Development of Semi-Automatic Dental Image Segmentation Workflows with Root Canal Recognition for Faster Ground Tooth Acquisition
Abstract
1. Introduction
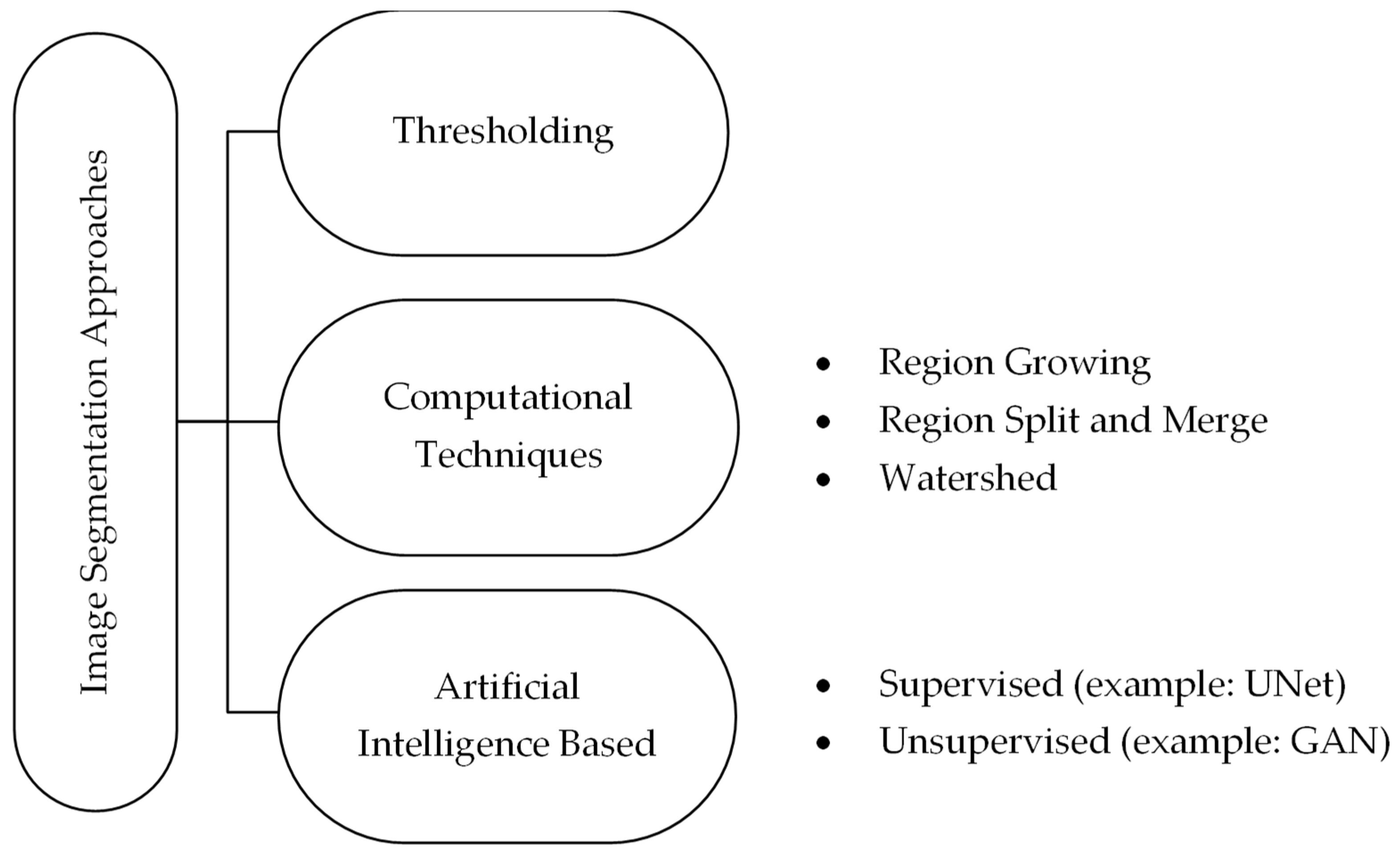
