Platelets Image Classification Through Data Augmentation: A Comparative Study of Traditional Imaging Augmentation and GAN-Based Synthetic Data Generation Techniques Using CNNs
Abstract
1. Introduction
2. Materials and Methods
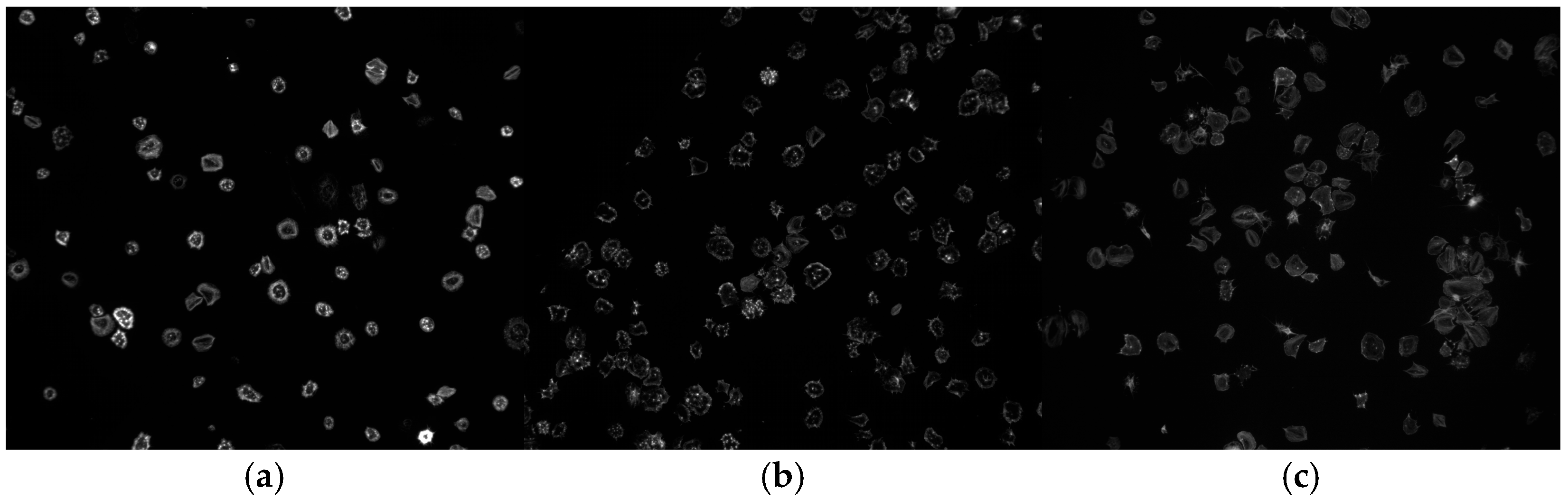
2.1. Data Description
2.2. Data Preparation and Organization
2.3. Transfer Learning Models
- Custom Model 1: Incorporating Conv2D, BatchNormalization, MaxPooling, Dropout, and two Dense layers.
- Custom Model 2: A simpler architecture with Conv2D and MaxPooling, followed by a single Dense layer.
