Gated Attention-Augmented Double U-Net for White Blood Cell Segmentation
Abstract
1. Introduction
- Attention-augmented convolutions are integrated to selectively emphasize informative features across channels and spatial scales.
- Gating mechanisms are incorporated to suppress irrelevant regions and enhance focus on meaningful anatomical structures.
- Unlike recent studies, this method was tested on both individual WBCs (cropped cell images) or multiple cells within one image. Our approach handles both scenarios effectively.
- The proposed method achieves accurate and efficient segmentation without relying on additional support inputs or specialized preprocesses.
2. Related Work
3. Method
3.1. Background Knowledge
3.1.1. Double U-Net Baseline
3.1.2. Attention-Augmented Convolution (AAC) Block
- is a standard 2D convolution on the input tensor x, producing a feature map , where is the number of output channels for the convolutional branch.
- is the self-attention on x that calculates attention weights to obtain global spatial relationships. The attention module produces a feature map , where is the number of output channels of the attention branch.
- concatenates the output of the attention and convolution branches along the channel axis to produce the final output .
- are the query, key, and value matrices, respectively, obtained from linear transformations of the input tensor x. These matrices are computed aswhere are learnable weight matrices ( is the query projection matrix, is the key projection matrix, and is the value projection matrix).
- The dimensionality of the key vectors is , which is used to scale the dot product attention scores and prevent vanishing gradients.
- normalizes the attention scores to produce a probability distribution over the spatial locations.
3.1.3. Gating Mechanism
- is a 1 × 1 convolution that reduces the number of channels from C to , producing an intermediate feature map .
- BatchNorm applies batch normalization to , resulting in .
- ReLU applies the rectified linear unit activation function to , producing .
- is a 1 × 1 convolution that expands the number of channels back from to C, yielding .
- The sigmoid function is denoted by , which generates the gating mask .
- Element-wise (Hadamard) multiplication is signified by ⊙, scaling the input x with the gating mask g to produce the final output .
3.2. Architecture
3.2.1. GAAD-U-Net First Phase
3.2.2. GAAD-U-Net Second Phase
4. Experimental Results and Analysis
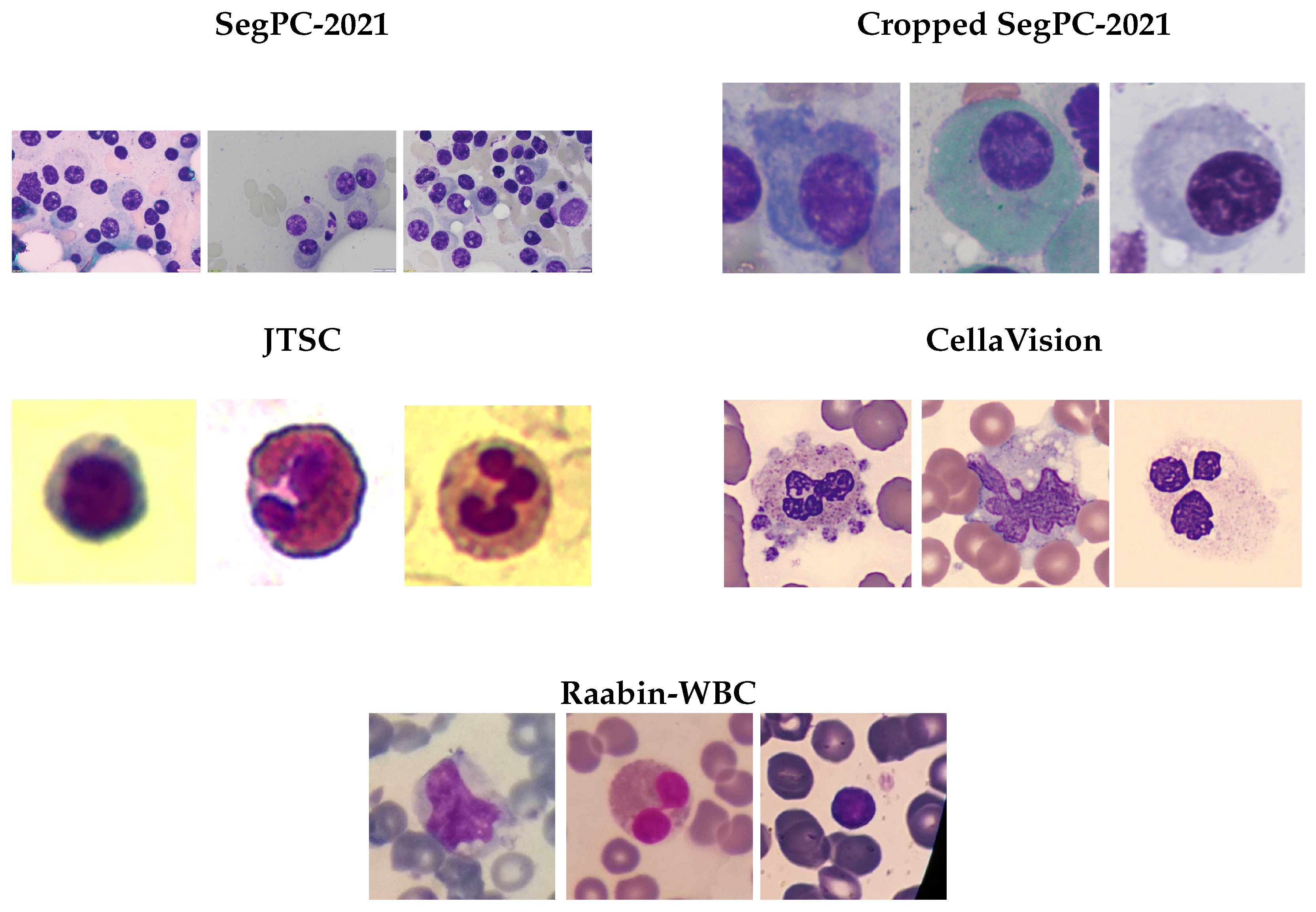
4.1. Datasets
- Cell density: Ranges from single cells (JTSC) to dense aggregates (SegPC-2021 and CellaVision).
- Staining variation: Uniform in SegPC-2021 and more inconsistent in Raabin-WBC.
- Background: Clean in JTSC and SegPC-2021 and more intricate with RBCs in CellaVision.
- Magnification: Varies from low (SegPC-2021) to high (JTSC and CellaVision).
4.2. Evaluation Metric
4.2.1. Intersection over Union (IoU)
- is the intersection of A and B.
- is the union of A and B.
4.2.2. Dice Similarity Coefficient (DSC)
- represents true positives.
- represents false positives.
- represents false negatives.
4.2.3. Accuracy
- represents true negatives.
4.2.4. Surface Distance Metrics
- A is the set of surface points of the ground truth segmentation.
- B is the set of surface points of the predicted segmentation.
- a is an individual point belonging to set A ().
- b is an individual point belonging to set B ().
- is the operator that calculates the 95th percentile from a set of distances.