1.1. Current State of Segmentation and Problems
1.2. Related Work
1.2.1. Semi-Automated Workflows
1.2.2. Generative Adversarial Networks
2. Materials and Methods
2.1. Datasets
2.2. Methodology
- Complete manual segmentation
- GFS algorithm
- WS algorithm
- Automated DentalSegmentator
2.3. Pre-Processing
2.3.1. Volume Cropping
2.3.2. Histogram Adjustment
2.3.3. Thresholding Masking
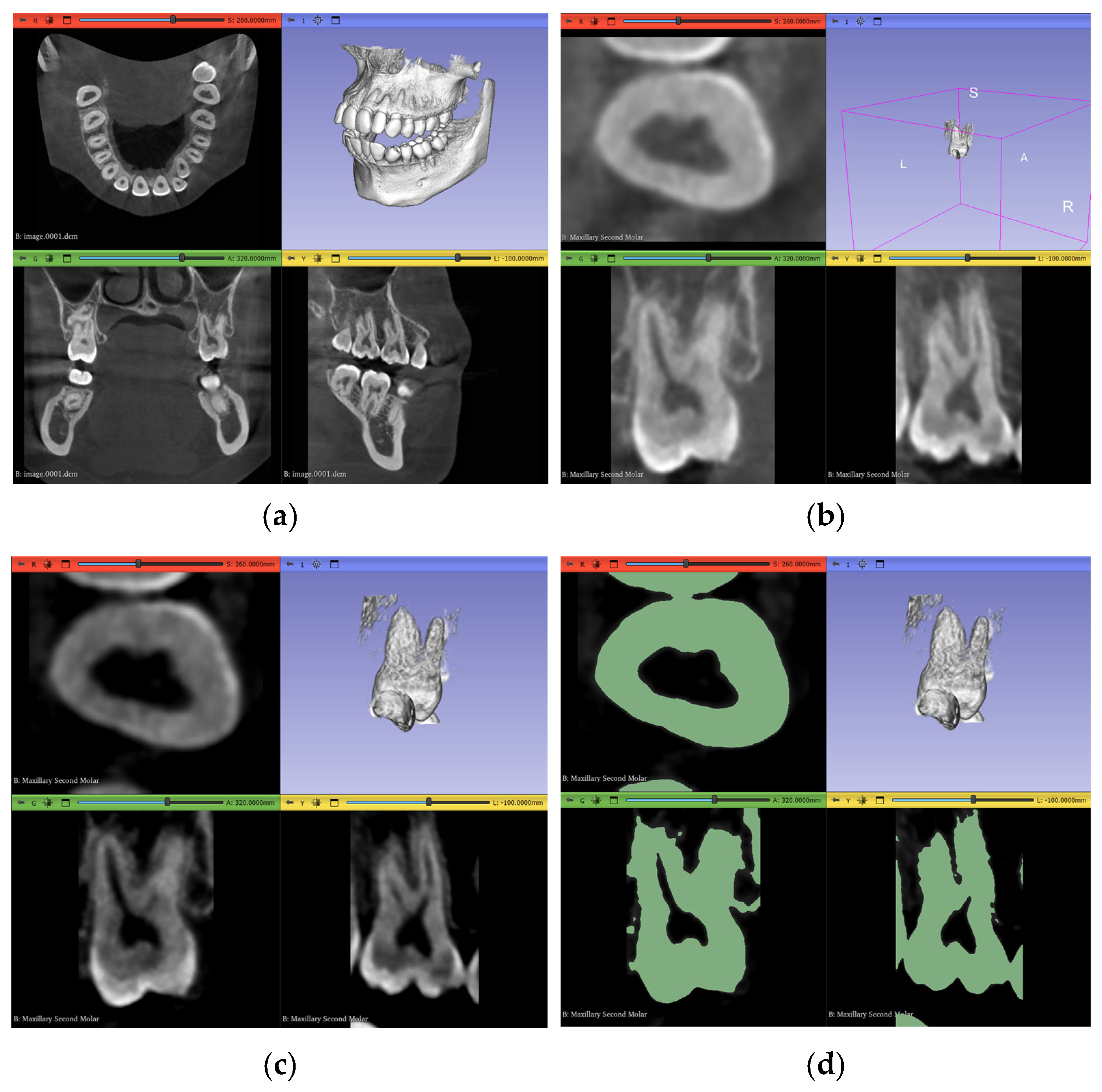
2.4. Workflow 1: Complete Manual Segmentation
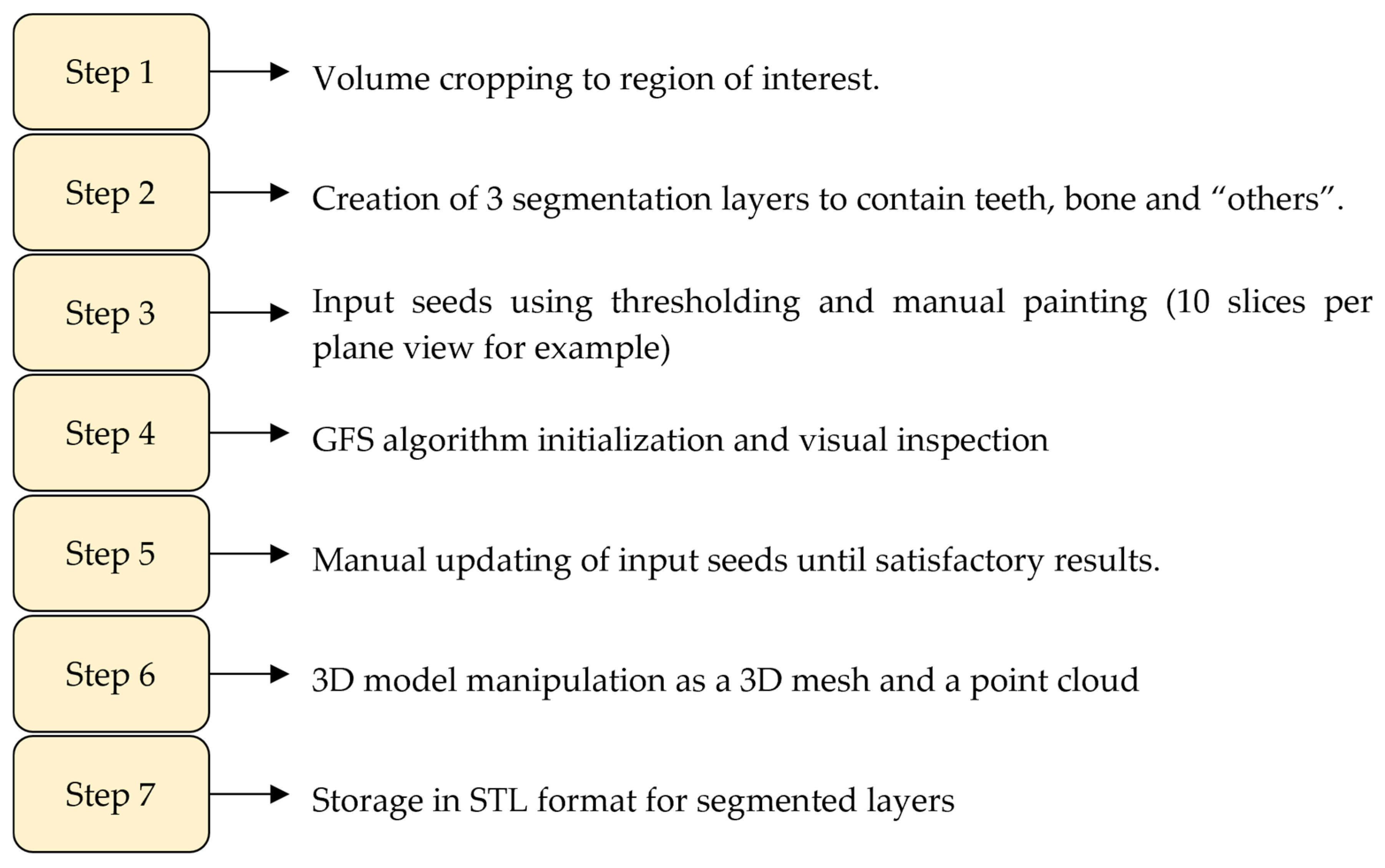
2.5. Workflow 2: GFS
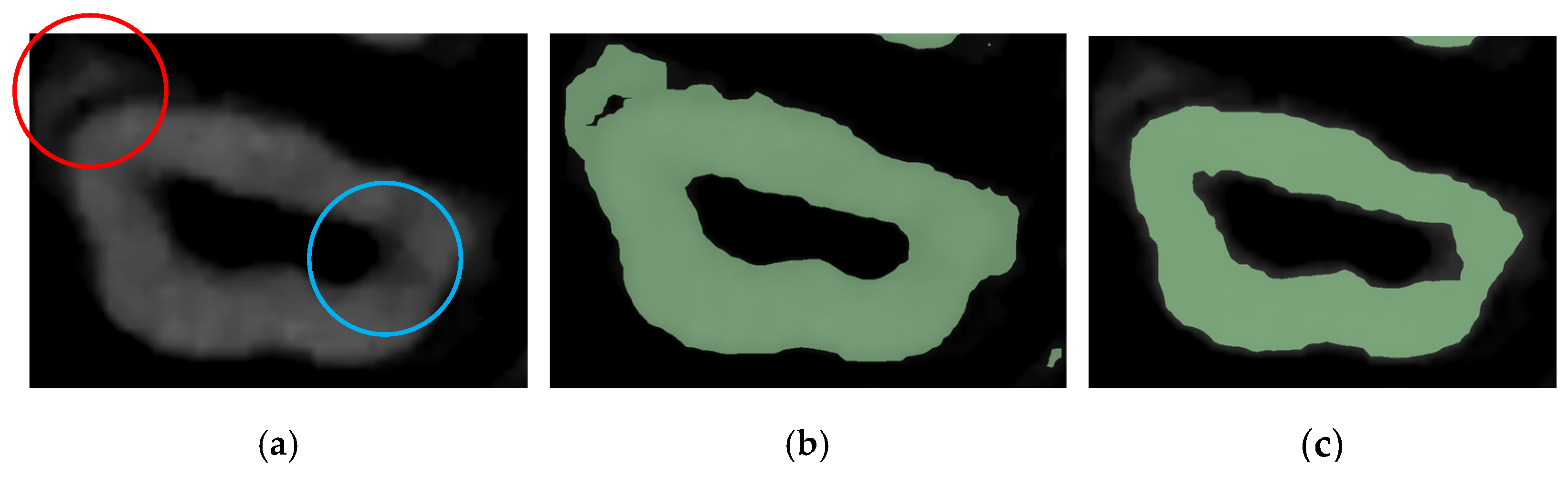