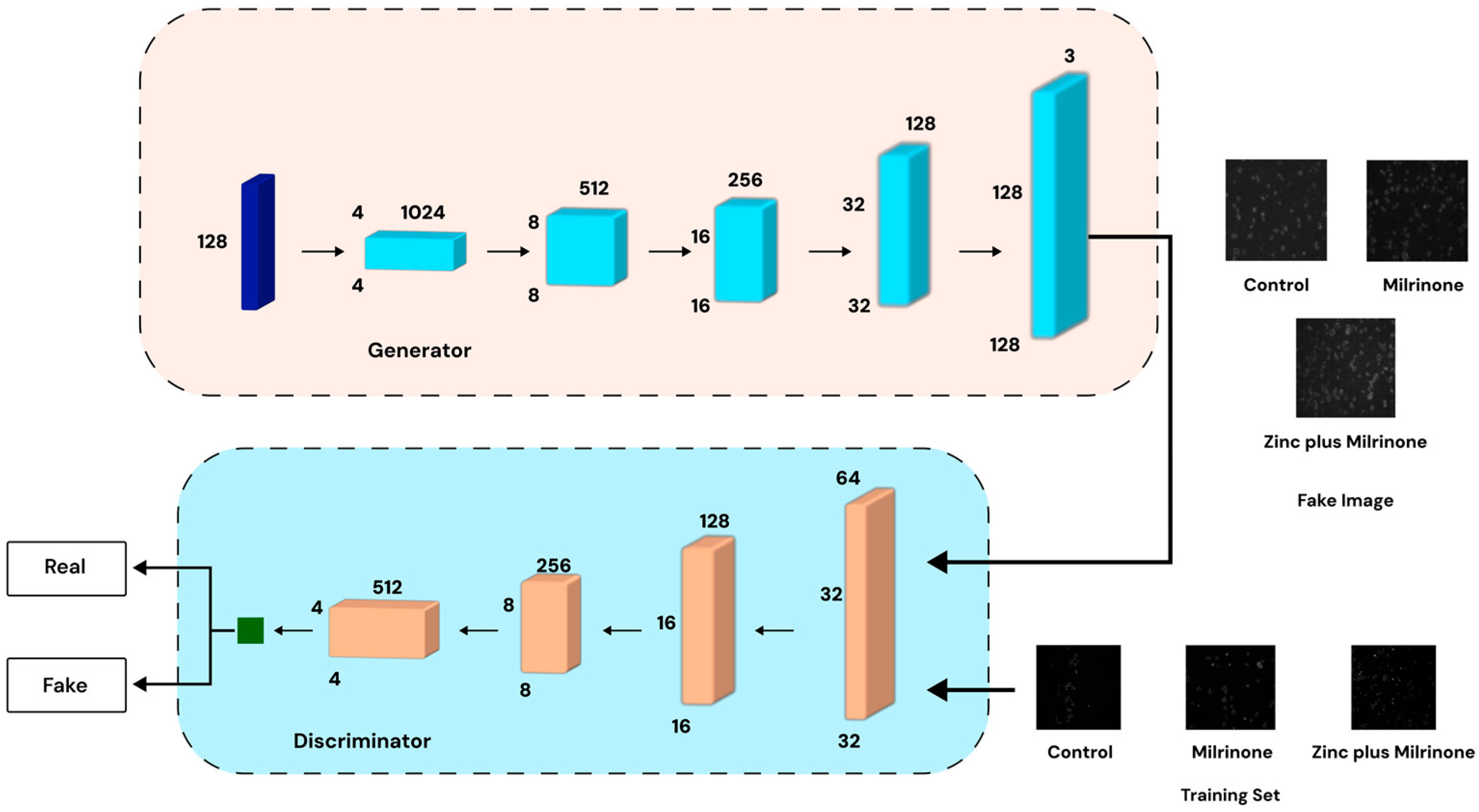
2.4. Data Augmentation (GAN Data Generation)
- Generator: This network transforms a latent noise vector into high-resolution (128 × 128 or 256 × 256 pixels) synthetic images using transpose convolutions, batch normalization, and LeakyReLU activations to ensure realistic feature generation.
- Critic (Discriminator): The discriminator evaluates both real and generated images through convolutional layers, outputting a scalar “realness” score to guide the generator’s improvement. To maintain training stability, the critic undergoes multiple updates per generator update before reaching convergence.
2.5. Model Training and Evaluation
2.5.1. Model Training
- Optimizer: Adam with a learning rate of 0.001.
- Batch Size: 32 (128 tested in some trials).
- Loss Function: Categorical cross-entropy for the multi-class classification.
