MedSAM/MedSAM2 Feature Fusion: Enhancing nnUNet for 2D TOF-MRA Brain Vessel Segmentation
Abstract
1. Introduction
- We propose a hybrid nnUNet-MedSAM/MedSAM2 architecture that integrates nnUNet with MedSAM/MedSAM2 to synergize their complementary strengths: MedSAM/MedSAM2 provides generalized feature representation through large-scale pretraining, while nnUNet offers task-specific adaptation. The hybrid design enhances multi-scale feature extraction and improves robustness against imaging noise and artifacts [14,15].
- We propose an enhanced nnUNet training strategy by incorporating Focal Loss to address the extreme class imbalance (e.g., <5% vascular pixels in TOF-MRA volumes). This effectively mitigates the severe imbalance between cerebrovascular structures and background tissues in TOF-MRA neuroimaging, enabling more balanced feature representation learning for both dominant background regions and sparse vascular targets.
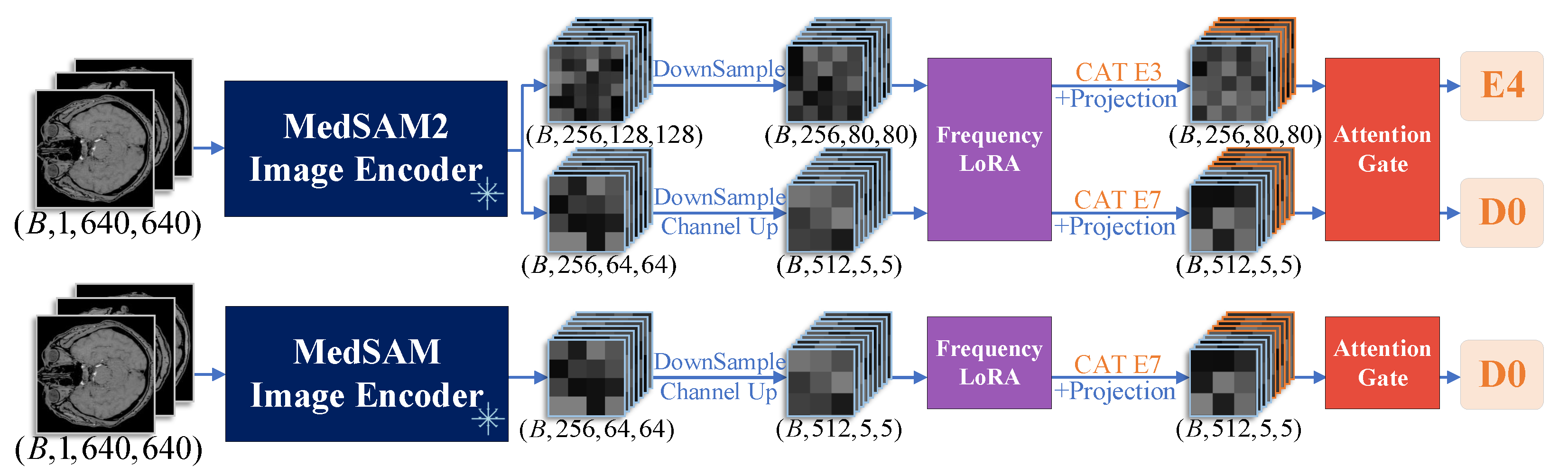
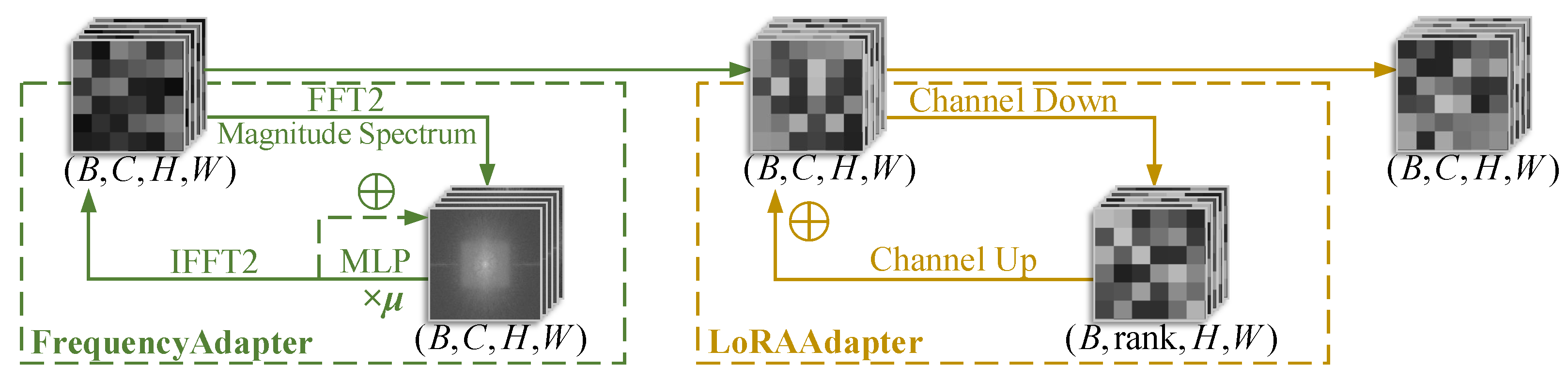
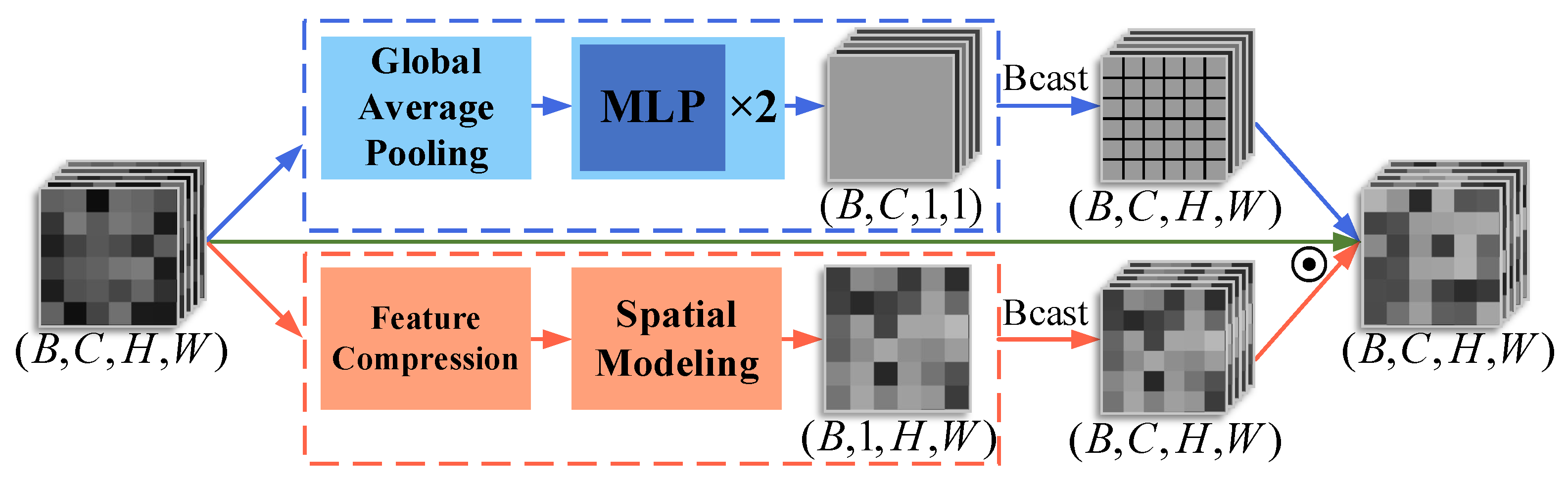
- We introduce a FrequencyLoRA module before feature fusion between nnUNet and MedSAM/MedSAM2, which captures global structures through spectral enhancement while achieving efficient local feature optimization via low-rank bottleneck compression, balancing noise robustness with computational efficiency. After feature fusion, we employ an AttentionGate module that synergistically combines channel attention (global context modeling) with spatial attention (local structure perception) to enhance feature selection and noise suppression simultaneously.