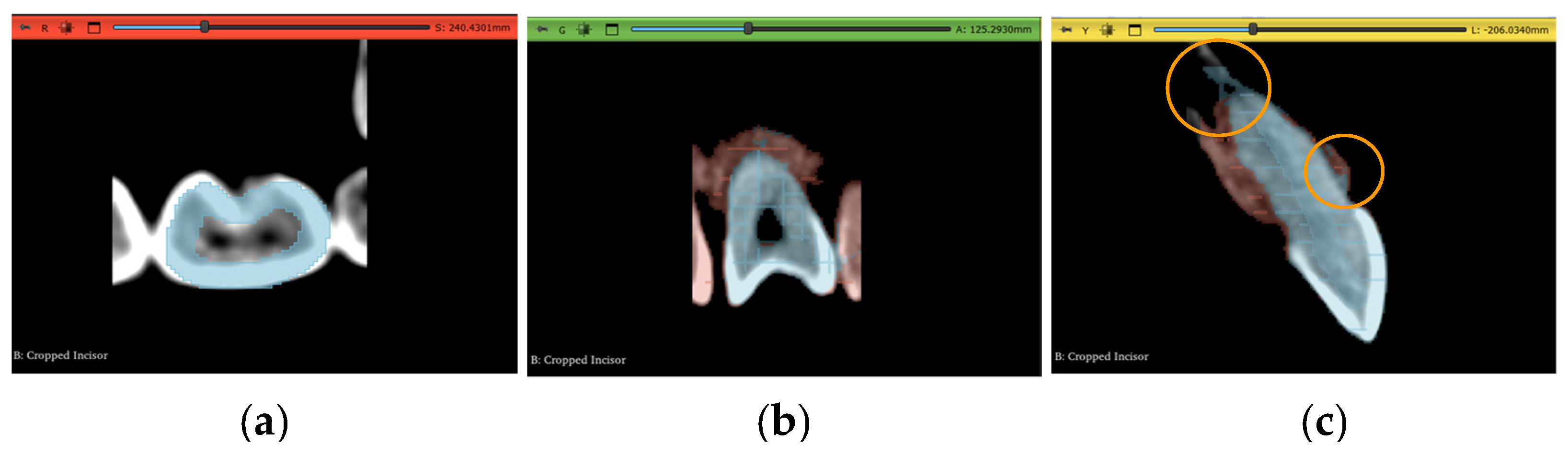
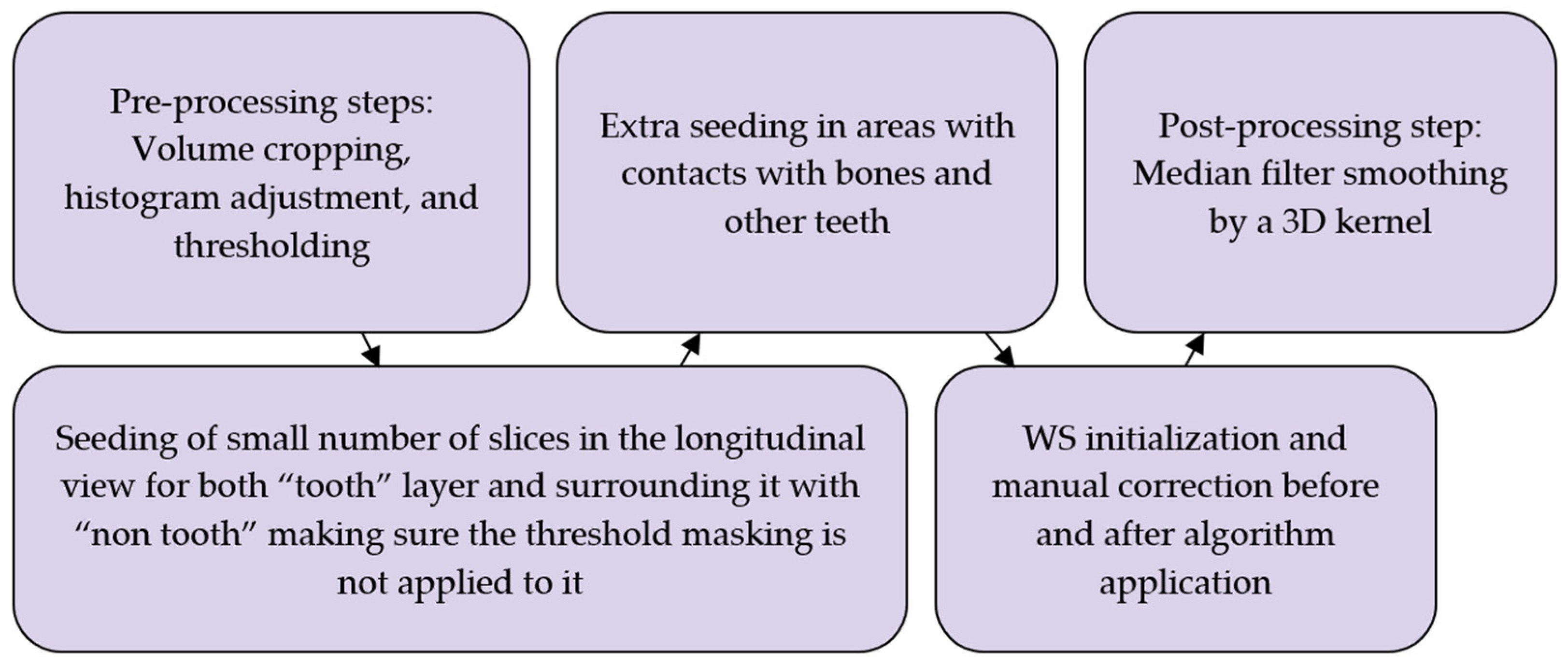
2.6. Workflow 3: WS
2.7. Workflow 4: Automated DentalSegmentator
- Maxilla and upper skull
