GCNet: A Deep Learning Framework for Enhanced Grape Cluster Segmentation and Yield Estimation Incorporating Occluded Grape Detection with a Correction Factor for Indoor Experimentation
Abstract
1. Introduction
- Grape Counting Network (GCNet): A novel deep learning framework designed to address the issue of occluded berry counting using object segmentation and a correction factor.
- GrapeSet: A new dataset for indoor experimentation having annotated images of grape clusters, including grape count and weight data, to enable more accurate yield estimation.
- Ablation study: An evaluation of the role of segmentation in grape yield estimation, showing its necessity and impact on accuracy.
- Efficacy study: Analysis of how image resolution, foliage density, and grape color affect the performance of GCNet in estimating grape yield.
2. Related Works
2.1. Feature-Based Segmentation Methods
2.2. Learning-Based Segmentation Methods
2.3. Occlusion in Cluster Segmentation
2.4. Existing Grape Datasets
| Dataset | Grape Color | Foliage | Background | Acquisition | Num. of Images in Dataset | Num. of Grape Clusters in Each Image | Ground Truth | Supplemental Feature Data |
|---|---|---|---|---|---|---|---|---|
| Grape CS—ML Datasets (1–4) [3] | Blue, Green, and Purple | Low | Green Natural | Outdoor | 2016 | 1 | X | Volume and/or color references |
| Grape CS—ML Dataset (5) [3] | Green | Low | Green Natural | Outdoor | 62 | 1–3 | ✓ | Num. of berries, volume, pH, hue, bunch weight, TSS, etc. |
| Grapevine Bunch Detection Dataset [51] | Blue and Green | Low, Medium | Green Natural | Outdoor | 910 | 1–3 | X | Annotation and condition of grape bunch |
| Embrapa WGISD [53] | Green | Medium | Green Natural | Outdoor | 300 | 5–25 | X | Boxed and masked cluster annotations |
| AI4Agriculture Grape Dataset [67] | Blue | High | Green Natural | Outdoor | 250 | 5–25 | X | Annotations of bounding boxes |
| wGrapeUNIPD-DL [54] | Green | High | Green Natural | Outdoor | 373 | 5–25 | X | Color reference |
| GrapeNet Dataset (3) [68] | Green | Low | Black, Coral, and Green Natural | Outdoor | 1705 | 1–2 | X | Augmented images |
| Segmentation of Wine Berries [50] | Green | High | White | Outdoor | 42 | 5–25 | ✓ | Labels of berries |
| GrapeSet (ours) | Blue, Green, and Purple | Low, Medium, and High | White and Green Bokeh | Indoor | 2160 | 3 | ✓ | Weight of grape bunches, actual count of berries |
3. Proposed Method
3.1. Problem Definition
- denote the set of visible grapes in image
- denote the set of occluded (hidden) grapes in image .
- Segmentation of Grape Clusters: A function segments the imge to produce a mask that identifies grape clusters, thereby isolating from background elements.
- Mask Overlay: The segmented mask is refined to reduce background noise and highlight grape clusters.
- Generating Final Count: A correction factor is applied via a regression model that estimates , adjusting to account for occluded grapes.