4.3. Implementation Details
4.4. Data Augmentation
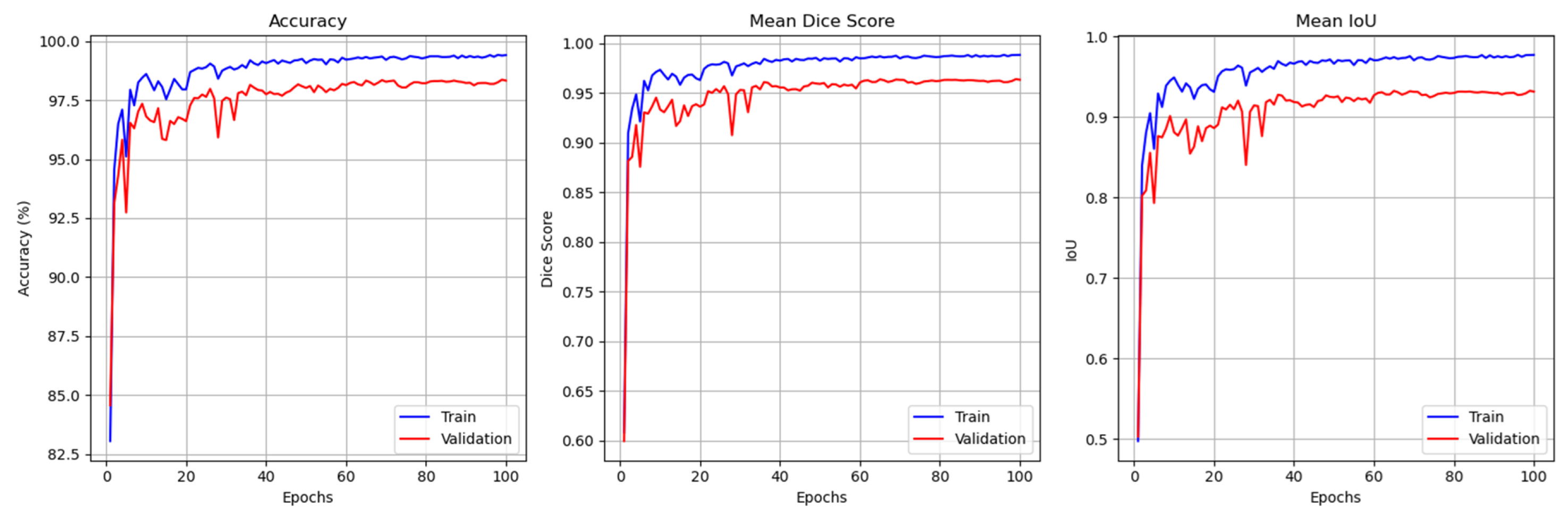
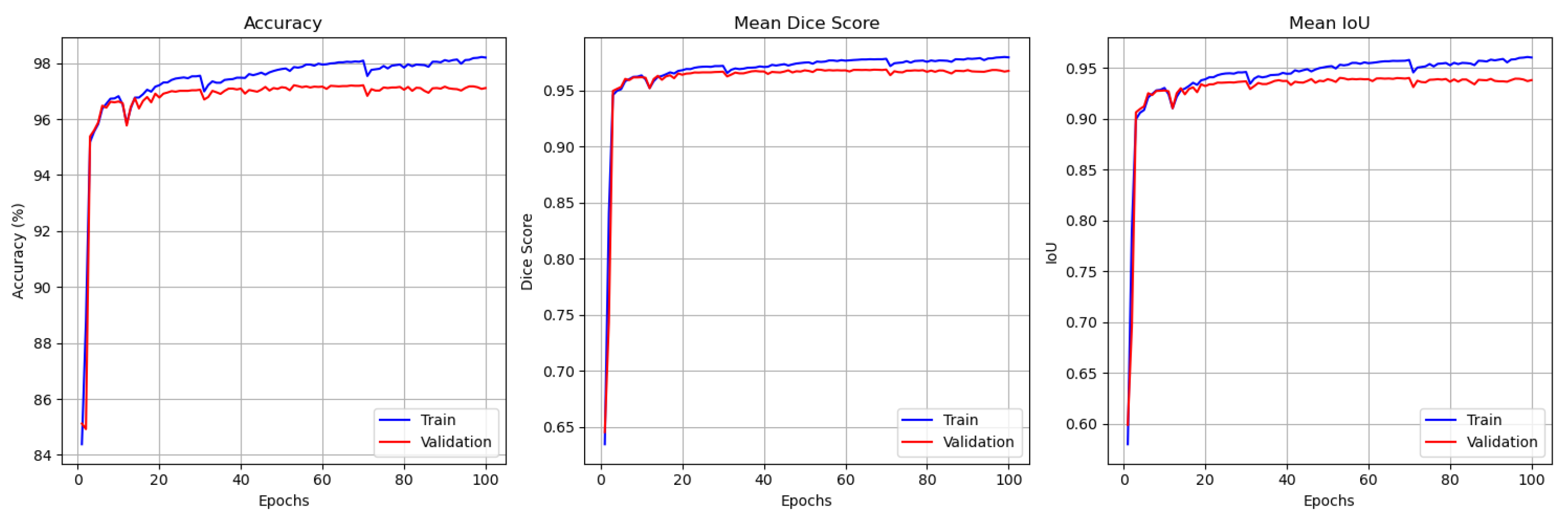
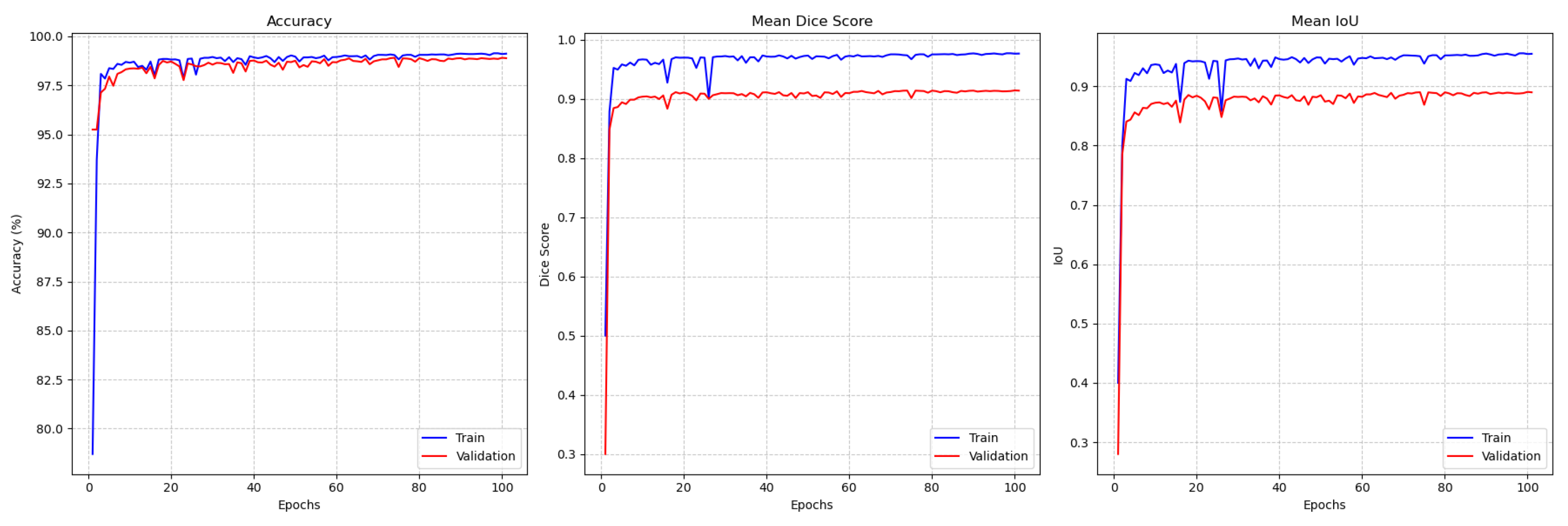
4.5. Results
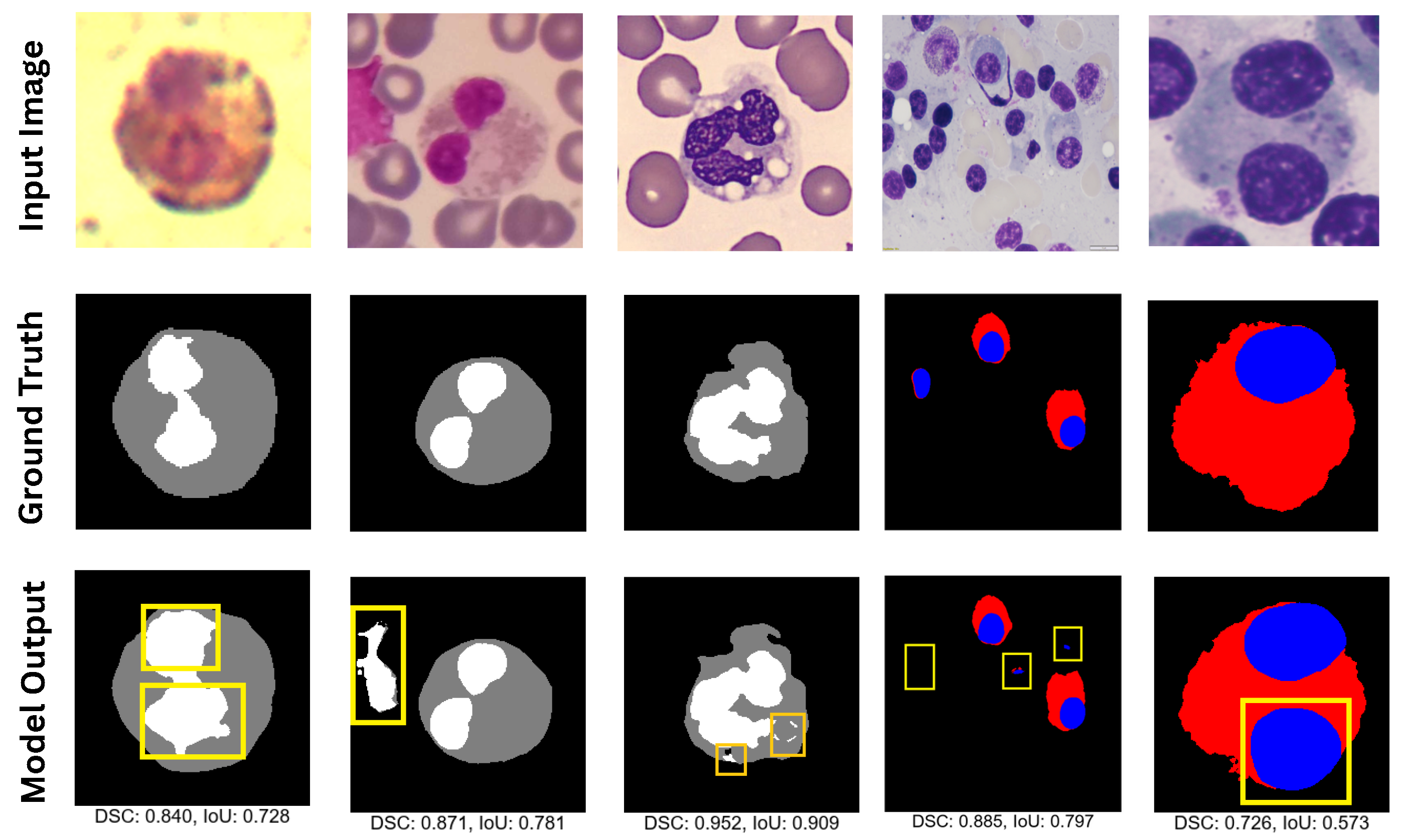
4.6. Prediction Visualizations
4.7. Ablation Study
4.7.1. AAC Integration Impact
4.7.2. Gating Module Significance
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Architecture
| Block | Tensor Size | Parameters |
|---|---|---|
| Input | [4, 3, 224, 224] | - |
| VGG19_block1 | [4, 64, 224, 224] | 38,720 |
| VGG19_block2 | [4, 128, 112, 112] | 221,440 |
| VGG19_block3 | [4, 256, 56, 56] | 2,065,408 |
| VGG19_block4 | [4, 512, 28, 28] | 8,259,584 |
| AAC | [4, 512, 28, 28] | 2,656,720 |
| Gating Module | [4, 512, 28, 28] | 263,424 |
| ASPP | [4, 1024, 28, 28] | 20,453,376 |
| Decoder1_block1 | [4, 512, 28, 28] | 9,177,600 |
| Decoder1_block2 | [4, 256, 56, 56] | 2,295,040 |
| Decoder1_block3 | [4, 128, 112, 112] | 574,080 |
| Decoder1_block4 | [4, 64, 224, 224] | 143,680 |
| Output_1 | [4, 3, 224, 224] | 195 |
| matmul | [4, 3, 224, 224] | - |
| Encoder2_block1 | [4, 64, 224, 224] | 39,360 |
| Encoder2_block2 | [4, 128, 112, 112] | 223,744 |
| Encoder2_block3 | [4, 256, 56, 56] | 893,952 |
| Encoder2_block4 | [4, 512, 28, 28] | 3,573,760 |
| AAC * | [4, 512, 28, 28] | 2,656,720 |
| Gating Module * | [4, 512, 28, 28] | 263,424 |
| ASPP * | [4, 1024, 28, 28] | 20,453,376 |
| Decoder2_block1 | [4, 512, 28, 28] | 11,569,664 |
| Decoder2_block2 | [4, 256, 56, 56] | 2,893,056 |
| Decoder2_block3 | [4, 128, 112, 112] | 723,584 |
| Decoder2_block4 | [4, 64, 224, 224] | 181,056 |
| Final_output | [4, 3, 224, 224] | 195 |
Appendix B. Dataset
| Dataset | Train | Val | Test | Total | Input Size | Augmentations | Links |
|---|---|---|---|---|---|---|---|
| SegPC-2021 | 360 | 89 | 49 | 493 | 512 × 512 | Horizontal flip Rotation Cutout probability = 0.25 | https://www.kaggle.com/datasets/sbilab/segpc2021dataset (accessed on: 20 April 2025) |
| Cropped SegPC-2021 | 1843 | 263 | 527 | 2633 | 224 × 224 | Horizontal flip Vertical flip Scaling probability = 0.5 | https://www.kaggle.com/datasets/sbilab/segpc2021dataset (accessed on: 20 April 2025) |
| CellaVision | 75 | – | 25 | 100 | 224 × 224 | Vertical flip Horizontal flip Rotation Gaussian noise Brightness alteration probability = 0.5 | https://github.com/zxaoyou/segmentation_WBC (accessed on: 20 April 2025) |
| JTSC | 225 | – | 75 | 300 | 224 × 224 | Vertical flip Horizontal flip Rotation Gaussian noise Brightness alteration probability = 0.5 | https://github.com/zxaoyou/segmentation_WBC (accessed on: 20 April 2025) |
| Raabin-WBC | 800 | 112 | 233 | 1145 | 224 × 224 | Vertical flip Horizontal flip Rotation Gaussian noise Brightness alteration probability = 0.5 | https://raabindata.com/ (accessed on: 20 April 2025) |
Appendix C. Hyperparameter
| (a) AAC Module Parameters. | |
| Parameter | Value |
| In Channels | 512 |
| Out Channels | 512 |
| Kernel Size | 3 |
| Attention Key Dim. | 32 |
| Value Key Dim. | 32 |
| Number of Heads | 4 |
| Relative | True |
| Stride | 1 |
| Shape | 28 |
| (b) Hyperparameter Settings. | |
| Hyperparameter | Value |
| Epochs | 100 (150 for segPC-2021) |
| Batch Size | 4 |
| Optimizer | Adam Optimizer |
| Initial Learning Rate | 1 × 10−4 |
| Weight Decay | 1 × 10−4 |
| Momentum | 0.9 |
| Gamma | 0.5 |
Appendix D. Computational Profile
| Stage | Kernel Sizes | Channels | Params | GFLOPs | VRAM (MB) | Latency (ms) |
|---|---|---|---|---|---|---|
| Phase1 Encoder1 VGG1 | 3 × 3 | 3 → 64 | 38,720 | 10.12 | 1417.7 | 6.54 |
| Phase1 Encoder1 VGG2 | 3 × 3 | 64 → 128 | 221,440 | 14.50 | 1385.9 | 6.40 |
| Phase1 Encoder1 VGG3 | 3 × 3 | 128 → 256 | 2,065,408 | 33.82 | 1339.6 | 12.41 |
| Phase1 Encoder1 VGG4 | 3 × 3 | 256 → 512 | 8,259,584 | 33.82 | 1323.4 | 12.56 |