2.5.2. Evaluation Metrics
3. Results
3.1. Original Dataset (71 Images)
| Models (Batch Size 32) | Accuracy (%) | F1-Score (%) | Precision (%) | Recall (%) |
|---|---|---|---|---|
| Custom Model 1 | 62 | 56 | 68 | 62 |
| Custom Model 2 | 57 | 42 | 33 | 57 |
| DenseNet121 | 81 | 79 | 84 | 81 |
| DenseNet169 | 71 | 67 | 77 | 71 |
| DenseNet201 | 76 | 74 | 83 | 76 |
| VGG16 | 57 | 47 | 43 | 57 |
| VGG19 | 52 | 45 | 40 | 52 |
| VGG19-FF | 62 | 59 | 63 | 62 |
| InceptionV3 | 62 | 51 | 46 | 62 |
| InceptionResNetV2 | 71 | 69 | 76 | 71 |
| AlexNet | 62 | 56 | 59 | 62 |
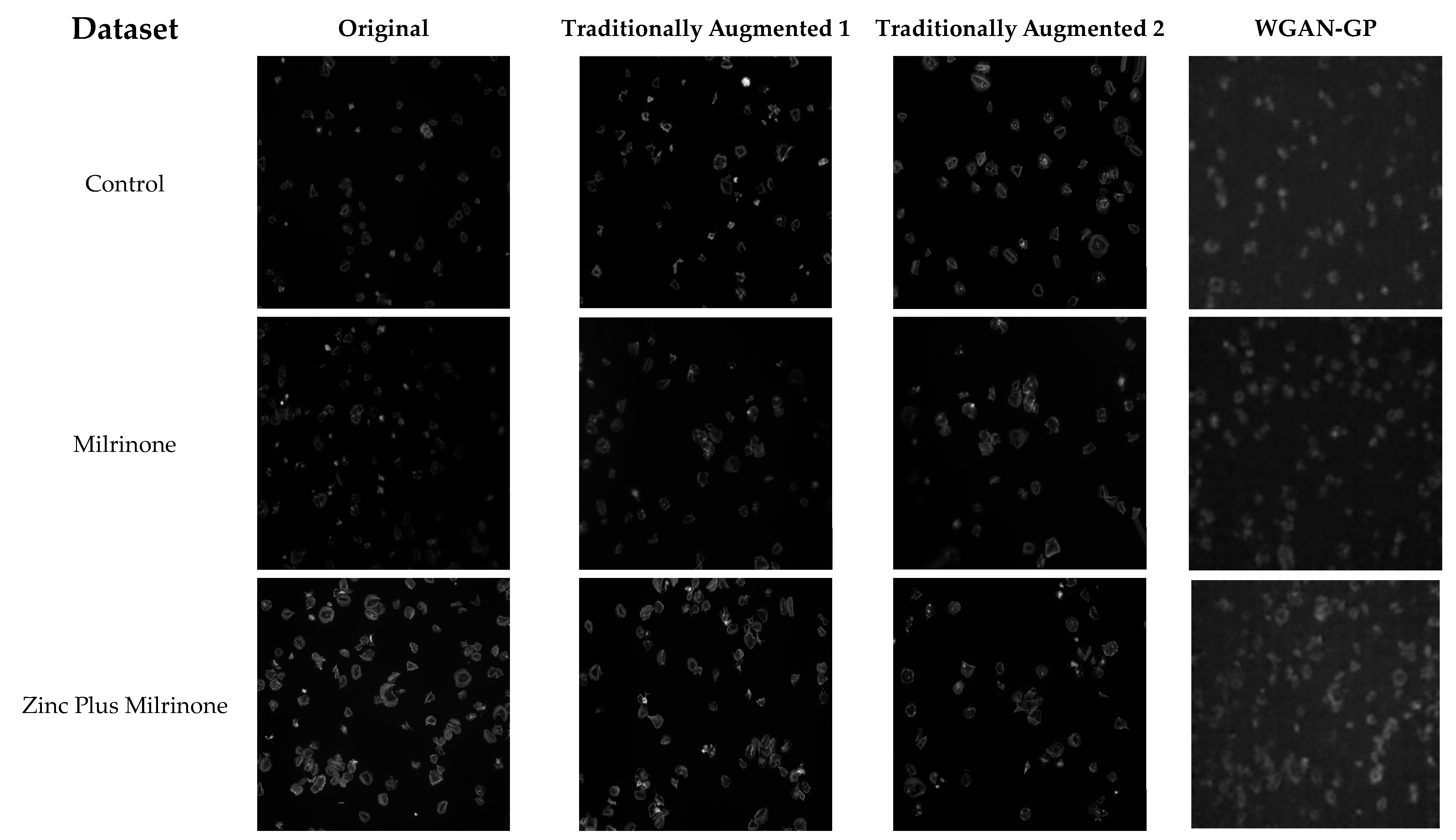
3.2. Augmented Dataset Level 1 (141 Images)
3.3. Augmented Dataset Level 2 (1463 Images)
3.4. GAN-Augmented Dataset (300 Images)
3.5. Synthetic Dataset Result
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CNNs | Convolutional Neural Networks |
| GAN | Generative Adversarial Network |
| WGAN-GP | Wasserstein GAN with Gradient Penalty |
| FID | Fréchet Inception Distance |
| IS | Inception Score |
| FF | Fine-Tuning |
| ReLU | Rectified Linear Unit |
| TPUs | Tensor Processing Units |
| GPUs | Graphics Processing Units |
| cGAN | Conditional Generative Adversarial Network |
| CycleGAN | Cycle-Consistent Generative Adversarial Network |
| MRI | Magnetic Resonance Imaging |
| TP | True Positive |
| FP | False Positive |
| TN | True Negative |
| FN | False Negative |
References
- Vinholt, P. The Role of Platelets in Bleeding in Patients with Thrombocytopenia and Hematological Disease. Clin. Chem. Lab. Med. 2019, 57, 1808–1817. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yasumoto, A.; Lei, C.; Huang, C.J.; Kobayashi, H.; Wu, Y.; Yan, S.; Sun, C.W.; Yatomi, Y.; Goda, K. Intelligent Classification of Platelet Aggregates by Agonist Type. eLife 2020, 9, e52938. [Google Scholar] [CrossRef]