| Algorithm 1 Proposed Grape Counting Network (GCNet) | |
| 1: | Input: where . |
| 2: | Output: The number of grapes where is the generated count for the image . Stage 1: Segmentation of Grape Clusters |
| 3: | begin |
| 4: | for = 1 to do |
| 5: | |
| 6: | end for |
| 7: | return |
| 8: | end Stage 2: Overlaying |
| 9: | begin |
| 10: | for = 1 to do |
| 11: | |
| 12: | end for |
| 13: | return |
| 14: | end Stage 3: Generating Final Count |
| 15: | begin |
| 16: | for = 1 to do |
| 17: | |
| 18: | end for |
| 19: | return |
| 20: | end |
3.2. Overview
3.2.1. CNN-Based U-Net Model
3.2.2. Stage 1: Segmentation of Grape Clusters
3.2.3. Stage 2: Mask Overlay
3.2.4. Stage 3: Generating Final Count
3.3. Implementation
4. Proposed Dataset
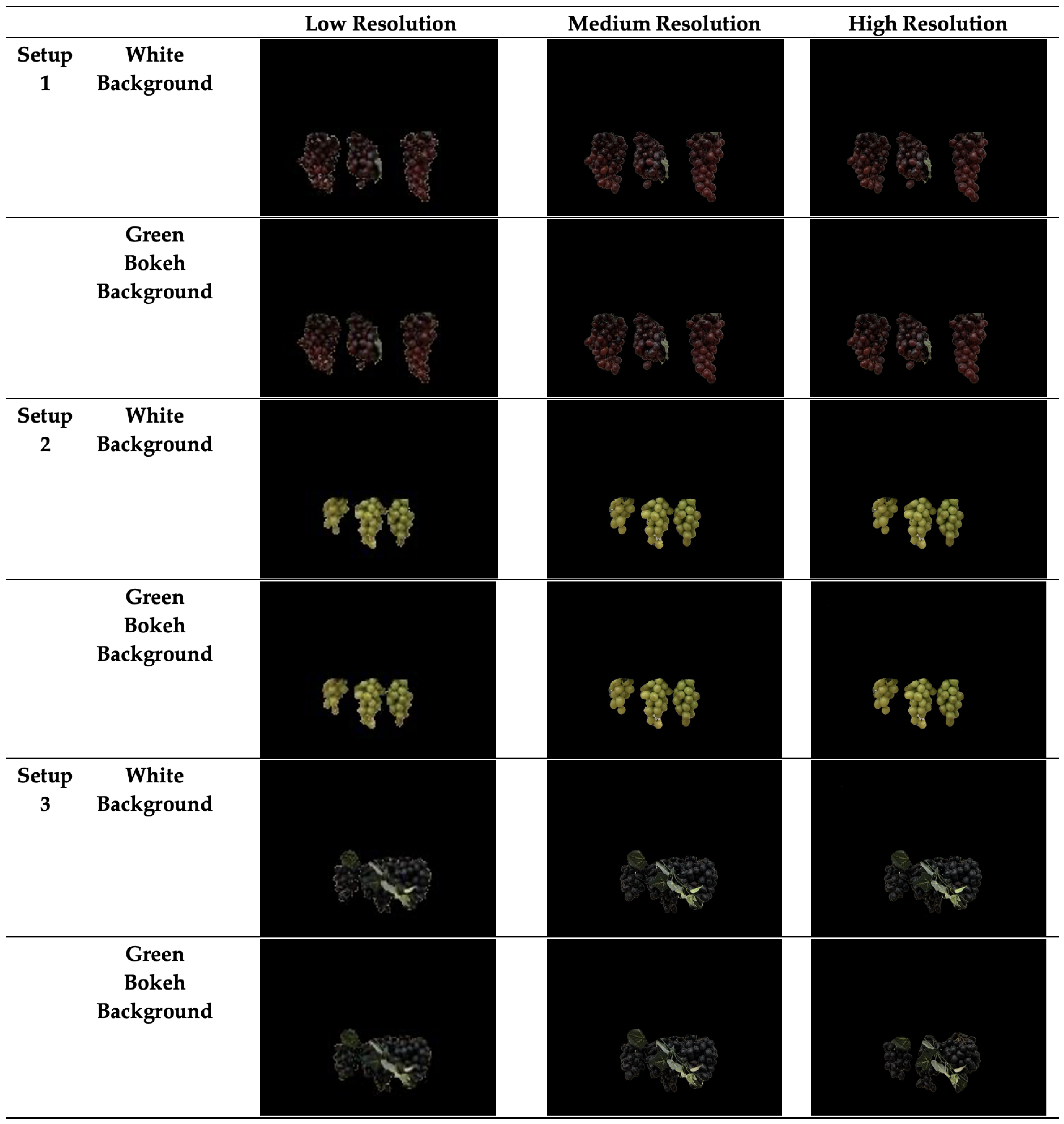
4.1. Imaging Setup
4.2. Dataset Organization
- Setup 1 (Low Foliage): Minimal foliage coverage, with grapes clearly visible and non-overlapping.
- Setup 2 (Medium Foliage): Grapes are closer together, overlapping slightly, and covered by more foliage, increasing the complexity of segmentation.
- Setup 3 (High Foliage): An additional layer of foliage is introduced, particularly with blue grapes, significantly increasing the difficulty of detecting and segmenting the clusters.