| Phase1 AAC1 | 3 × 3 | 512 → 512 | 2,656,848 | 2.72 | 1358.0 | 2.62 |
| Phase1 Gating1 | 1 × 1 | 512 → 512 | 263,424 | 0.27 | 1293.3 | 0.48 |
| Phase1 ASPP1 | 1 × 1, 3 × 3, 3 × 3, 1 × 1 | 512 → 1024 | 20,453,376 | 20.41 | 1334.4 | 7.93 |
| Phase1 Decoder1 Block1 | 2 × 2 (up), 3 × 3, 1 × 1 | 1024 → 512 | 9,177,600 | 15.84 | 1321.4 | 4.14 |
| Phase1 Decoder1 Block2 | 2 × 2 (up), 3 × 3, 1 × 1 | 512 → 256 | 2,295,040 | 31.17 | 1389.9 | 11.00 |
| Phase1 Decoder1 Block3 | 2 × 2 (up), 3 × 3, 1 × 1 | 256 → 128 | 574,080 | 37.65 | 1578.5 | 13.91 |
| Phase1 Decoder1 Block4 | 2 × 2 (up), 3 × 3, 1 × 1 | 128 → 64 | 143,680 | 37.72 | 1673.6 | 19.62 |
| Phase1 output | 1 × 1 | 64 → 3 | 195 | 0.00 | 50 | 0.01 |
| Phase2 Encoder2 Block1 | 3 × 3 | 3 → 64 | 39,360 | 10.27 | 1417.7 | 8.03 |
| Phase2 Encoder2 Block2 | 3 × 3 | 64 → 128 | 223,744 | 14.57 | 1401.9 | 5.64 |
| Phase2 Encoder2 Block3 | 3 × 3 | 128 → 256 | 893,952 | 14.53 | 1348.6 | 5.33 |
| Phase2 Encoder2 Block4 | 3 × 3 | 256 → 512 | 3,573,760 | 14.51 | 1326.4 | 3.12 |
| Phase2 AAC2 | 3 × 3 | 512 → 512 | 2,656,848 | 2.72 | 1358.0 | 2.81 |
| Phase2 Gating2 | 1 × 1 | 512 → 512 | 263,424 | 0.27 | 1293.3 | 0.27 |
| Phase2 ASPP2 | 1 × 1, 3 × 3, 3 × 3, 1 × 1 | 512 → 1024 | 20,453,376 | 20.41 | 1334.4 | 8.20 |
| Phase2 Decoder2 Block1 | 2 × 2 (up), 3 × 3, 1 × 1 | 1024 → 512 | 11,569,664 | 18.26 | 1334.4 | 5.31 |
| Phase2 Decoder2 Block2 | 2 × 2 (up), 3 × 3, 1 × 1 | 512 → 256 | 2,893,056 | 40.84 | 1440.1 | 13.79 |
| Phase2 Decoder2 Block3 | 2 × 2 (up), 3 × 3, 1 × 1 | 256 → 128 | 723,584 | 47.32 | 1707.3 | 18.02 |
| Phase2 Decoder2 Block4 | 2 × 2 (up), 3 × 3, 1 × 1 | 128 → 64 | 181,056 | 47.40 | 1801.8 | 21.44 |
| Final output | 1 × 1 | 64 → 3 | 195 | 0.00 | 50 | 0.01 |
| Total/Peak | - | - | 89,621,024 | 469.14 | 1801.8 | 189.57 |
| Pipeline | Time (ms) 512 × 512 | Time (ms) 224 × 224 |
|---|---|---|
| Augmentation time per sample | 550.73 | 103.86 |
| Initialization and checkpoint loading | 1400.13 | 1399.34 |
| GPU batch loading time | 1.85 (batch of 4) | 1.42 (batch of 16) |
| Inference | 190.83 | 66.48 |
| Throughput | 9.15 images/second | 28.98 images/second |
| Average latency | 179.34 ms per image | 64.51 ms per image |
Appendix E. Extended Ablation Study
| Dataset | Metric | 1 Head | 2 Heads | 4 Heads | 8 Heads | 16 Heads | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Lower | Upper | Mean | Lower | Upper | Mean | Lower | Upper | Mean | Lower | Upper | Mean | Lower | Upper | ||
| Cropped SegPC-2021 | DSC (%) | 0.9607 | 0.9578 | 0.9635 | 0.9594 | 0.9574 | 0.9621 | 0.9614 | 0.9534 | 0.9694 | 0.8884 | 0.8786 | 0.8981 | 0.6444 | 0.6197 | 0.6690 |
| IoU (%) | 0.9257 | 0.9205 | 0.9308 | 0.9323 | 0.9279 | 0.9366 | 0.9330 | 0.9229 | 0.9431 | 0.7333 | 0.7221 | 0.7444 | 0.5343 | 0.5170 | 0.5516 | |
| SegPC-2021 | DSC (%) | 0.8862 | 0.8615 | 0.9109 | 0.8890 | 0.8632 | 0.9149 | 0.9011 | 0.8761 | 0.9261 | 0.8790 | 0.8571 | 0.9109 | 0.8849 | 0.8581 | 0.9116 |
| IoU (%) | 0.8044 | 0.7646 | 0.8442 | 0.8198 | 0.7686 | 0.8509 | 0.8301 | 0.7964 | 0.8638 | 0.8023 | 0.7599 | 0.8446 | 0.8035 | 0.7614 | 0.8455 | |
| CellaVision | DSC (%) | 0.8914 | 0.8532 | 0.9295 | 0.9136 | 0.8936 | 0.8936 | 0.9589 | 0.9564 | 0.9614 | 0.9515 | 0.9490 | 0.9540 | 0.9251 | 0.9156 | 0.9346 |
| IoU (%) | 0.8143 | 0.7566 | 0.8721 | 0.8500 | 0.8320 | 0.8680 | 0.9214 | 0.9158 | 0.9270 | 0.9282 | 0.9226 | 0.9338 | 0.8714 | 0.8594 | 0.8834 | |
| JTSC | DSC (%) | 0.9695 | 0.9640 | 0.9749 | 0.9712 | 0.9662 | 0.9764 | 0.9714 | 0.9689 | 0.9739 | 0.9719 | 0.9668 | 0.9770 | 0.9720 | 0.9671 | 0.9770 |
| IoU (%) | 0.9415 | 0.9314 | 0.9516 | 0.9449 | 0.9354 | 0.9543 | 0.9674 | 0.9618 | 0.9730 | 0.9460 | 0.9365 | 0.9556 | 0.9462 | 0.9369 | 0.9556 | |
| Raabin-WBC | DSC (%) | 0.8643 | 0.8618 | 0.8668 | 0.9002 | 0.8972 | 0.9032 | 0.9119 | 0.9099 | 0.9139 | 0.9170 | 0.9144 | 0.9194 | 0.9152 | 0.9128 | 0.9176 |