2. Methods
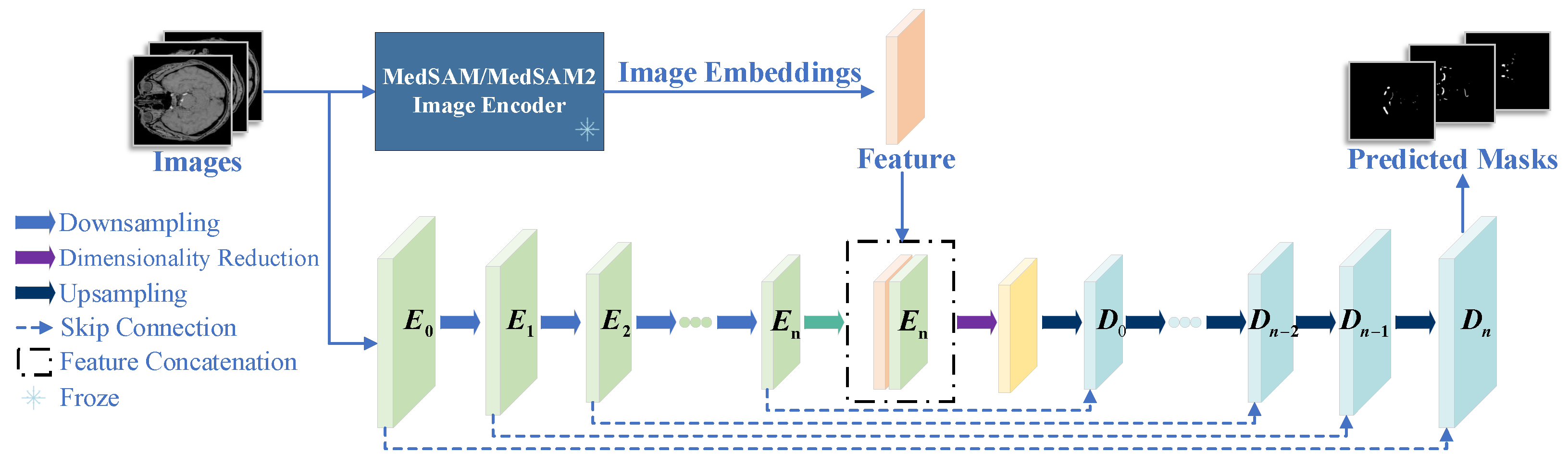
2.1. Architecture Overview
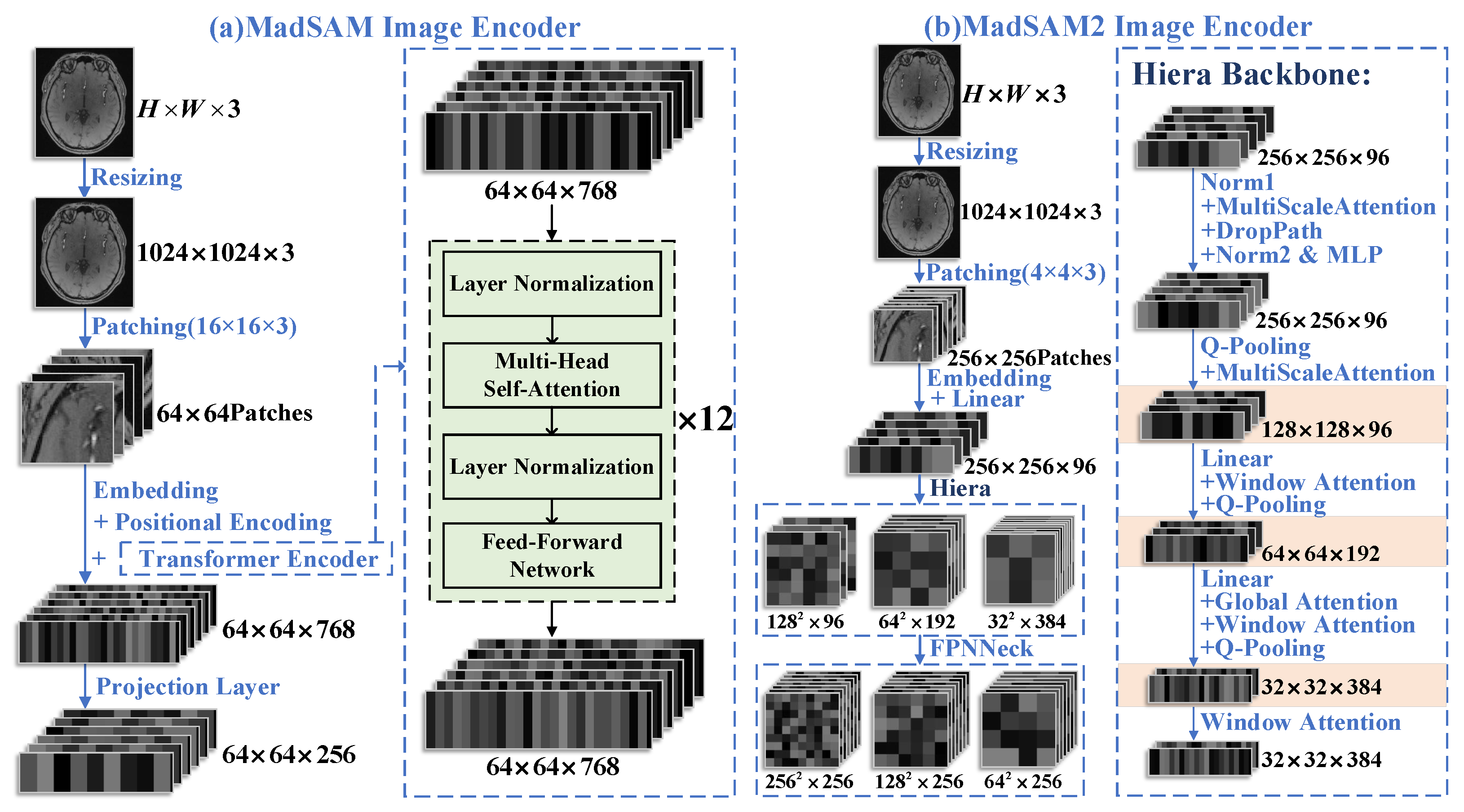
2.2. Feature Extraction Network in MedSAM/MedSAM2
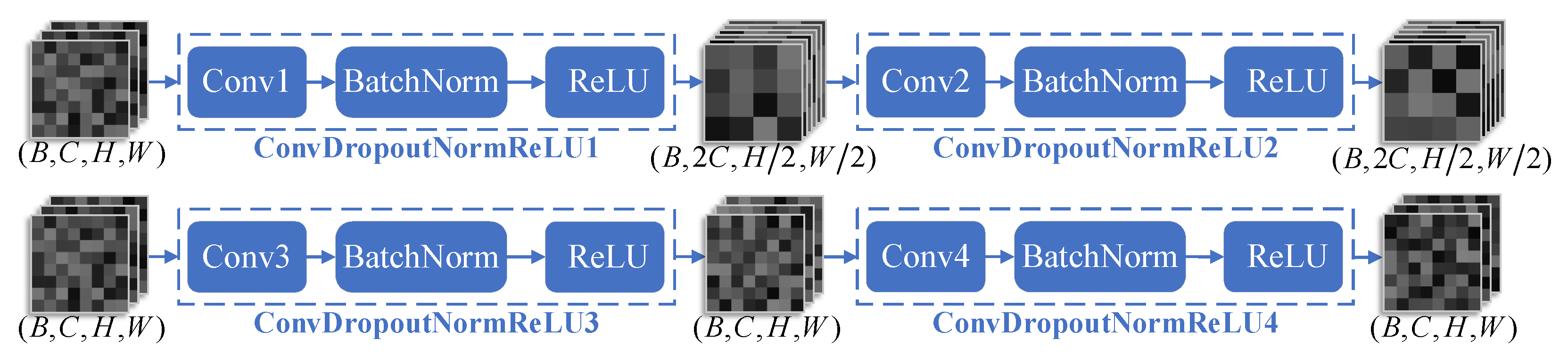
2.3. nnUNet Segmentation Framework
2.4. Feature Extraction Network in MedSAM/MedSAM2
2.5. Focal Loss-Based Optimization for nnUNet
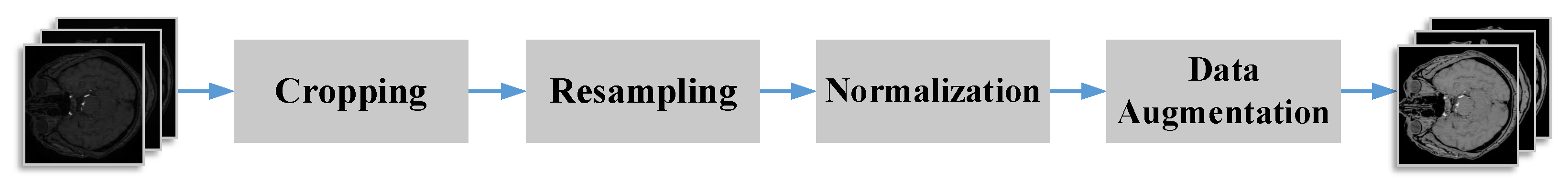
2.6. Dataset and Preprocessing
2.7. Computational Environment and Parameters
3. Results
3.1. Evaluation Metrics
3.2. Comparison with Baseline and Competing Methods
3.3. Ablation Study on Loss Functions
3.4. Ablation Study on Feature Fusion
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feigin, V.L.; Abate, M.D.; Abate, Y.H.; ElHafeez, S.A.; Abd-Allah, F.; Abdelalim, A.; Abdelkader, A.; Abdelmasseh, M.; Abd-Elsalam, S.; Abdi, P.; et al. Global, regional, and national burden of stroke and its risk factors, 1990–2021: A systematic analysis for the Global Burden of Disease Study 2021. Lancet Neurol. 2024, 23, 973–1003. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wang, D.; Qi, C.; Zou, M.; Song, J.; Li, L.; Xie, H.; Ren, H.; Hao, H.; Yang, G.; et al. Brain ischemia causes systemic Notch1 activity in endothelial cells to drive atherosclerosis. Immunity 2024, 57, 2157–2172. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Zhao, Q.; Wang, T.; Yang, Y.; Luo, J.; Zhang, X.; Feng, Y.; Ma, Y.; Dmytriw, A.A.; Yang, G.; et al. Optical coherence tomography in cerebrovascular disease: Open up new horizons. Transl. Stroke Res. 2023, 14, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Coppenrath, E.M.; Lummel, N.; Linn, J.; Lenz, O.; Habs, M.; Nikolaou, K.; Reiser, M.F.; Dichgans, M.; Pfefferkorn, T.; Saam, T. Time-of-flight angiography: A viable alternative to contrast-enhanced MR angiography and fat-suppressed T1w images for the diagnosis of cervical artery dissection? Eur. Radiol. 2013, 23, 2784–2792. [Google Scholar] [CrossRef] [PubMed]