5. Experiment Setup
5.1. Ablation Study
5.1.1. Resolution Experiment
5.1.2. Color Experiment
5.2. Segmentation Study
5.2.1. Resolution Experiment
5.2.2. Color Experiment
6. Experimental Results
6.1. Ablation Study
6.1.1. Resolution Experiment
6.1.2. Color Experiment
6.2. Segmentation Study
6.2.1. Resolution Experiment
6.2.2. Color Experiment
7. Discussion
7.1. A Comparative Study with Prior Studies
7.2. Impact of Segmentation on Model Performance
7.3. Influence of Image Resolution on Segmentation Accuracy
7.4. Influence of Grape Color on Segmentation Performance
7.5. Future Prospects for Grape Counting
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alston, J.M.; Sambucci, O. Grapes in the World Economy. In The Grape Genome; Compendium of Plant Genomes; Cantu, D., Walker, M.A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–24. [Google Scholar] [CrossRef]
- Pinheiro, I.; Moreira, G.; Queirós da Silva, D.; Magalhães, S.; Valente, A.; Moura Oliveira, P.; Cunha, M.; Santos, F. Deep learning YOLO-based solution for grape bunch detection and assessment of biophysical lesions. Agronomy 2023, 13, 1120. [Google Scholar] [CrossRef]
- Seng, K.P.; Ang, L.-M.; Schmidtke, L.M.; Rogiers, S.Y. Computer vision and machine learning for viticulture technology. IEEE Access 2018, 6, 67494–67510. [Google Scholar] [CrossRef]
- Sozzi, M.; Cantalamessa, S.; Cogato, A.; Kayad, A.; Marinello, F. Automatic bunch detection in white grape varieties using YOLOv3, YOLOv4, and YOLOv5 deep learning algorithms. Agronomy 2022, 12, 319. [Google Scholar] [CrossRef]
- Razouk, H.; Kern, R.; Mischitz, M.; Moser, J.; Memic, M.; Liu, L.; Burmer, C.; Safont-Andreu, A. AI-Based Knowledge Management System for Risk Assessment and Root Cause Analysis in Semiconductor Industry. In Artificial Intelligence for Digitising Industry–Applications; River Publishers: Aalborg, Denmark, 2022; pp. 113–129. Available online: https://www.taylorfrancis.com/chapters/oa-edit/10.1201/9781003337232-11/ai-based-knowledge-management-system-risk-assessment-root-cause-analysis-semiconductor-industry-houssam-razouk-roman-kern-martin-mischitz-josef-moser-mirhad-memic-lan-liu-christian-burmer-anna-safont-andreu (accessed on 2 October 2024).
- Zabawa, L.; Kicherer, A.; Klingbeil, L.; Milioto, A.; Topfer, R.; Kuhlmann, H.; Roscher, R. Detection of single grapevine berries in images using fully convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Long Beach, CA, USA, 16–17 June 2019. [Google Scholar]
- Bruni, V.; Dominijanni, G.; Vitulano, D. A Machine-Learning Approach for Automatic Grape-Bunch Detection Based on Opponent Colors. Sustainability 2023, 15, 4341. [Google Scholar] [CrossRef]
- Palacios, F.; Melo-Pinto, P.; Diago, M.P.; Tardaguila, J. Deep learning and computer vision for assessing the number of actual berries in commercial vineyards. Biosyst. Eng. 2022, 218, 175–188. [Google Scholar] [CrossRef]
- Chen, S.W.; Shivakumar, S.S.; Dcunha, S.; Das, J.; Okon, E.; Qu, C.; Taylor, C.J.; Kumar, V. Counting apples and oranges with deep learning: A data-driven approach. IEEE Robot. Autom. Lett. 2017, 2, 781–788. [Google Scholar] [CrossRef]
- Xiao, F.; Wang, H.; Li, Y.; Cao, Y.; Lv, X.; Xu, G. Object detection and recognition techniques based on digital image processing and traditional machine learning for fruit and vegetable harvesting robots: An overview and review. Agronomy 2023, 13, 639. [Google Scholar] [CrossRef]
- Santos, T.T.; De Souza, L.L.; Santos, A.A.D.; Avila, S. Grape detection, segmentation, and tracking using deep neural networks and three-dimensional association. Comput. Electron. Agric. 2020, 170, 105247. [Google Scholar] [CrossRef]