| IoU (%) | 0.7886 | 0.7830 | 0.7942 | 0.8795 | 0.8740 | 0.8850 | 0.8873 | 0.8804 | 0.8967 | 0.8988 | 0.8944 | 0.9032 | 0.8911 | 0.8865 | 0.8957 | |
| Phase 1 | Phase 2 | SegPC-2021 | Cropped SegPC-2021 | CellaVision | JTSC | Raabin-WBC | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DSC | IoU | DSC | IoU | DSC | IoU | DSC | IoU | DSC | IoU | ||
| ✗ | ✗ | 0.8580 | 0.7630 | 0.8980 | 0.7991 | 0.9495 | 0.9096 | 0.9621 | 0.9319 | 0.9091 | 0.8832 |
| ✓ | ✗ | 0.8796 | 0.7966 | 0.9620 | 0.9755 | 0.9523 | 0.9129 | 0.9672 | 0.9526 | 0.9124 | 0.9008 |
| ✗ | ✓ | 0.8807 | 0.8054 | 0.9589 | 0.9733 | 0.9542 | 0.9155 | 0.9630 | 0.9437 | 0.9099 | 0.8844 |
| ✓ | ✓ | 0.9011 | 0.8301 | 0.9614 | 0.9740 | 0.9589 | 0.9214 | 0.9714 | 0.9674 | 0.9119 | 0.8885 |
Appendix F. Generalization Tests
Appendix F.1. Cross-Dataset Evaluation
| Model | Train Dataset | Test: SegPC-2021 | Test: Cropped SegPC-2021 | Test: CellaVision | Test: JTSC | Test: Raabin-WBC | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DSC | IoU | DSC | IoU | DSC | IoU | DSC | IoU | DSC | IoU | ||
| DCSAU-Net | SegPC-2021 | 0.8860 | 0.8060 | 0.2689 | 0.2056 | 0.2990 | 0.2638 | 0.3050 | 0.2509 | 0.3033 | 0.2778 |
| Cropped SegPC-2021 | 0.1546 | 0.1170 | 0.8860 | 0.8060 | 0.3931 | 0.3310 | 0.4206 | 0.3250 | 0.4875 | 0.4167 | |
| CellaVision | 0.0031 | 0.0025 | 0.1889 | 0.1206 | 0.9460 | 0.9018 | 0.3390 | 0.2569 | 0.5521 | 0.4632 | |
| JTSC | 0.0578 | 0.0349 | 0.6722 | 0.5344 | 0.3362 | 0.2790 | 0.9592 | 0.9235 | 0.3678 | 0.3214 | |
| Raabin-WBC | 0.0475 | 0.0204 | 0.5330 | 0.4451 | 0.5567 | 0.4697 | 0.3563 | 0.2979 | 0.9102 | 0.8867 | |
| GA2Net | SegPC-2021 | 0.8770 | 0.7930 | 0.3748 | 0.2800 | 0.5712 | 0.5042 | 0.4536 | 0.3145 | 0.4536 | 0.3875 |
| Cropped SegPC-2021 | 0.2437 | 0.1838 | 0.9274 | 0.9254 | 0.4383 | 0.3582 | 0.7649 | 0.6611 | 0.4304 | 0.3754 | |
| CellaVision | 0.0207 | 0.0113 | 0.2276 | 0.1525 | 0.8989 | 0.8403 | 0.4978 | 0.4109 | 0.6583 | 0.5581 | |
| JTSC | 0.0373 | 0.0284 | 0.5739 | 0.4425 | 0.3741 | 0.3275 | 0.9680 | 0.9389 | 0.3849 | 0.3507 | |
| Raabin-WBC | 0.0499 | 0.0276 | 0.5172 | 0.4314 | 0.7162 | 0.6175 | 0.4374 | 0.3837 | 0.9055 | 0.8700 | |
| GAAD-UNet | SegPC-2021 | 0.9011 | 0.8301 | 0.3397 | 0.2444 | 0.6059 | 0.5376 | 0.2719 | 0.2299 | 0.3024 | 0.2769 |
| Cropped SegPC-2021 | 0.2667 | 0.1955 | 0.9614 | 0.9330 | 0.4300 | 0.3798 | 0.7381 | 0.6396 | 0.4380 | 0.3784 | |
| CellaVision | 0.0376 | 0.0293 | 0.3278 | 0.2569 | 0.9589 | 0.9214 | 0.4369 | 0.3611 | 0.7839 | 0.7176 | |
| JTSC | 0.0422 | 0.0318 | 0.7427 | 0.6231 | 0.3524 | 0.3043 | 0.9714 | 0.9674 | 0.3937 | 0.3592 | |
| Raabin-WBC | 0.0513 | 0.0265 | 0.5408 | 0.4574 | 0.7822 | 0.6881 | 0.3575 | 0.3060 | 0.9119 | 0.8885 | |
Appendix F.2. Scale and Color Sensitivity
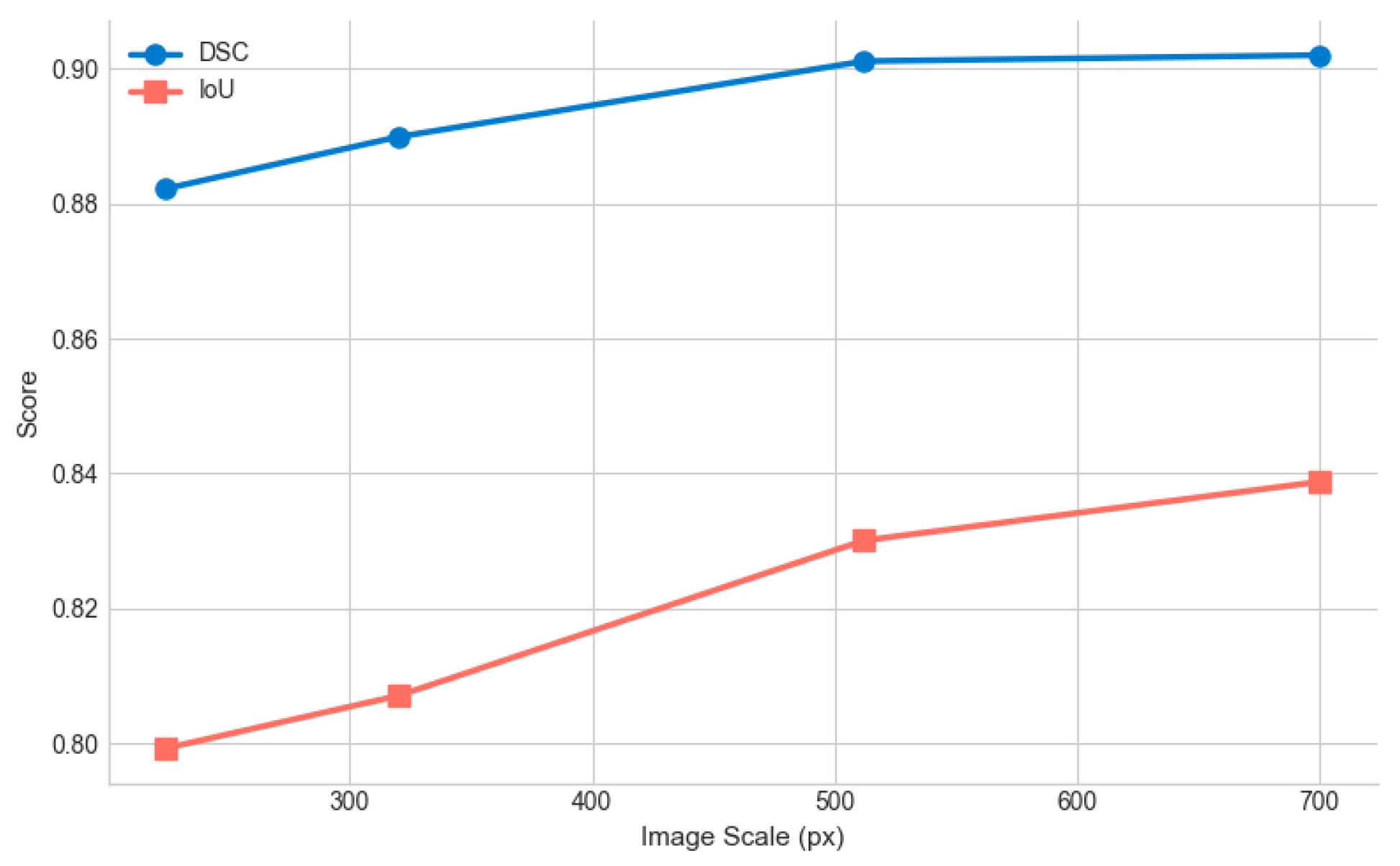
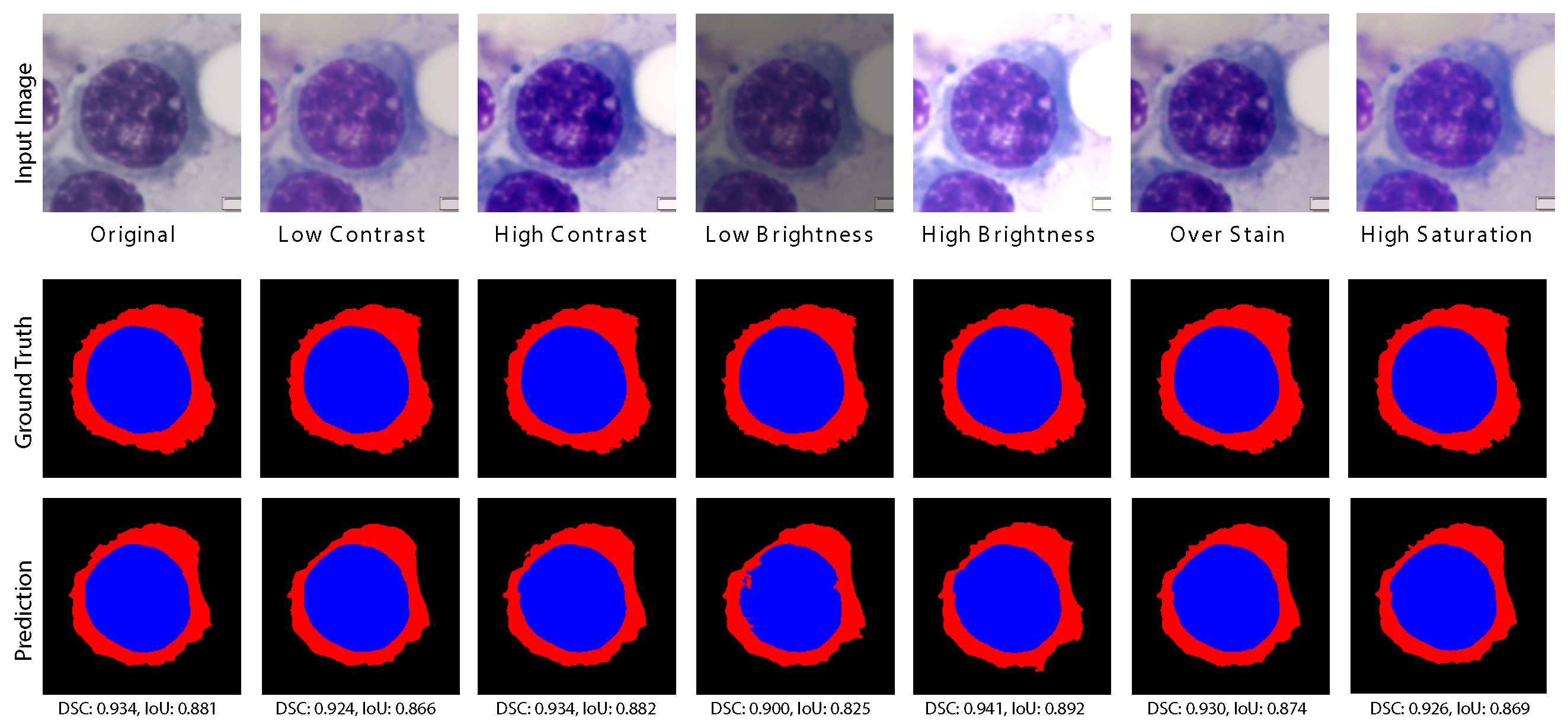
Appendix G. Gating Mechanism Evaluation

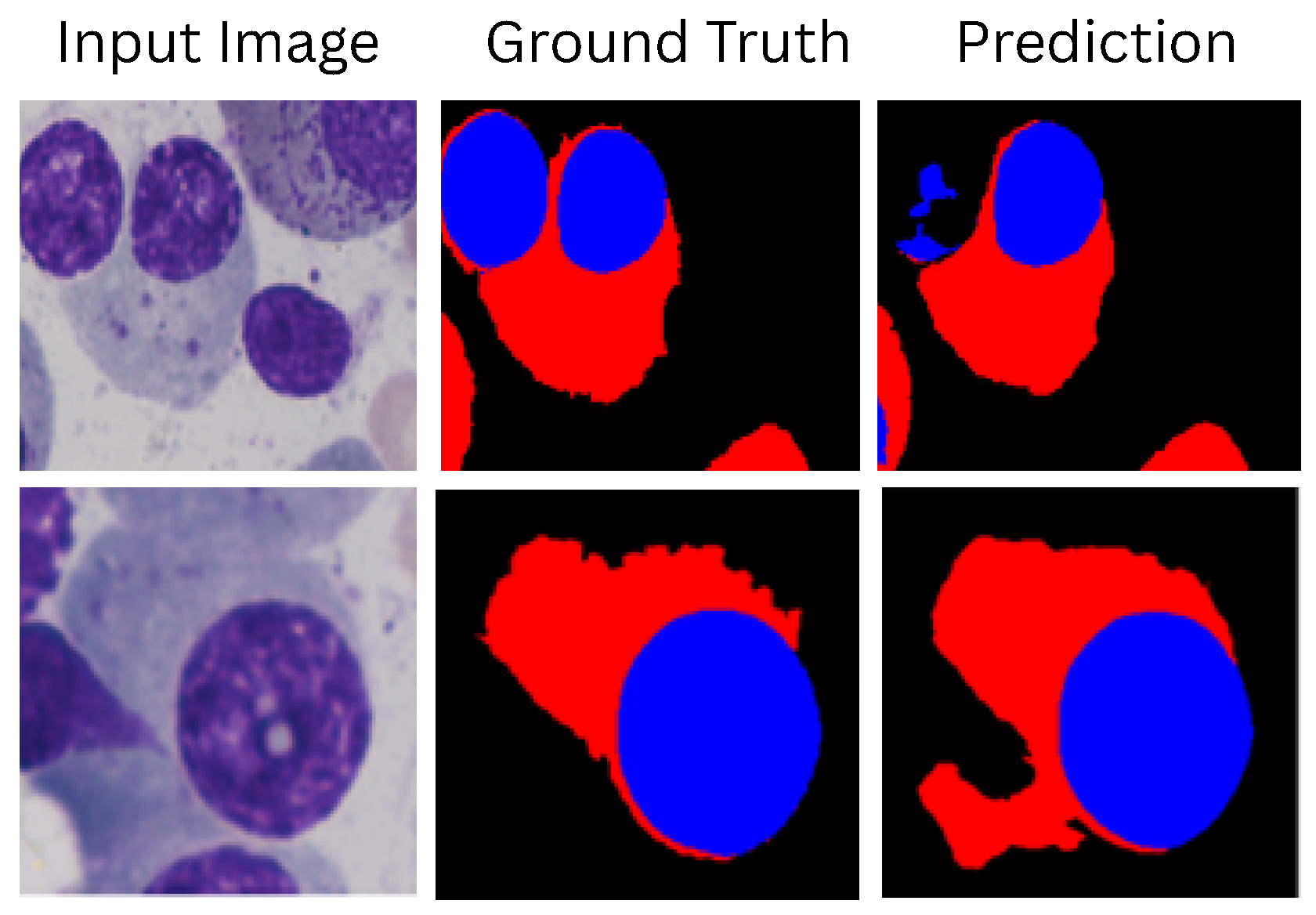
Appendix H. Fail Cases


References
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.Y.; et al. Segment anything. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 2–3 October 2023; pp. 4015–4026. [Google Scholar]
- Ma, J.; He, Y.; Li, F.; Han, L.; You, C.; Wang, B. Segment anything in medical images. Nat. Commun. 2024, 15, 654. [Google Scholar] [CrossRef] [PubMed]
- Madni, H.A.; Umer, R.M.; Zottin, S.; Marr, C.; Foresti, G.L. FL-W3S: Cross-domain federated learning for weakly supervised semantic segmentation of white blood cells. Int. J. Med. Inform. 2025, 195, 105806. [Google Scholar] [CrossRef] [PubMed]
- Sutton, R.T.; Pincock, D.; Baumgart, D.C.; Sadowski, D.C.; Fedorak, R.N.; Kroeker, K.I. An overview of clinical decision support systems: Benefits, risks, and strategies for success. NPJ Digit. Med. 2020, 3, 17. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Hong, S.; Li, Q.; Li, P.; Coats, T.; Zou, B.; Kong, G. Cross-modal similar clinical case retrieval using a modular model based on contrastive learning and k-nearest neighbor search. Int. J. Med. Inform. 2025, 193, 105680. [Google Scholar] [CrossRef]