- Bash, S.; Villablanca, J.P.; Jahan, R.; Duckwiler, G.; Tillis, M.; Kidwell, C.; Saver, J.; Sayre, J. Intracranial vascular stenosis and occlusive disease: Evaluation with CT angiography, MR angiography, and digital subtraction angiography. Am. J. Neuroradiol. 2005, 26, 1012–1021. [Google Scholar] [PubMed]
- Anzalone, N.; Scomazzoni, F.; Cirillo, M.; Righi, C.; Simionato, F.; Cadioli, M.; Iadanza, A.; Kirchin, M.; Scotti, G. Follow-up of coiled cerebral aneurysms at 3T: Comparison of 3D time-of-flight MR angiography and contrast-enhanced MR angiography. Am. J. Neuroradiol. 2008, 29, 1530–1536. [Google Scholar] [CrossRef] [PubMed]
- D’Layla, A.W.C.; De La Croix, N.J.; Ahmad, T.; Han, F. EHR-Protect: A Steganographic Framework Based on Data-Transformation to Protect Electronic Health Records. Intell. Syst. Appl. 2025, 26, 200493. [Google Scholar]
- Wang, Y.; Zhao, X.; Liu, L.; Soo, Y.O.; Pu, Y.; Pan, Y.; Wang, Y.; Zou, X.; Leung, T.W.; Cai, Y.; et al. Prevalence and Outcomes of Symptomatic Intracranial Large Artery Stenoses and Occlusions in China: The Chinese Intracranial Atherosclerosis (CICAS) Study. Stroke 2014, 45, 663–669. [Google Scholar] [CrossRef] [PubMed]
- Kiruluta, A.J.M.; González, R.G. Magnetic resonance angiography: Physical principles and applications. In Handbook of Clinical Neurology; Elsevier: Amsterdam, The Netherland, 2016; Volume 135, pp. 137–149. [Google Scholar]
- Ghouri, M.A.; Gupta, N.; Bhat, A.P.; Thimmappa, N.D.; Saboo, S.S.; Khandelwal, A.; Nagpal, P. CT and MR imaging of the upper extremity vasculature: Pearls, pitfalls, and challenges. Cardiovasc. Diagn. Ther. 2019, 9, S152. [Google Scholar] [CrossRef] [PubMed]