- Babker, A.M.; Suliman, R.S.; Elshaikh, R.H.; Boboyorov, S.; Lyashenko, V. Sequence of Simple Digital Technologies for Detection of Platelets in Medical Images. Biomed. Pharmacol. J. 2024, 17, 141–152. [Google Scholar] [CrossRef]
- Hamid, G.A. Clinical Hematology; Deenanath Mangeshkar Hospital & Research Center: Pune, India, 2013; Available online: https://www.dmhospital.org/specialty-details/CLINICAL-HAEMATOLOGY (accessed on 20 June 2024).
- Coupland, C.A.; Naylor-Adamson, L.; Booth, Z.; Price, T.W.; Gil, H.M.; Firth, G.; Avery, M.; Ahmed, Y.; Stasiuk, G.J.; Calaminus, S.D.J. Platelet Zinc Status Regulates Prostaglandin-Induced Signaling, Altering Thrombus Formation. J. Thromb. Haemost. 2023, 21, 2545–2558. [Google Scholar] [CrossRef]
- Costa, M.I.; Sarmento-Ribeiro, A.B.; Gonçalves, A.C. Zinc: From Biological Functions to Therapeutic Potential. Int. J. Mol. Sci. 2023, 24, 4822. [Google Scholar] [CrossRef]
- Gaydos, L.A.; Freireich, E.J.; Mantel, N. The Quantitative Relation between Platelet Count and Hemorrhage in Patients with Acute Leukemia. N. Engl. J. Med. 1962, 266, 905–909. [Google Scholar] [CrossRef]
- Mustafa, M.E.; Mansoor, M.M.; Mohammed, A.; Babker, A.A. Evaluation of Platelets Count and Coagulation Parameters among Patients with Liver Disease. World J. Pharm. Res. 2015, 4, 360–368. [Google Scholar]
- Isbister, J.P. Common Presentations of Haematological Diseases. Available online: https://journals.co.za/doi/pdf/10.10520/AJA02599333_2849 (accessed on 20 June 2024).