- Koirala, A.; Walsh, K.B.; Wang, Z. Attempting to estimate the unseen—Correction for occluded fruit in tree fruit load estimation by machine vision with deep learning. Agronomy 2021, 11, 347. [Google Scholar] [CrossRef]
- Häni, N.; Roy, P.; Isler, V. A comparative study of fruit detection and counting methods for yield mapping in apple orchards. J. Field Robot. 2020, 37, 263–282. [Google Scholar] [CrossRef]
- Taner, A.; Mengstu, M.T.; Selvi, K.Ç.; Duran, H.; Kabaş, Ö.; Gür, İ.; Karaköse, T.; Gheorghiță, N.E. Multiclass apple varieties classification using machine learning with histogram of oriented gradient and color moments. Appl. Sci. 2023, 13, 7682. [Google Scholar] [CrossRef]
- Taner, A.; Mengstu, M.T.; Selvi, K.Ç.; Duran, H.; Gür, İ.; Ungureanu, N. Apple Varieties Classification Using Deep Features and Machine Learning. Agriculture 2024, 14, 252. [Google Scholar] [CrossRef]
- Torras, C. Computer Vision: Theory and Industrial Applications; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Szeliski, R. Computer Vision: Algorithms and Applications; Springer Nature: London, UK, 2022; Available online: https://books.google.com/books?hl=en&lr=&id=QptXEAAAQBAJ&oi=fnd&pg=PR9&dq=Szeliski,+R.+(2022).+Computer+Vision:+Algorithms+and+Applications.+Switzerland:+Springer+International+Publishing.+&ots=BNwhC4Sytm&sig=113JWJn4mMroBkqzURVwKmE33ng (accessed on 2 October 2024).
- Tian, H.; Wang, T.; Liu, Y.; Qiao, X.; Li, Y. Computer vision technology in agricultural automation—A review. Inf. Process. Agric. 2020, 7, 1–19. [Google Scholar] [CrossRef]
- Wu, G.; Li, B.; Zhu, Q.; Huang, M.; Guo, Y. Using color and 3D geometry features to segment fruit point cloud and improve fruit recognition accuracy. Comput. Electron. Agric. 2020, 174, 105475. [Google Scholar] [CrossRef]
- Zhang, C.; Zou, K.; Pan, Y. A method of apple image segmentation based on color-texture fusion feature and machine learning. Agronomy 2020, 10, 972. [Google Scholar] [CrossRef]
- Lin, G.; Tang, Y.; Zou, X.; Wang, C. Three-dimensional reconstruction of guava fruits and branches using instance segmentation and geometry analysis. Comput. Electron. Agric. 2021, 184, 106107. [Google Scholar] [CrossRef]
- Wang, X.; Kang, H.; Zhou, H.; Au, W.; Chen, C. Geometry-aware fruit grasping estimation for robotic harvesting in apple orchards. Comput. Electron. Agric. 2022, 193, 106716. [Google Scholar] [CrossRef]
- Pothen, Z.S.; Nuske, S. Texture-based fruit detection via images using the smooth patterns on the fruit. In Proceedings of the 2016 IEEE International Conference on Robotics and Automation (ICRA), Stokholm, Sweden, 16–21 May 2016; pp. 5171–5176. Available online: https://ieeexplore.ieee.org/abstract/document/7487722/ (accessed on 7 October 2024).
- Qureshi, W.S.; Satoh, S.; Dailey, M.N.; Ekpanyapong, M. Dense segmentation of textured fruits in video sequences. In Proceedings of the 2014 International Conference on Computer Vision Theory and Applications (VISAPP), Lisbon, Portugal, 5–8 January 2014; pp. 441–447. Available online: https://ieeexplore.ieee.org/abstract/document/7294963/ (accessed on 7 October 2024).
- Zele, H. Fruit and Vegetable Segmentation with Decision Trees. In Data Management, Analytics and Innovation; Lecture Notes in Networks and Systems; Sharma, N., Goje, A.C., Chakrabarti, A., Bruckstein, A.M., Eds.; Springer Nature: Singapore, 2024; Volume 997, pp. 333–343. [Google Scholar] [CrossRef]
- Dávila-Rodríguez, I.-A.; Nuño-Maganda, M.-A.; Hernández-Mier, Y.; Polanco-Martagón, S. Decision-tree based pixel classification for real-time citrus segmentation on FPGA. In Proceedings of the 2019 International Conference on ReConFigurable Computing and FPGAs (ReConFig), Cancun, Mexico, 9–11 December 2019; pp. 1–8. Available online: https://ieeexplore.ieee.org/abstract/document/8994792/ (accessed on 7 October 2024).