- Fourcade, A.; Khonsari, R.H. Deep Learning in Medical Image Analysis: A Third Eye for Doctors. J. Stomatol. Oral Maxillofac. Surg. 2019, 120, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, P.; Kossen, T.; Ritter, K.; Hennemuth, A.; Hildebrand, K.; Hilbert, A.; Sobesky, J.; Livne, M.; Galinovic, I.; Khalil, A.A. Generating 3D TOF-MRA volumes and segmentation labels using generative adversarial networks. Med. Image Anal. 2022, 78, 102396. [Google Scholar] [CrossRef] [PubMed]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; He, Y.; Li, F.; Han, L.; You, C.; Wang, B. Segment anything in medical images. Nat. Commun. 2024, 15, 654. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Hamdi, A.; Qi, Y.; Jin, Y.; Wu, J. Medical sam 2: Segment medical images as video via segment anything model 2. arXiv 2024, arXiv:2408.00874. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 2117–2125. [Google Scholar]
- Ryali, C.; Hu, Y.T.; Bolya, D.; Wei, C.; Fan, H.; Huang, P.Y.; Aggarwal, V.; Chowdhury, A.; Poursaeed, O.; Hoffman, J.; et al. Hiera: A hierarchical vision transformer without the bells-and-whistles. In Proceedings of the 40th International Conference on Machine Learning, PMLR, Honolulu, HI, USA, 23–29 July 2023; Volume 202, pp. 29441–29454. [Google Scholar]
- Lee, C.Y.; Xie, S.; Gallagher, P.; Zhang, Z.; Tu, Z. Deeply-supervised nets. In Proceedings of the 18th International Conference on Artificial Intelligence and Statistics, PMLR, San Diego, CA, USA, 9–12 May 2015; Volume 38, pp. 562–570. [Google Scholar]
- Zeiler, M.D.; Krishnan, D.; Taylor, G.W.; Fergus, R. Deconvolutional networks. In Proceedings of the 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Francisco, CA, USA, 13–18 June 2010; IEEE: Piscataway, NJ, USA, 2010; pp. 2528–2535. [Google Scholar]
- Hu, E.J.; Shen, Y.; Wallis, P.; Allen-Zhu, Z.; Li, Y.; Wang, S.; Wang, L.; Chen, W. Lora: Low-rank adaptation of large language models. ICLR 2022, 1, 3. [Google Scholar]