- Zhang, C.; Xiao, X.; Li, X.; Chen, Y.J.; Zhen, W.; Chang, J.; Zheng, C.; Liu, Z. White Blood Cell Segmentation by Color-Space-Based K-Means Clustering. Sensors 2014, 14, 16128–16147. [Google Scholar] [CrossRef]
- Suganyadevi, S.; Seethalakshmi, V.; Balasamy, K. A review on deep learning in medical image analysis. Int. J. Multimed. Inf. Retr. 2022, 11, 19–38. [Google Scholar] [CrossRef]
- Çınar, A.; Tuncer, S.A. Classification of lymphocytes, monocytes, eosinophils, and neutrophils on white blood cells using hybrid Alexnet-GoogleNet-SVM. SN Appl. Sci. 2021, 3, 503. [Google Scholar] [CrossRef]
- Meng, J.; Lu, Y.; He, W.; Fan, X.; Zhou, G.; Wei, H. Leukocyte segmentation based on DenseREU-Net. J. King Saud Univ.—Comput. Inf. Sci. 2024, 36, 102236. [Google Scholar] [CrossRef]
- Putzu, L.; Porcu, S.; Loddo, A. Distributed collaborative machine learning in real-world application scenario: A white blood cell subtypes classification case study. Image Vis. Comput. 2025, 162, 105673. [Google Scholar] [CrossRef]
- Sellam, A.Z.; Benlamoudi, A.; Cid, C.A.; Dobelle, L.; Slama, A.; Hillali, Y.E.; Taleb-Ahmed, A. Deep Learning Solution for Quantification of Fluorescence Particles on a Membrane. Sensors 2023, 23, 1794. [Google Scholar] [CrossRef]
- Escobar, F.I.F.; Alipo-on, J.R.T.; Novia, J.L.U.; Tan, M.J.T.; Abdul Karim, H.; AlDahoul, N. Automated counting of white blood cells in thin blood smear images. Comput. Electr. Eng. 2023, 108, 108710. [Google Scholar] [CrossRef]
- Zhao, M.; Yang, H.; Shi, F.; Zhang, X.; Zhang, Y.; Sun, X.; Wang, H. MSS-WISN: Multiscale Multistaining WBCs Instance Segmentation Network. IEEE Access 2022, 10, 65598–65610. [Google Scholar] [CrossRef]
- Fu, L.; Chen, J.; Zhang, Y.; Huang, X.; Sun, L. CNN and Transformer-based deep learning models for automated white blood cell detection. Image Vis. Comput. 2025, 161, 105631. [Google Scholar] [CrossRef]
- Liu, Y.; Mazumdar, S.; Bath, P.A. An unsupervised learning approach to diagnosing Alzheimer’s disease using brain magnetic resonance imaging scans. Int. J. Med. Inform. 2023, 173, 105027. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Millana, A.; Saez-Saez, A.; Tornero-Costa, R.; Azzopardi-Muscat, N.; Traver, V.; Novillo-Ortiz, D. Artificial intelligence and its impact on the domains of universal health coverage, health emergencies and health promotion: An overview of systematic reviews. Int. J. Med. Inform. 2022, 166, 104855. [Google Scholar] [CrossRef]
- Aletti, G.; Benfenati, A.; Naldi, G. A Semiautomatic Multi-Label Color Image Segmentation Coupling Dirichlet Problem and Colour Distances. J. Imaging 2021, 7, 208. [Google Scholar] [CrossRef]
- Wang, R.; Lei, T.; Cui, R.; Zhang, B.; Meng, H.; Nandi, A.K. Medical image segmentation using deep learning: A survey. IET Image Process. 2022, 16, 1243–1267. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Bougourzi, F.; Distante, C.; Dornaika, F.; Taleb-Ahmed, A.; Hadid, A.; Chaudhary, S.; Yang, W.; Qiang, Y.; Anwar, T.; Breaban, M.E.; et al. COVID-19 Infection Percentage Estimation from Computed Tomography Scans: Results and Insights from the International Per-COVID-19 Challenge. Sensors 2024, 24, 1557. [Google Scholar] [CrossRef]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Trans. Med. Imaging 2019, 39, 1856–1867. [Google Scholar] [CrossRef]
- Jha, D.; Riegler, M.A.; Johansen, D.; Halvorsen, P.; Johansen, H.D. DoubleU-Net: A Deep Convolutional Neural Network for Medical Image Segmentation. arXiv 2020, arXiv:2006.04868. [Google Scholar]
- Rahil, M.; Anoop, B.N.; Girish, G.N.; Kothari, A.R.; Koolagudi, S.G.; Rajan, J. A Deep Ensemble Learning-Based CNN Architecture for Multiclass Retinal Fluid Segmentation in OCT Images. IEEE Access 2023, 11, 17241–17251. [Google Scholar] [CrossRef]