- Pham, V.H.; Lee, B.R. An image segmentation approach for fruit defect detection using k-means clustering and graph-based algorithm. Vietnam J. Comput. Sci. 2015, 2, 25–33. [Google Scholar] [CrossRef]
- Dubey, S.R.; Dixit, P.; Singh, N.; Gupta, J.P. Infected fruit part detection using K-means clustering segmentation technique. Int. J. Interact. Multimed. Artif. Intell. 2013, 2. Available online: https://reunir.unir.net/handle/123456789/9723 (accessed on 7 October 2024). [CrossRef][Green Version]
- Henila, M.; Chithra, P. Segmentation using fuzzy cluster-based thresholding method for apple fruit sorting. IET Image Process. 2020, 14, 4178–4187. [Google Scholar] [CrossRef]
- Tanco, M.M.; Tejera, G.; Di Martino, M. Computer Vision based System for Apple Detection in Crops. In Proceedings of the 13th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2018), Funchal, Madeira, Portugal, 27–29 January 2018; pp. 239–249. Available online: https://pdfs.semanticscholar.org/4fd2/ffb430ab3338c6d7599e68f7f5ae800bce6c.pdf (accessed on 2 October 2024).
- Mizushima, A.; Lu, R. An image segmentation method for apple sorting and grading using support vector machine and Otsu’s method. Comput. Electron. Agric. 2013, 94, 29–37. [Google Scholar] [CrossRef]
- Schölkopf, B. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond; The MIT Press: Cambridge, MA, USA, 2002; pp. 239–249. Available online: https://direct.mit.edu/books/monograph/1821/bookpreview-pdf/2411769 (accessed on 4 December 2024).
- Liu, S.; Whitty, M. Automatic grape bunch detection in vineyards with an SVM classifier. J. Appl. Log. 2015, 13, 643–653. [Google Scholar] [CrossRef]
- Dhiman, B.; Kumar, Y.; Hu, Y.-C. A general purpose multi-fruit system for assessing the quality of fruits with the application of recurrent neural network. Soft Comput. 2021, 25, 9255–9272. [Google Scholar] [CrossRef]
- Itakura, K.; Saito, Y.; Suzuki, T.; Kondo, N.; Hosoi, F. Estimation of citrus maturity with fluorescence Spectroscopy Using Deep Learning. Horticulturae 2019, 5, 2. [Google Scholar] [CrossRef]
- Vaviya, H.; Yadav, A.; Vishwakarma, V.; Shah, N. Identification of Artificially Ripened Fruits Using Machine Learning. In Proceedings of the 2nd International Conference on Advances in Science & Technology (ICAST), Bahir Dar, Ethiopia, 2–4 August 2019; Available online: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3368903 (accessed on 7 October 2024).
- Marani, R.; Milella, A.; Petitti, A.; Reina, G. Deep neural networks for grape bunch segmentation in natural images from a consumer-grade camera. Precision Agric. 2021, 22, 387–413. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, A.; Liu, J.; Faheem, M. A comparative study of semantic segmentation models for identification of grape with different varieties. Agriculture 2021, 11, 997. [Google Scholar] [CrossRef]
- Peng, J.; Ouyang, C.; Peng, H.; Hu, W.; Wang, Y.; Jiang, P. MultiFuseYOLO: Redefining Wine Grape Variety Recognition through Multisource Information Fusion. Sensors 2024, 24, 2953. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhang, D.; Luo, L.; Yi, T. PointResNet: A grape bunches point cloud semantic segmentation model based on feature enhancement and improved PointNet++. Comput. Electron. Agric. 2024, 224, 109132. [Google Scholar] [CrossRef]
- Liang, J.; Wang, S. Key Components Design of the Fresh Grape Picking Robot in Equipment Greenhouse. In Proceedings of the 2023 International Conference on Service Robotics (ICoSR), Shanghai, China, 21–23 July 2023; pp. 16–21. Available online: https://ieeexplore.ieee.org/abstract/document/10429043/ (accessed on 5 December 2024).