- Cooley, J.W.; Tukey, J.W. An Algorithm for the Machine Calculation of Complex Fourier Series. Math. Comput. 1965, 19, 297–301. [Google Scholar] [CrossRef]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.-S. CBAM: Convolutional Block Attention Module. In Computer Vision—ECCV 2018; Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y., Eds.; Springer: Cham, Switzerland, 2018; Volume 11211, pp. 3–19. [Google Scholar]
- Lin, T.-Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Chen, H.; Zhao, X.; Sun, H.; Dou, J.; Du, C.; Yang, R.; Lin, X.; Yu, S.; Liu, J.; Yuan, C.; et al. Cerebral artery segmentation challenge. In Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Vancouver, BC, Canada, 8–12 October 2023. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W., Frangi, A., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-UNet: UNet-like Pure Transformer for Medical Image Segmentation. In European Conference on Computer Vision; Springer Nature: Cham, Switzerland, 2022; pp. 205–218. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 565–571. [Google Scholar]
- Salehi, S.S.M.; Erdogmus, D.; Gholipour, A. Tversky loss function for image segmentation using 3D fully convolutional deep networks. In International Workshop on Machine Learning in Medical Imaging; Springer International Publishing: Cham, Switzerland, 2017; pp. 379–387. [Google Scholar]



| Component | Specification |
|---|---|
| Operating System | Ubuntu 22.04.4 LTS |
| CPU | 13th Gen Intel® Core™ i9-13900KF |
| GPU | NVIDIA® GeForce RTX 4090 |
| RAM | 64 GB DDR5 4800 MT/s |
| Method | Dice (%) | IoU (%) | HD95 (mm) | ASD (mm) |
|---|---|---|---|---|
| UNet | 81.92 ± 1.18 | 70.84 ± 1.70 | 51.13 ± 11.21 | 5.49 ± 1.03 |
| SwinUNet | 82.09 ± 1.00 | 71.04 ± 1.49 | 50.63 ± 11.82 | 5.46 ± 0.81 |
| TransUNet | 82.21 ± 1.08 | 71.19 ± 1.57 | 50.37 ± 12.13 | 5.40 ± 0.92 |
| nnUNet | 83.89 ± 0.92 | 73.86 ± 1.39 | 48.20 ± 10.89 | 5.33 ± 0.76 |
| Ours (MS) | 84.42 ± 0.97 | 74.55 ± 1.46 | 46.51 ± 12.02 | 5.03 ± 0.77 |
| Ours (MS2) | 84.49 ± 0.99 | 74.60 ± 1.49 | 46.30 ± 11.67 | 4.97 ± 0.80 |
| Method | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|