- Benaissa, I.; Zitouni, A.; Sbaa, S. Using Multiclass Semantic Segmentation for Close Boundaries and Overlapping Blood Cells. In Proceedings of the 2024 8th International Conference on Image and Signal Processing and their Applications (ISPA), Biskra, Algeria, 21–22 April 2024; pp. 1–5. [Google Scholar] [CrossRef]
- Meng, W.; Liu, S.; Wang, H. AFC-Unet: Attention-fused full-scale CNN-transformer unet for medical image segmentation. Biomed. Signal Process. Control 2025, 99, 106839. [Google Scholar] [CrossRef]
- Rajamani, K.T.; Rani, P.; Siebert, H.; ElagiriRamalingam, R.; Heinrich, M.P. Attention-augmented U-Net (AA-U-Net) for semantic segmentation. Signal Image Video Process. 2023, 17, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Tao, H.; Stojanovic, V. End-to-end multi-scale residual network with parallel attention mechanism for fault diagnosis under noise and small samples. ISA Trans. 2024, 157, 419–433. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Shi, Y.; Li, S. Self-attention and long-range relationship capture network for underwater object detection. J. King Saud Univ.—Comput. Inf. Sci. 2024, 36, 101971. [Google Scholar] [CrossRef]
- Zeng, Z.; Liu, J.; Huang, X.; Luo, K.; Yuan, X.; Zhu, Y. Efficient Retinal Vessel Segmentation with 78K Parameters. J. Imaging 2025, 11, 306. [Google Scholar] [CrossRef]
- Lu, Y.; Qin, X.; Fan, H.; Lai, T.; Li, Z. WBC-Net: A white blood cell segmentation network based on UNet++ and ResNet. Appl. Soft Comput. 2021, 101, 107006. [Google Scholar] [CrossRef]
- Guo, Y.; Shahin, A.I.; Garg, H. An indeterminacy fusion of encoder-decoder network based on neutrosophic set for white blood cells segmentation. Expert Syst. Appl. 2024, 246, 123156. [Google Scholar] [CrossRef]
- Roy, R.M.; Ameer, P.M. Segmentation of leukocyte by semantic segmentation model: A deep learning approach. Biomed. Signal Process. Control 2021, 65, 102385. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, F.; Zhang, W.; Wang, Q.; Liu, Y.; Jiang, Z. RotU-Net: An Innovative U-Net With Local Rotation for Medical Image Segmentation. IEEE Access 2024, 12, 21114–21128. [Google Scholar] [CrossRef]
- Li, C.; Tan, Y.; Chen, W.; Luo, X.; He, Y.; Gao, Y.; Li, F. ANU-Net: Attention-based nested U-Net to exploit full resolution features for medical image segmentation. Comput. Graph. 2020, 90, 11–20. [Google Scholar] [CrossRef]
- Li, D.; Yin, S.; Lei, Y.; Qian, J.; Zhao, C.; Zhang, L. Segmentation of White Blood Cells Based on CBAM-DC-UNet. IEEE Access 2023, 11, 1074–1082. [Google Scholar] [CrossRef]
- Xu, Q.; Ma, Z.; HE, N.; Duan, W. DCSAU-Net: A deeper and more compact split-attention U-Net for medical image segmentation. Comput. Biol. Med. 2023, 154, 106626. [Google Scholar] [CrossRef] [PubMed]
- Hayat, M.; Gupta, M.; Suanpang, P.; Nanthaamornphong, A. Super-Resolution Methods for Endoscopic Imaging: A Review. In Proceedings of the 2024 12th International Conference on Internet of Everything, Microwave, Embedded, Communication and Networks (IEMECON), lJaipur, India, 24 August 2024; pp. 1–6. [Google Scholar] [CrossRef]
- Fiaz, M.; Noman, M.; Cholakkal, H.; Anwer, R.M.; Hanna, J.; Khan, F.S. Guided-attention and gated-aggregation network for medical image segmentation. Pattern Recognit. 2024, 156, 110812. [Google Scholar] [CrossRef]
- Patil, P.S.; Holambe, R.S.; Waghmare, L.M. An Attention Augmented Convolution-Based Tiny-Residual UNet for Road Extraction. IEEE Trans. Artif. Intell. 2024, 5, 3951–3964. [Google Scholar] [CrossRef]