- Huang, X.; Peng, D.; Qi, H.; Zhou, L.; Zhang, C. Detection and Instance Segmentation of Grape Clusters in Orchard Environments Using an Improved Mask R-CNN Model. Agriculture 2024, 14, 918. [Google Scholar] [CrossRef]
- Yi, X.; Zhou, Y.; Wu, P.; Wang, G.; Mo, L.; Chola, M.; Fu, X.; Qian, P.U. U-Net with Coordinate Attention and VGGNet: A Grape Image Segmentation Algorithm Based on Fusion Pyramid Pooling and the Dual-Attention Mechanism. Agronomy 2024, 14, 925. [Google Scholar] [CrossRef]
- Wang, J.; Lin, X.; Luo, L.; Chen, M.; Wei, H.; Xu, L.; Luo, S. Cognition of grape cluster picking point based on visual knowledge distillation in complex vineyard environment. Comput. Electron. Agric. 2024, 225, 109216. [Google Scholar] [CrossRef]
- Jiang, T.; Li, Y.; Feng, H.; Wu, J.; Sun, W.; Ruan, Y. Research on a Trellis Grape Stem Recognition Method Based on YOLOv8n-GP. Agriculture 2024, 14, 1449. [Google Scholar] [CrossRef]
- Slaviček, P.; Hrabar, I.; Kovačić, Z. Generating a Dataset for Semantic Segmentation of Vine Trunks in Vineyards Using Semi-Supervised Learning and Object Detection. Robotics 2024, 13, 20. [Google Scholar] [CrossRef]
- Mohimont, L.; Steffenel, L.A.; Roesler, M.; Gaveau, N.; Rondeau, M.; Alin, F.; Pierlot, C.; de Oliveira, R.O.; Coppola, M. Ai-driven yield estimation using an autonomous robot for data acquisition. In Artificial Intelligence for Digitising Industry–Applications; River Publishers: Aalborg, Denmark, 2022; pp. 279–288. Available online: https://www.taylorfrancis.com/chapters/oa-edit/10.1201/9781003337232-24/ai-driven-yield-estimation-using-autonomous-robot-data-acquisition-lucas-mohimont-luiz-angelo-steffenel-mathias-roesler-nathalie-gaveau-marine-rondeau-fran%C3%A7ois-alin-cl%C3%A9ment-pierlot-rachel-ouvinha-de-oliveira-marcello-coppola (accessed on 2 October 2024).
- Pisciotta, A.; Barone, E.; Di Lorenzo, R. Table-Grape Cultivation in Soil-Less Systems: A Review. Horticulturae 2022, 8, 553. [Google Scholar] [CrossRef]
- El-Masri, I.Y.; Samaha, C.; Sassine, Y.N.; Assadi, F. Effects of Nano-Fertilizers and Greenhouse Cultivation on Phenological Development-Stages and Yield of Seedless Table Grapes Varieties. In Proceedings of the IX International Scientific Agriculture Symposium “AGROSYM 2018”, Jahorina, Bosnia and Herzegovina, 4–7 October 2018; pp. 192–196. Available online: https://www.cabidigitallibrary.org/doi/full/10.5555/20193108816 (accessed on 5 December 2024).
- Zabawa, L.; Kicherer, A. Segmentation of Wine Berries. 2021. [Google Scholar] [CrossRef]
- Pinheiro, I. Grapevine Bunch Detection Dataset. Zenodo 2023. [Google Scholar] [CrossRef]
- Seng, J.; Ang, K.; Schmidtke, L.; Rogiers, S. Grape Image Database–Charles Sturt University Research Output. Available online: https://researchoutput.csu.edu.au/en/datasets/grape-image-database (accessed on 5 December 2024).
- Santos, T.; De Souza, L.; Santos, A.D.; Sandra, A. Embrapa wine grape instance segmentation dataset–embrapa wgisd. Zenodo. 2019. Available online: https://zenodo.org/records/3361736 (accessed on 5 December 2024).