| UNet | 86.12 ± 1.55 | 81.52 ± 1.33 | 81.92 ± 1.18 |
| SwinUNet | 86.20 ± 1.37 | 81.61 ± 1.12 | 82.09 ± 1.00 |
| TransUNet | 86.22 ± 1.39 | 81.69 ± 1.21 | 82.21 ± 1.08 |
| nnUNet | 87.76 ± 1.31 | 83.51 ± 1.06 | 83.89 ± 0.92 |
| Ours (MS) | 88.06 ± 1.37 | 83.99 ± 1.08 | 84.42 ± 0.97 |
| Ours (MS2) | 87.83 ± 1.35 | 84.37 ± 1.13 | 84.49 ± 0.99 |
| Method | Dice (%) | IoU (%) | HD95 (mm) | ASD (mm) |
|---|---|---|---|---|
| Full (MS) 1 | 84.42 ± 0.97 | 74.55 ± 1.46 | 46.51 ± 12.02 | 5.03 ± 0.77 |
| w/o FL (MS) 2 | 84.27 ± 0.94 | 74.35 ± 1.40 | 47.26 ± 11.39 | 5.31 ± 0.76 |
| Full (MS2) 3 | 84.49 ± 0.99 | 74.60 ± 1.49 | 46.30 ± 11.67 | 4.97 ± 0.80 |
| w/o FL (MS2) 4 | 84.29 ± 0.93 | 74.38 ± 1.41 | 47.04 ± 11.62 | 5.26 ± 0.75 |
| Method | Dice (%) | IoU (%) | HD95 (mm) | ASD (mm) |
|---|---|---|---|---|
| Full (MS) 1 | 84.42 ± 0.97 | 74.55 ± 1.46 | 46.51 ± 12.02 | 5.03 ± 0.77 |
| Full (MS2) 2 | 84.49 ± 0.99 | 74.60 ± 1.49 | 46.30 ± 11.67 | 4.97 ± 0.80 |
| w/o FF 3 | 84.18 ± 0.97 | 74.27 ± 1.47 | 47.42 ± 11.46 | 5.29 ± 0.76 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, H.; Zhang, J.; Zhao, L. MedSAM/MedSAM2 Feature Fusion: Enhancing nnUNet for 2D TOF-MRA Brain Vessel Segmentation. J. Imaging 2025, 11, 202. https://doi.org/10.3390/jimaging11060202
Zhong H, Zhang J, Zhao L. MedSAM/MedSAM2 Feature Fusion: Enhancing nnUNet for 2D TOF-MRA Brain Vessel Segmentation. Journal of Imaging. 2025; 11(6):202. https://doi.org/10.3390/jimaging11060202
Chicago/Turabian StyleZhong, Han, Jiatian Zhang, and Lingxiao Zhao. 2025. "MedSAM/MedSAM2 Feature Fusion: Enhancing nnUNet for 2D TOF-MRA Brain Vessel Segmentation" Journal of Imaging 11, no. 6: 202. https://doi.org/10.3390/jimaging11060202
APA StyleZhong, H., Zhang, J., & Zhao, L. (2025). MedSAM/MedSAM2 Feature Fusion: Enhancing nnUNet for 2D TOF-MRA Brain Vessel Segmentation. Journal of Imaging, 11(6), 202. https://doi.org/10.3390/jimaging11060202