- Gupta, A.; Gehlot, S.; Goswami, S.; Motwani, S.; Gupta, R.; Faura, Á.G.; Štepec, D.; Martinčič, T.; Azad, R.; Merhof, D.; et al. SegPC-2021: A challenge & dataset on segmentation of Multiple Myeloma plasma cells from microscopic images. Med. Image Anal. 2023, 83, 102677. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, Y.; Wang, G.; Liu, J. Fast and robust segmentation of white blood cell images by self-supervised learning. Micron 2018, 107, 55–71. [Google Scholar] [CrossRef]
- Kouzehkanan, Z.M.; Saghari, S.; Tavakoli, S.; Rostami, P.; Abaszadeh, M.; Mirzadeh, F.; Satlsar, E.S.; Gheidishahran, M.; Gorgi, F.; Mohammadi, S.; et al. A large dataset of white blood cells containing cell locations and types, along with segmented nuclei and cytoplasm. Sci. Rep. 2022, 12, 1123. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2017, arXiv:1412.6980. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar] [CrossRef]
- Alom, M.Z.; Hasan, M.; Yakopcic, C.; Taha, T.M.; Asari, V.K. Recurrent Residual Convolutional Neural Network based on U-Net (R2U-Net) for Medical Image Segmentation. arXiv 2018, arXiv:1802.06955. [Google Scholar]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Johansen, D.; Lange, T.D.; Halvorsen, P.; Johansen, H.D. ResUNet++: An Advanced Architecture for Medical Image Segmentation. In Proceedings of the 2019 IEEE International Symposium on Multimedia (ISM), San Diego, CA, USA, 9–11 December 2019; pp. 225–2255. [Google Scholar] [CrossRef]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.W.; Wu, J. UNet 3+: A Full-Scale Connected UNet for Medical Image Segmentation. In Proceedings of the ICASSP 2020—2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1055–1059. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar] [CrossRef]
- Xu, G.; Wu, X.; Zhang, X.; He, X. LeViT-UNet: Make Faster Encoders with Transformer for Medical Image Segmentation. arXiv 2021, arXiv:2102.043062107.08623. [Google Scholar]
- Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [CrossRef]
- Huang, X.; Deng, Z.; Li, D.; Yuan, X.; Fu, Y. MISSFormer: An Effective Transformer for 2D Medical Image Segmentation. IEEE Trans. Med. Imaging 2023, 42, 1484–1494. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cao, P.; Wang, J.; Zaiane, O.R. UCTransNet: Rethinking the Skip Connections in U-Net from a Channel-Wise Perspective with Transformer. In Proceedings of the AAAI Conference on Artificial Intelligence, Virtual, 22 February–1 March 2022; Volume 36, pp. 2441–2449. [Google Scholar] [CrossRef]




| Feature | Double U-Net | AA-U-Net | DCSAU-Net | GAAD-U-Net (Ours) |
|---|---|---|---|---|
| Base | Double U-Net | U-Net | U-Net | Double U-Net |
| Attention | None | AAC | CSA | AAC |
| Mechanism | N/A | Multi-Head Self-Attention | Multi-Path Soft Attention | Multi-Head Self-Attention |
| Placement | N/A | Bottleneck | Encoder or Decoder skips | Both bottlenecks with Gating, ASPP |
| Attention Enhancement Modules | N/A | None | PFC | Gating, ASPP, Dual-Skip |
| Benefit | Dual-Phase Refinement | Wider Context | Multi-Scale Efficiency | Feature Refinement and Wider Context |
| Method | Accuracy | Precision | Recall | DSC | Mean IoU |
|---|---|---|---|---|---|
| U-Net (2015) [19] | 0.939 | 0.842 | 0.879 | 0.855 | 0.766 |
| Attention U-Net (2018) [44] | 0.940 | 0.845 | 0.866 | 0.849 | 0.757 |
| R2U-Net (2018 [45] | 0.933 | 0.852 | 0.831 | 0.834 | 0.744 |
| ResU-Net++ (2019) [46] | 0.934 | 0.838 | 0.858 | 0.840 | 0.736 |
| U-Net++ (2020) [34] | 0.942 | 0.855 | 0.876 | 0.857 | 0.770 |
| Double U-Net (2020) [22] | 0.937 | 0.833 | 0.896 | 0.858 | 0.763 |
| UNet3+ (2020) [47] | 0.939 | 0.848 | 0.866 | 0.852 | 0.766 |
| TransUNet (2021) [48] | 0.939 | 0.822 | 0.869 | 0.838 | 0.741 |
| LeViT-UNet (2021) [49] | 0.939 | 0.850 | 0.837 | 0.837 | 0.738 |
| DCSAU-Net (2023) [36] | 0.950 | 0.871 | 0.910 | 0.886 | 0.806 |
| GA2Net (2024) [38] | 0.953 | 0.866 | 0.793 | 0.877 | 0.793 |
| GAAD-U-Net (ours) | 0.960 | 0.873 | 0.924 | 0.901 | 0.830 |
| Model | Accuracy | DSC | Mean IoU |
|---|---|---|---|
| U-Net (2015) [19] | 0.9390 | 0.8808 | 0.8820 |