- Goliwas, K.F.; Richter, J.R.; Pruitt, H.C.; Araysi, L.M.; Anderson, N.R.; Samant, R.S.; Lobo-Ruppert, S.M.; Berry, J.L.; Frost, A.R. Methods to Evaluate Cell Growth, Viability, and Response to Treatment in a Tissue Engineered Breast Cancer Model. Sci. Rep. 2017, 7, 14167. [Google Scholar] [CrossRef]
- Mohammed, E.A.; Mohamed, M.M.; Far, B.H.; Naugler, C. Peripheral Blood Smear Image Analysis: A Comprehensive Review. J. Pathol. Inform. 2014, 5, 9. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, M.; Wu, G.; Yao, C.; Zhang, J. Recent Advances in Morphological Cell Image Analysis. Comput. Math. Methods Med. 2012, 2012, 101536. [Google Scholar] [CrossRef]
- Santos-Silva, M.A.; Sousa, N.; Sousa, J.C. Artificial Intelligence in Routine Blood Tests. Front. Med. Eng. 2024, 2, 1369265. [Google Scholar] [CrossRef]
- Farfour, E.; Clichet, V.; Péan de Ponfilly, G.; Carbonnelle, E.; Vasse, M. Impact of COVID-19 Pandemic on Blood Culture Practices and Bacteremia Epidemiology. Diagn. Microbiol. Infect. Dis. 2023, 107, 116002. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, X.; Cao, X.; Huang, C.; Liu, E.; Qian, S.; Liu, X.; Wu, Y.; Dong, F.; Qiu, C.-W.; et al. Artificial Intelligence: A Powerful Paradigm for Scientific Research. Innovation 2021, 2, 100179. [Google Scholar] [CrossRef] [PubMed]
- Maturana, C.R.; de Oliveira, A.D.; Nadal, S.; Bilalli, B.; Serrat, F.Z.; Soley, M.E.; Igual, E.S.; Bosch, M.; Lluch, A.V.; Abelló, A.; et al. Advances and Challenges in Automated Malaria Diagnosis Using Digital Microscopy Imaging with Artificial Intelligence Tools: A Review. Front. Microbiol. 2022, 13, 1006659. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, Y.; Wu, Q.; Xia, Y. Medical Image Classification Using Deep Learning. IEEE Trans. Med. Imaging 2018, 37, 1249–1258. [Google Scholar] [CrossRef]
- Li, M.; Jiang, Y.; Zhang, Y.; Zhu, H. Medical Image Analysis Using Deep Learning Algorithms. Front. Public Health 2023, 11, 1273253. [Google Scholar] [CrossRef]
- Berryman, S.; Matthews, K.; Lee, J.H.; Duffy, S.P.; Ma, H. Image-Based Phenotyping of Disaggregated Cells Using Deep Learning. Commun. Biol. 2020, 3, 1399. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.; Rochman, N.D.; Sun, S.X. Cell Type Classification and Unsupervised Morphological Phenotyping from Low-Resolution Images Using Deep Learning. Sci. Rep. 2019, 9, 50010. [Google Scholar] [CrossRef]
- Sandfort, V.; Yan, K.; Pickhardt, P.J.; Summers, R.M. Data Augmentation Using Generative Adversarial Networks (CycleGAN) to Improve Generalizability in CT Segmentation Tasks. Sci. Rep. 2019, 9, 16884. [Google Scholar] [CrossRef]
- Perez, L.; Wang, J. The Effectiveness of Data Augmentation in Image Classification Using Deep Learning. arXiv 2017, arXiv:1712.04621. [Google Scholar]
- Yi, X.; Walia, E.; Babyn, P. Generative Adversarial Network in Medical Imaging: A Review. Med. Image Anal. 2019, 58, 101552. [Google Scholar] [CrossRef]
- Frid-Adar, M.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. Synthetic Data Augmentation Using GAN for Improved Liver Lesion Classification. IEEE Trans. Med. Imaging 2018, 38, 809–818. [Google Scholar]
- Salehinejad, H.; Colak, E.; Dowdell, T.; Barfett, J.; Georgescu, B. Synthesizing Chest X-ray Pathology for Training Deep Convolutional Neural Networks. arXiv 2018, arXiv:1807.07514. [Google Scholar] [CrossRef]
- Abidoye, I.; Ikeji, F.; Sousa, E. Automatic Classification of Platelets Images: Augmented and Non-Augmented Comparison of Pre-Trained versus Custom Models [Poster]. Presented at ResearchGate. 2025. Available online: https://www.researchgate.net/publication/385884476_Automatic_Classification_of_Platelets_Images_Augmented_and_Non-augmented_Comparison_of_Pre-trained_Versus_Custom_Models (accessed on 20 June 2024).