- Sozzi, M.; Cantalamessa, S.; Cogato, A.; Kayad, A.; Marinello, F. wGrapeUNIPD-DL: An open dataset for white grape bunch detection. Data Brief 2022, 43, 108466. [Google Scholar] [CrossRef]
- Li, Z.; Huang, H.; Duan, Z.; Zhang, W. Control temperature of greenhouse for higher yield and higher quality grapes production by combining STB in situ service with on time sensor monitoring. Heliyon 2023, 9, e13521. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Xie, Y.; Li, B.; Wei, X.; Huang, R.; Liu, S.; Ma, L. To improve grape photosynthesis, yield and fruit quality by covering reflective film on the ground of a protected facility. Sci. Hortic. 2024, 327, 112792. [Google Scholar] [CrossRef]
- Sabir, A.; Sabir, F.; Yazar, K.; Kara, Z. Investigations on development of some grapevine cultivars (V. vinifera L.) in soilless culture under controlled glasshouse condition. Curr. Trends Technol. Sci. 2012, 5, 622–626. [Google Scholar]
- Zhu, S.; Liang, Y.; Gao, D. Study of soil respiration and fruit quality of table grape (Vitis vinifera L.) in response to different soil water content in a greenhouse. Commun. Soil Sci. Plant Anal. 2018, 49, 2689–2699. [Google Scholar] [CrossRef]
- Yuan, Y.; Xie, Y.; Li, B.; Wei, X.; Huang, R.; Liu, S.; Ma, L. Propagation, Establishment, and Early Fruit Production of Table Grape Microvines in an LED -Lit Hydroponics System: A Demonstration Case Study. Plant Enviro. Interact. 2024, 5, e70018. [Google Scholar] [CrossRef]
- Zhang, R.; Chen, L.; Niu, Z.; Song, S.; Zhao, Y. Water stress affects the frequency of Firmicutes, Clostridiales and Lysobacter in rhizosphere soils of greenhouse grape. Agric. Water Manag. 2019, 226, 105776. [Google Scholar] [CrossRef]
- Choudhury, S.D.; Stoerger, V.; Samal, A.; Schnable, J.C.; Liang, Z.; Yu, J.-G. Automated vegetative stage phenotyping analysis of maize plants using visible light images. In Proceedings of the KDD Workshop on Data Science for Food, Energy and Water, San Francisco, CA, USA, 13–17 August 2016; Available online: https://www.researchgate.net/profile/Zhikai-Liang/publication/317692319_Automated_Vegetative_Stage_Phenotyping_Analysis_of_Maize_Plants_using_Visible_Light_Images_DS-FEW_'/links/594955b14585158b8fd5aec3/Automated-Vegetative-Stage-Phenotyping-Analysis-of-Maize-Plants-using-Visible-Light-Images-DS-FEW.pdf (accessed on 5 December 2024).
- Choudhury, S.D.; Bashyam, S.; Qiu, Y.; Samal, A.; Awada, T. Holistic and component plant phenotyping using temporal image sequence. Plant Methods 2018, 14, 35. [Google Scholar] [CrossRef] [PubMed]
- Mazis, A.; Choudhury, S.D.; Morgan, P.B.; Stoerger, V.; Hiller, J.; Ge, Y.; Awada, T. Application of high-throughput plant phenotyping for assessing biophysical traits and drought response in two oak species under controlled environment. For. Ecol. Manag. 2020, 465, 118101. [Google Scholar] [CrossRef]
- Quiñones, R.; Munoz-Arriola, F.; Choudhury, S.D.; Samal, A. Multi-feature data repository development and analytics for image cosegmentation in high-throughput plant phenotyping. PLoS ONE 2021, 16, e0257001. [Google Scholar] [CrossRef] [PubMed]
- Quiñones, R.; Samal, A.; Choudhury, S.D.; Muñoz-Arriola, F. OSC-CO2: Coattention and cosegmentation framework for plant state change with multiple features. Front. Plant Sci. 2023, 14, 1211409. [Google Scholar] [CrossRef]
- Fahlgren, N.; Gehan, M.A.; Baxter, I. Lights, camera, action: High-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015, 24, 93–99. [Google Scholar] [CrossRef]
- Morros, J.R.; Lobo, T.P.; Salmeron-Majadas, S.; Villazan, J.; Merino, D.; Antunes, A.; Datcu, M.; Karmakar, C.; Guerra, E.; Pantazi, D.-A.; et al. AI4Agriculture Grape Dataset. Zenodo. 2021. Available online: https://zenodo.org/records/5660081 (accessed on 5 December 2024).
- Barbole, D.K.; Jadhav, P.M. GrapesNet: Indian RGB & RGB-D vineyard image datasets for deep learning applications. Data Brief 2023, 48, 109100. [Google Scholar] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, Munich, Germany, 5–9 October 2015. [Google Scholar]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Meyes, R.; Lu, M.; de Puiseau, C.W.; Meisen, T. Ablation Studies in Artificial Neural Networks. arXiv 2019, arXiv:1901.08644. [Google Scholar] [CrossRef]
- Aich, S.; Stavness, I. Leaf counting with deep convolutional and deconvolutional networks. In Proceedings of the IEEE International Conference on Computer Vision Workshops 2017; pp. 2080–2089. Available online: https://openaccess.thecvf.com/content_ICCV_2017_workshops/w29/html/Aich_Leaf_Counting_With_ICCV_2017_paper.html (accessed on 15 October 2024).