| U-Net++ (2020) [34] | 0.9420 | 0.9102 | 0.9092 |
| Double U-Net (2020) [22] | 0.9470 | 0.8941 | 0.7991 |
| MultiResUNet (2021) [50] | - | 0.8649 | 0.8676 |
| TransU-Net (2021) [48] | 0.9390 | 0.8233 | 0.8338 |
| MissFormer (2023) [51] | - | 0.8082 | 0.8209 |
| UCTransNet (2021) [52] | - | 0.9174 | 0.9159 |
| DCSAU-Net (2023) [36] | 0.9504 | 0.8860 | 0.8060 |
| GA2-Net (2024) [38] | - | 0.9274 | 0.9254 |
| GAAD-U-Net (Ours) | 0.9852 | 0.9614 | 0.9330 |
| Model | CellaVision | JTSC | Raabin-WBC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Metrics | Acc. | DSC | Mean IoU | Acc. | DSC | Mean IoU | Acc. | DSC | Mean IoU |
| U-Net (2015) [19] | 0.9422 | 0.8922 | 0.8215 | 0.9720 | 0.9519 | 0.9104 | 0.9860 | 0.9068 | 0.8800 |
| Double U-Net (2020) [22] | 0.9792 | 0.9495 | 0.9096 | 0.9803 | 0.9621 | 0.9319 | 0.9843 | 0.9091 | 0.8832 |
| TransUNet (2021) [48] | 0.9790 | 0.9534 | 0.9145 | 0.9737 | 0.9603 | 0.9249 | 0.9870 | 0.9094 | 0.8840 |
| DCSAU-Net (2023) [36] | 0.9737 | 0.9460 | 0.9018 | 0.9767 | 0.9592 | 0.9235 | 0.9865 | 0.9102 | 0.8867 |
| GA2Net (2024) [38] | 0.9560 | 0.8989 | 0.8403 | 0.9812 | 0.9680 | 0.9389 | 0.9865 | 0.9055 | 0.8700 |
| GAAD-U-Net (Ours) | 0.9830 | 0.9589 | 0.9214 | 0.9832 | 0.9714 | 0.9674 | 0.9884 | 0.9119 | 0.8885 |
| Model | SegPC-2021 | Cropped SegPC-2021 | CellaVision | JTSC | Raabin-WBC | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| HD95 | ASSD | HD95 | ASSD | HD95 | ASSD | HD95 | ASSD | HD95 | ASSD | |
| DCSAU-Net | 11.8905 | 1.7358 | 0.5435 | 0.1148 | 1.8626 | 0.3877 | 0.6720 | 0.0942 | 0.3380 | 0.0681 |
| GA2Net | 20.2653 | 4.0739 | 0.5830 | 0.1092 | 2.1052 | 0.3864 | 0.4634 | 0.0663 | 0.3653 | 0.0693 |
| GAAD-U-Net | 10.5024 | 1.5632 | 0.4453 | 0.0943 | 1.1974 | 0.1912 | 0.4211 | 0.0689 | 0.3470 | 0.0706 |
| Dataset | Metric | Base | Base+AAC | Base+Gating | Base+AAC+Gating |
|---|---|---|---|---|---|
| SegPC-2021 | Accuracy | 0.9370 | 0.9470 | 0.9320 | 0.9597 |
| DSC | 0.8580 | 0.8780 | 0.8509 | 0.9011 | |
| Mean IoU | 0.7630 | 0.7824 | 0.7598 | 0.8301 | |
| Cropped SegPC-2021 | Accuracy | 0.9470 | 0.9722 | 0.9722 | 0.9852 |
| DSC | 0.8980 | 0.9585 | 0.9587 | 0.9614 | |
| Mean IoU | 0.7991 | 0.9112 | 0.9112 | 0.9330 | |
| JTSC | Accuracy | 0.9285 | 0.9831 | 0.9300 | 0.9832 |
| DSC | 0.9621 | 0.9713 | 0.9646 | 0.9714 | |
| Mean IoU | 0.9319 | 0.9645 | 0.9444 | 0.9674 | |
| CellaVision | Accuracy | 0.9792 | 0.9845 | 0.9792 | 0.9830 |
| DSC | 0.9495 | 0.9611 | 0.9496 | 0.9589 | |
| Mean IoU | 0.9096 | 0.9189 | 0.9100 | 0.9214 | |
| Raabin-WBC | Accuracy | 0.9843 | 0.9870 | 0.9851 | 0.9884 |
| DSC | 0.9091 | 0.9112 | 0.9088 | 0.9119 | |
| Mean IoU | 0.8832 | 0.8846 | 0.8822 | 0.8885 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benaissa, I.; Zitouni, A.; Sbaa, S.; Aydin, N.; Megherbi, A.C.; Sellam, A.Z.; Taleb-Ahmed, A.; Distante, C. Gated Attention-Augmented Double U-Net for White Blood Cell Segmentation. J. Imaging 2025, 11, 386. https://doi.org/10.3390/jimaging11110386
Benaissa I, Zitouni A, Sbaa S, Aydin N, Megherbi AC, Sellam AZ, Taleb-Ahmed A, Distante C. Gated Attention-Augmented Double U-Net for White Blood Cell Segmentation. Journal of Imaging. 2025; 11(11):386. https://doi.org/10.3390/jimaging11110386
Chicago/Turabian StyleBenaissa, Ilyes, Athmane Zitouni, Salim Sbaa, Nizamettin Aydin, Ahmed Chaouki Megherbi, Abdellah Zakaria Sellam, Abdelmalik Taleb-Ahmed, and Cosimo Distante. 2025. "Gated Attention-Augmented Double U-Net for White Blood Cell Segmentation" Journal of Imaging 11, no. 11: 386. https://doi.org/10.3390/jimaging11110386
APA StyleBenaissa, I., Zitouni, A., Sbaa, S., Aydin, N., Megherbi, A. C., Sellam, A. Z., Taleb-Ahmed, A., & Distante, C. (2025). Gated Attention-Augmented Double U-Net for White Blood Cell Segmentation. Journal of Imaging, 11(11), 386. https://doi.org/10.3390/jimaging11110386