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. In Proceedings of the AAAI Conference on Artificial Intelligence, Phoenix, AZ, USA, 12–17 February 2016. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Arjovsky, M.; Chintala, S.; Bottou, L. Wasserstein GAN. arXiv 2017, arXiv:1701.07875. [Google Scholar]
- Arjovsky, M.; Bottou, L. Towards Principled Methods for Training Generative Adversarial Networks. arXiv 2017, arXiv:1701.04862. [Google Scholar]
- Shahinfar, S.; Meek, P.; Falzon, G. How Many Images Do I Need? Understanding How Sample Size per Class Affects Deep Learning Model Performance Metrics for Balanced Designs in Autonomous Wildlife Monitoring. Ecol. Inform. 2020, 57, 101085. [Google Scholar] [CrossRef]
| Models (Batch Size 32) | Accuracy (%) | F1-Score (%) | Precision (%) | Recall (%) |
|---|---|---|---|---|
| Custom Model 1 | 67 | 66 | 69 | 67 |
| Custom Model 2 | 38 | 21 | 15 | 38 |
| DenseNet121 | 79 | 79 | 79 | 79 |
| DenseNet169 | 79 | 78 | 83 | 79 |
| DenseNet201 | 86 | 86 | 88 | 86 |
| VGG16 | 62 | 62 | 72 | 62 |
| VGG19 | 64 | 64 | 68 | 64 |
| VGG19-FF | 76 | 76 | 80 | 76 |
| InceptionV3 | 76 | 76 | 79 | 76 |
| InceptionResNetV2 | 71 | 69 | 80 | 71 |
| AlexNet | 67 | 65 | 66 | 67 |
| Models (Batch Size 32) | Accuracy (%) | F1-Score (%) | Precision (%) | Recall (%) |
|---|---|---|---|---|
| Custom Model 1 | 97 | 97 | 97 | 97 |
| Custom Model 2 | 88 | 87 | 91 | 88 |
| DenseNet121 | 97 | 97 | 98 | 97 |
| DenseNet169 | 97 | 97 | 97 | 97 |
| DenseNet201 | 98 | 98 | 98 | 98 |
| VGG16 | 97 | 97 | 97 | 97 |
| VGG19 | 94 | 94 | 94 | 94 |
| VGG19-FF | 95 | 95 | 95 | 95 |
| InceptionV3 | 99 | 99 | 99 | 99 |
| InceptionResNetV2 | 99 | 99 | 99 | 99 |
| AlexNet | 30 | 14 | 9 | 30 |
| Models (Batch Size 32) | Accuracy (%) | F1-Score (%) | Precision (%) | Recall (%) |
|---|---|---|---|---|
| Custom Model 1 | 97 | 94 | 95 | 94 |
| Custom Model 2 | 87 | 87 | 87 | 87 |
| DenseNet121 | 97 | 97 | 97 | 97 |
| DenseNet169 | 91 | 91 | 93 | 91 |
| DenseNet201 | 96 | 96 | 96 | 96 |
| VGG16 | 83 | 83 | 85 | 83 |
| VGG19 | 89 | 89 | 89 | 89 |
| VGG19-FF | 88 | 88 | 89 | 88 |
| InceptionV3 | 94 | 94 | 95 | 94 |
| InceptionResNetV2 | 90 | 90 | 90 | 90 |
| AlexNet | 74 | 75 | 75 | 74 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abidoye, I.; Ikeji, F.; Coupland, C.A.; Calaminus, S.D.J.; Sander, N.; Sousa, E. Platelets Image Classification Through Data Augmentation: A Comparative Study of Traditional Imaging Augmentation and GAN-Based Synthetic Data Generation Techniques Using CNNs. J. Imaging 2025, 11, 183. https://doi.org/10.3390/jimaging11060183
Abidoye I, Ikeji F, Coupland CA, Calaminus SDJ, Sander N, Sousa E. Platelets Image Classification Through Data Augmentation: A Comparative Study of Traditional Imaging Augmentation and GAN-Based Synthetic Data Generation Techniques Using CNNs. Journal of Imaging. 2025; 11(6):183. https://doi.org/10.3390/jimaging11060183
Chicago/Turabian StyleAbidoye, Itunuoluwa, Frances Ikeji, Charlie A. Coupland, Simon D. J. Calaminus, Nick Sander, and Eva Sousa. 2025. "Platelets Image Classification Through Data Augmentation: A Comparative Study of Traditional Imaging Augmentation and GAN-Based Synthetic Data Generation Techniques Using CNNs" Journal of Imaging 11, no. 6: 183. https://doi.org/10.3390/jimaging11060183
APA StyleAbidoye, I., Ikeji, F., Coupland, C. A., Calaminus, S. D. J., Sander, N., & Sousa, E. (2025). Platelets Image Classification Through Data Augmentation: A Comparative Study of Traditional Imaging Augmentation and GAN-Based Synthetic Data Generation Techniques Using CNNs. Journal of Imaging, 11(6), 183. https://doi.org/10.3390/jimaging11060183








