A.K.; Dyrmann, M.; Karstoft, H.; Jørgensen, R.N.; Gislum, R. Semantic Segmentation of Mixed Crops using Deep Convolutional Neural Network. In Proceedings of the International Conference on Agricultural Engineering 2016, Aarhus, Danmark, 26–29 June 2016. [Google Scholar]
- Dyrmann, M.; Mortensen, A.K.; Midtiby, H.S.; Jørgensen, R.N. Pixel-wise classification of weeds and crop in images by using a Fully Convolutional neural network. In Proceedings of the International Conference on Agricultural Engineering 2016, Aarhus, Danmark, 26–29 June 2016. [Google Scholar]
- Meyer, G.E.; Neto, J.C. Verification of color vegetation indices for automated crop imaging applications. Comput. Electron. Agric. 2008, 63, 282–293. [Google Scholar] [CrossRef]
- Beucher, S.; Lantuéjoul, C. Use of Watersheds in Contour Detection. In Proceedings of the International Workshop on Image Processing: Real-Time Edge and Motion Detection/Estimation, Rennes, France, 17–21 September 1979. [Google Scholar]
- MATLAB R2015a (8.5.0); The MathWorks Inc.: Natick, MA, USA, 2015.
- Mortensen, A.K.; Gislum, R.; Larsen, R.; Nyholm Jørgensen, R. Estimation of above-ground dry matter and nitrogen uptake in catch crops using images acquired from an octocopter. In Proceedings of the 10th European Conference on Precision Agriculture, Volcani Center, Israel, 12–16 July 2015; pp. 127–134. [Google Scholar]
- Jensen, K.; Laursen, M.S.; Midtiby, H.; Jørgensen, R.N. Autonomous Precision Spraying Trials Using a Novel Cell Spray Implement Mounted on an Armadillo Tool Carrier. In Proceedings of the XXXV CIOSTA & CIGR V Conference, Billund, Denmark, 3–5 July 2013. [Google Scholar]
- Overskeid, O.; Hoeg, A.; Overeng, S.; Stavlund, H. System for Controlled Application of Herbicides. U.S. Patent 8,454,245, 4 June 2013. [Google Scholar]
- Harris, S.L.; Thom, E.R.; Clark, D.A. Effect of high rates of nitrogen fertiliser on perennial ryegrass growth and morphology in grazed dairy pasture in northern New Zealand. N. Z. J. Agric. Res. 1996, 39, 159–169. [Google Scholar] [CrossRef]



| 1st Occasion | 2nd Occasion | 3rd Occasion | |
|---|---|---|---|
| Photo Date | 25 June | 10 September | 11 October |
| Cutting Date | 25 June | 11 September | 14 October |
| Samples | 10 | 5 | 30 |
| Cut Number | 2 | 4 | 4, 5 |
| Solar Azimuth | 29–33° | 36–37° | 24–25° |
| Solar Altitude | 90–97° | 199–203° | 155–164° |
| Global | Individual | |||||
|---|---|---|---|---|---|---|
| Method | Both | Indirect | Direct | Both | Indirect | Direct |
| Bonesmo | 40.4% | 48.2% | 27.5% | 40.3% | 48.7% | 29.0% |
| Bonesmo + | 40.3% | 48.1% | 27.4% | 40.2% | 48.7% | 29.1% |
| Bonesmo + | 42.2% | 48.5% | 31.4% | 42.1% | 49.4% | 33.6% |
| Bonesmo + Flood-fill | 38.1% | 40.7% | 34.1% | 40.9% | 46.1% | 34.5% |
| Bonesmo + Watershed | 40.8% | 47.1% | 30.0% | 42.1% | 48.6% | 33.3% |
| Bonesmo + Watershed + Flood-fill | 39.1% | 46.4% | 27.0% | 39.6% | 46.9% | 31.1% |
| Bonesmo + + Watershed | 41.2% | 46.8% | 32.7% | 42.6% | 48.6% | 34.8% |
| Bonesmo + + Watershed | 43.2% | 47.8% | 35.5% | 43.7% | 49.2% | 37.7% |
| Bonesmo + + Watershed + Flood-fill | 39.1% | 46.4% | 27.0% | 41.9% | 46.9% | 36.8% |
| Bonesmo + + Watershed + Flood-fill | 40.3% | 45.0% | 32.7% | 41.5% | 47.6% | 37.7% |
| Variable | Estimate | SE | t | p |
|---|---|---|---|---|
| Intercept | −205 | 884 | −0.242 | 0.810 |
| 845 | 389 | 2.17 | 0.0390 | |
| S | −0.585 | 6.40 | −0.0915 | 0.928 |
| T | −0.147 | 0.0790 | −1.86 | 0.0739 |
| M | 409 | 233 | 1.76 | 0.0901 |
| S:T | 0.00181 | 0.000611 | 2.96 | 0.00636 |
| S:M | −3.98 | 1.4 | −2.84 | 0.000848 |
| Dataset | RMSE | NRMSE | MAE | MARE |
|---|---|---|---|---|
| Training set | 125 kg ha | 11% | 90 kg ha | 9.8% |
| Test set | 210 kg ha | 17.5% | 171 kg ha | 19% |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mortensen, A.K.; Karstoft, H.; Søegaard, K.; Gislum, R.; Jørgensen, R.N. Preliminary Results of Clover and Grass Coverage and Total Dry Matter Estimation in Clover-Grass Crops Using Image Analysis. J. Imaging 2017, 3, 59. https://doi.org/10.3390/jimaging3040059
Mortensen AK, Karstoft H, Søegaard K, Gislum R, Jørgensen RN. Preliminary Results of Clover and Grass Coverage and Total Dry Matter Estimation in Clover-Grass Crops Using Image Analysis. Journal of Imaging. 2017; 3(4):59. https://doi.org/10.3390/jimaging3040059
Chicago/Turabian StyleMortensen, Anders K., Henrik Karstoft, Karen Søegaard, René Gislum, and Rasmus N. Jørgensen. 2017. "Preliminary Results of Clover and Grass Coverage and Total Dry Matter Estimation in Clover-Grass Crops Using Image Analysis" Journal of Imaging 3, no. 4: 59. https://doi.org/10.3390/jimaging3040059
APA StyleMortensen, A. K., Karstoft, H., Søegaard, K., Gislum, R., & Jørgensen, R. N. (2017). Preliminary Results of Clover and Grass Coverage and Total Dry Matter Estimation in Clover-Grass Crops Using Image Analysis. Journal of Imaging, 3(4), 59. https://doi.org/10.3390/jimaging3040059