- Ji, Z.; Mu, J.; Liu, J.; Zhang, H.; Dai, C.; Zhang, X.; Ganchev, I. ASD-Net: A novel U-Net based asymmetric spatial-channel convolution network for precise kidney and kidney tumor image segmentation. Med. Biol. Eng. Comput. 2024, 62, 1673–1687. [Google Scholar] [CrossRef]
- Ji, Z.; Wang, X.; Liu, C.; Wang, Z.; Yuan, N.; Ganchev, I. EFAM-Net: A Multi-Class Skin Lesion Classification Model Utilizing Enhanced Feature Fusion and Attention Mechanisms. IEEE Access 2024. Available online: https://ieeexplore.ieee.org/abstract/document/10695064/ (accessed on 15 January 2025). [CrossRef]



| Resolution | Without Segmentation | With Segmentation | ||
|---|---|---|---|---|
| MAE | R2 | MAE | R2 | |
| Low | 29 | 0.85 | 26 | 0.88 |
| Medium | 22 | 0.89 | 21 | 0.91 |
| High | 20 | 0.91 | 19 | 0.92 |
| Average | 24 | 0.88 | 22 | 0.90 |
| Grape Color | Without Segmentation | With Segmentation | ||
|---|---|---|---|---|
| MAE | R2 | MAE | R2 | |
| Blue | 13 | 0.95 | 13 | 0.96 |
| Green | 33 | 0.82 | 29 | 0.85 |
| Purple | 21 | 0.91 | 20 | 0.91 |
| Average | 22 | 0.89 | 21 | 0.91 |
| Resolution | IoU | F-1 Score |
|---|---|---|
| Low | 0.83 | 0.93 |
| Medium | 0.87 | 0.95 |
| High | 0.90 | 0.96 |
| Average | 0.87 | 0.95 |
| Grape Color | IoU | F-1 Score |
|---|---|---|
| Blue | 0.93 | 0.97 |
| Green | 0.82 | 0.92 |
| Purple | 0.87 | 0.95 |
| Average | 0.87 | 0.95 |
| Study | Approach | IoU | F1 Score |
|---|---|---|---|
| Zabawa et al. [6] | Deep learning segmentation | 0.89 | |
| Santos et al. [11] | Deep learning segmentation | 0.89 | |
| Sozzi et al. [4] | Traditional image processing | 0.77 | |
| Marani et al. [37] | CNN-based segmentation | 0.88 | |
| Peng et al. [39] | CNN-based segmentation | 0.88 | |
| (Ours) GCNet | U-Net-based segmentation + correction factor | 0.93 | 0.97 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quiñones, R.; Banu, S.M.; Gultepe, E. GCNet: A Deep Learning Framework for Enhanced Grape Cluster Segmentation and Yield Estimation Incorporating Occluded Grape Detection with a Correction Factor for Indoor Experimentation. J. Imaging 2025, 11, 34. https://doi.org/10.3390/jimaging11020034
Quiñones R, Banu SM, Gultepe E. GCNet: A Deep Learning Framework for Enhanced Grape Cluster Segmentation and Yield Estimation Incorporating Occluded Grape Detection with a Correction Factor for Indoor Experimentation. Journal of Imaging. 2025; 11(2):34. https://doi.org/10.3390/jimaging11020034
Chicago/Turabian StyleQuiñones, Rubi, Syeda Mariah Banu, and Eren Gultepe. 2025. "GCNet: A Deep Learning Framework for Enhanced Grape Cluster Segmentation and Yield Estimation Incorporating Occluded Grape Detection with a Correction Factor for Indoor Experimentation" Journal of Imaging 11, no. 2: 34. https://doi.org/10.3390/jimaging11020034
APA StyleQuiñones, R., Banu, S. M., & Gultepe, E. (2025). GCNet: A Deep Learning Framework for Enhanced Grape Cluster Segmentation and Yield Estimation Incorporating Occluded Grape Detection with a Correction Factor for Indoor Experimentation. Journal of Imaging, 11(2), 34. https://doi.org/10.3390/jimaging11020034