- Mandible
- Upper teeth
- Lower teeth
- Mandibular canal
2.8. Post-Processing
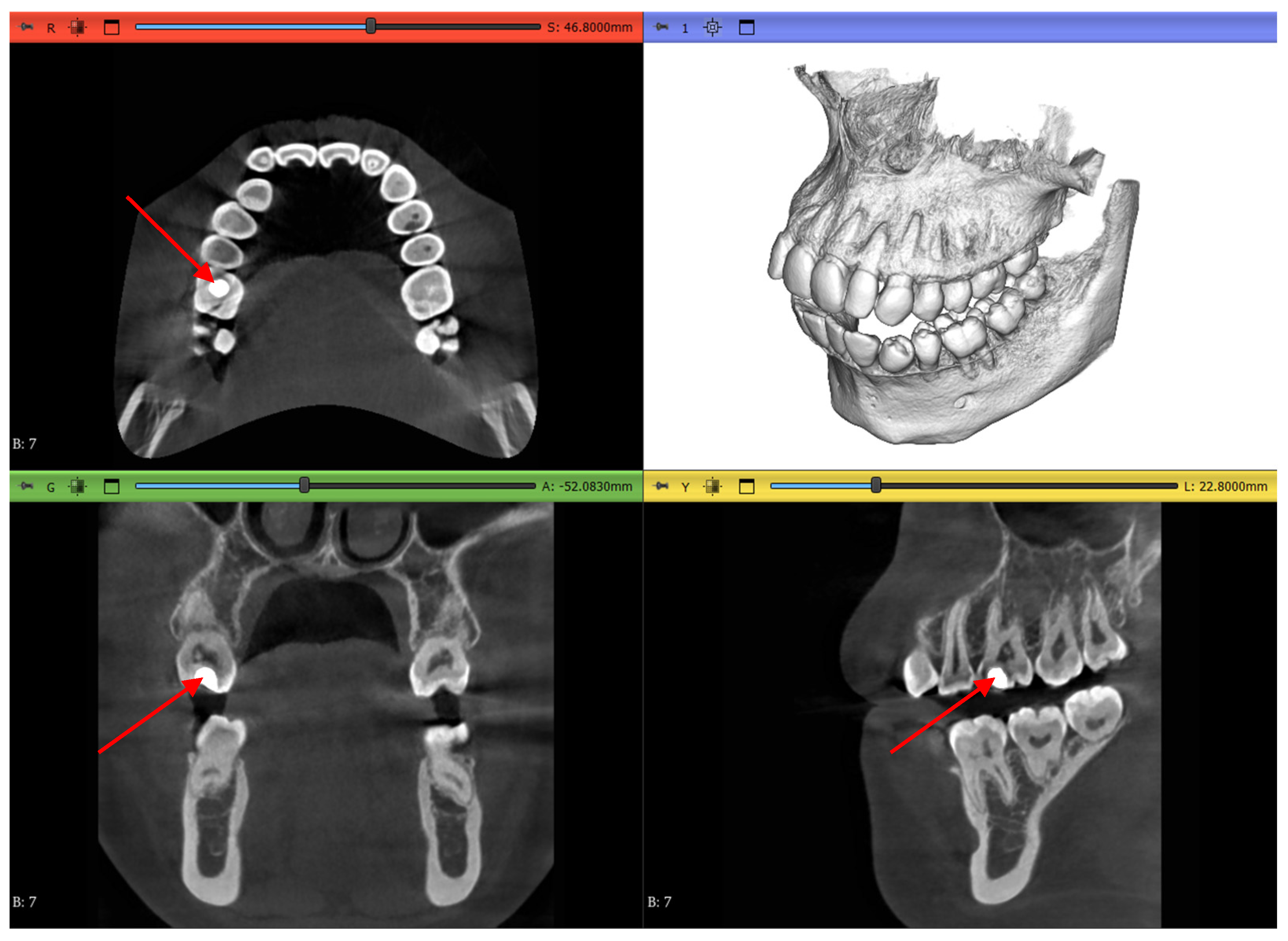
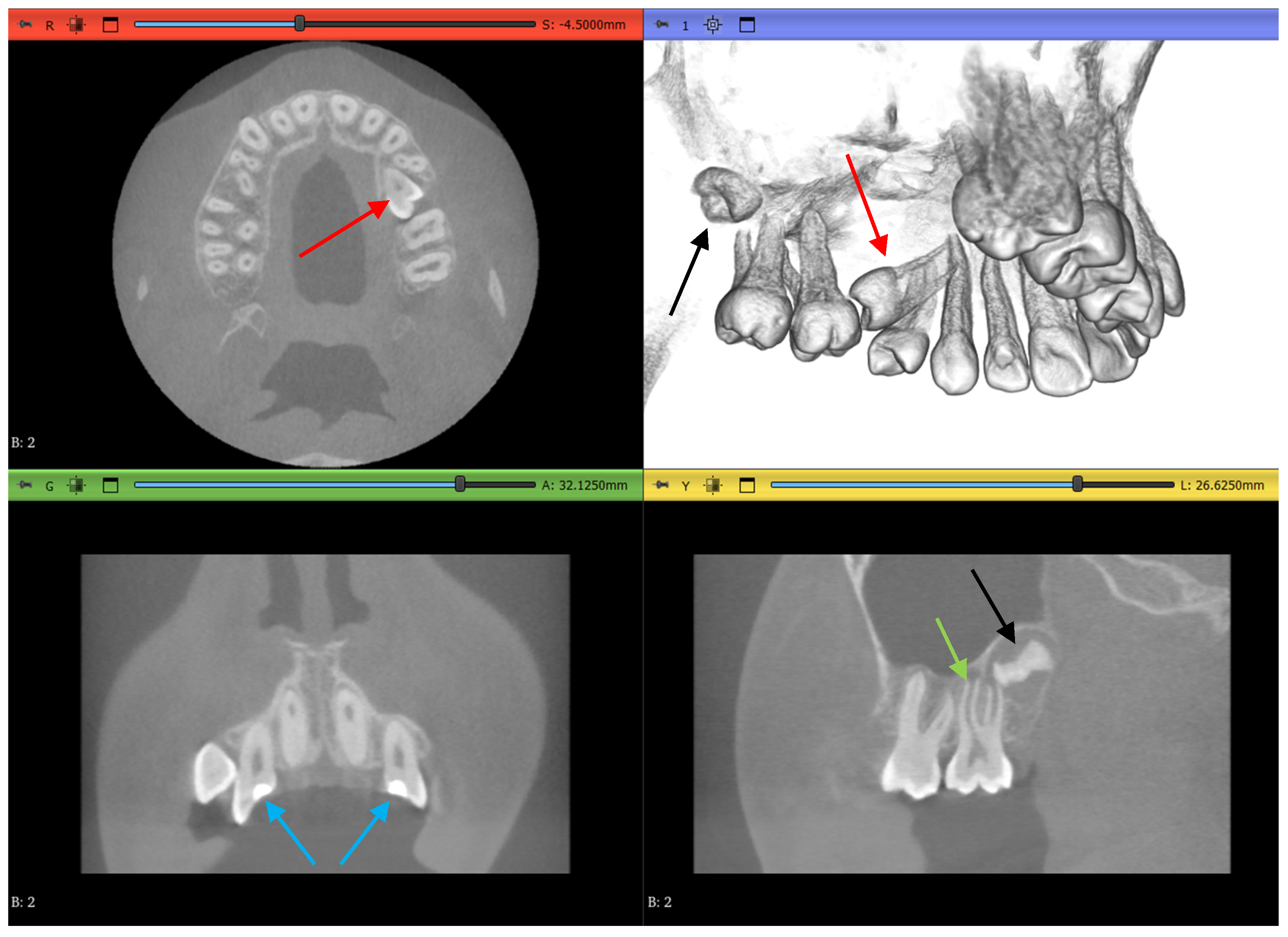
3. Results
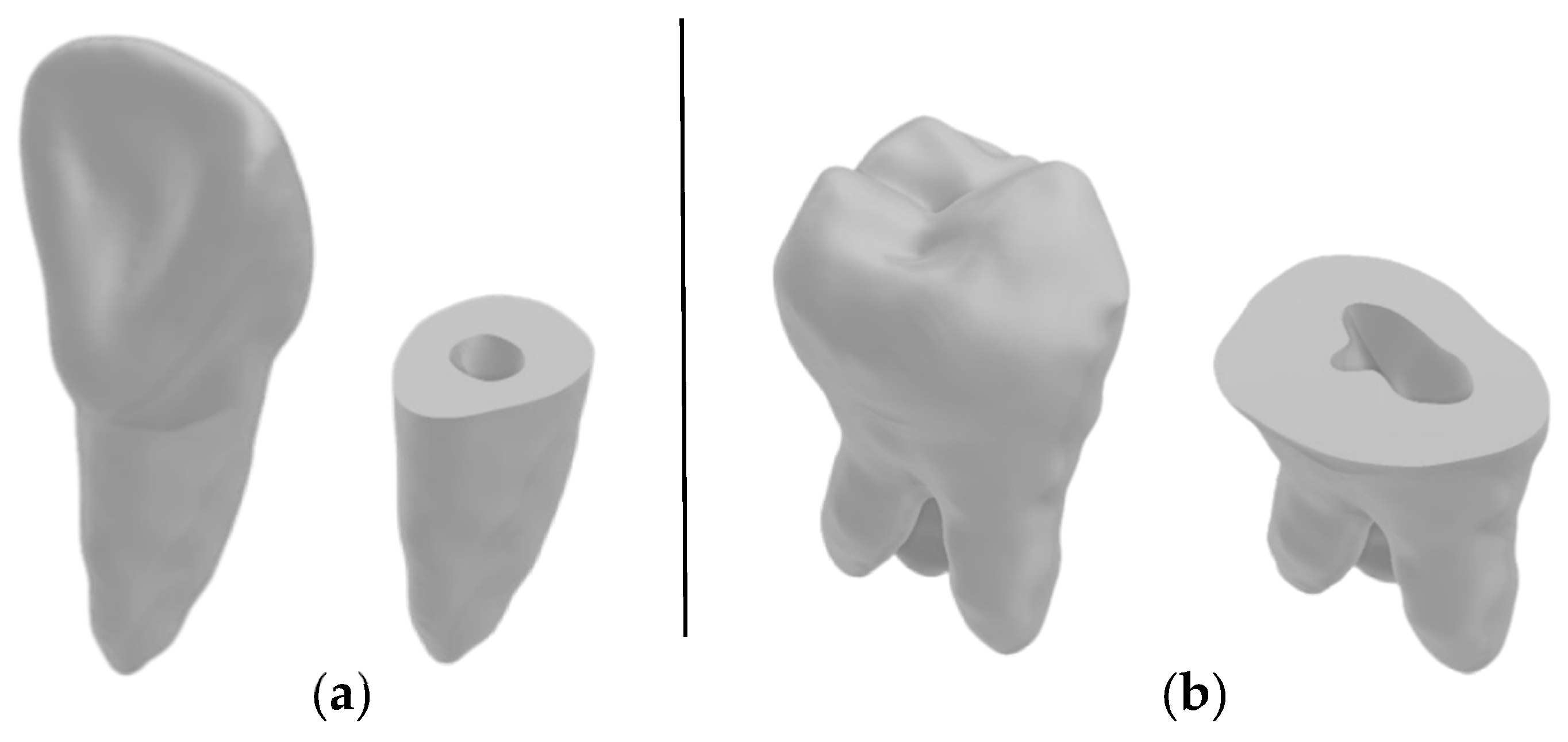
3.1. Three-Dimensional Model Comparisons
- Incisor model from all four workflows against the benchmark incisor model,
- Incisor root model from all four workflows against the benchmark incisor root model,
- Molar model from all four workflows against the benchmark molar model,
- Molar root model from all four workflows against the benchmark molar root model.
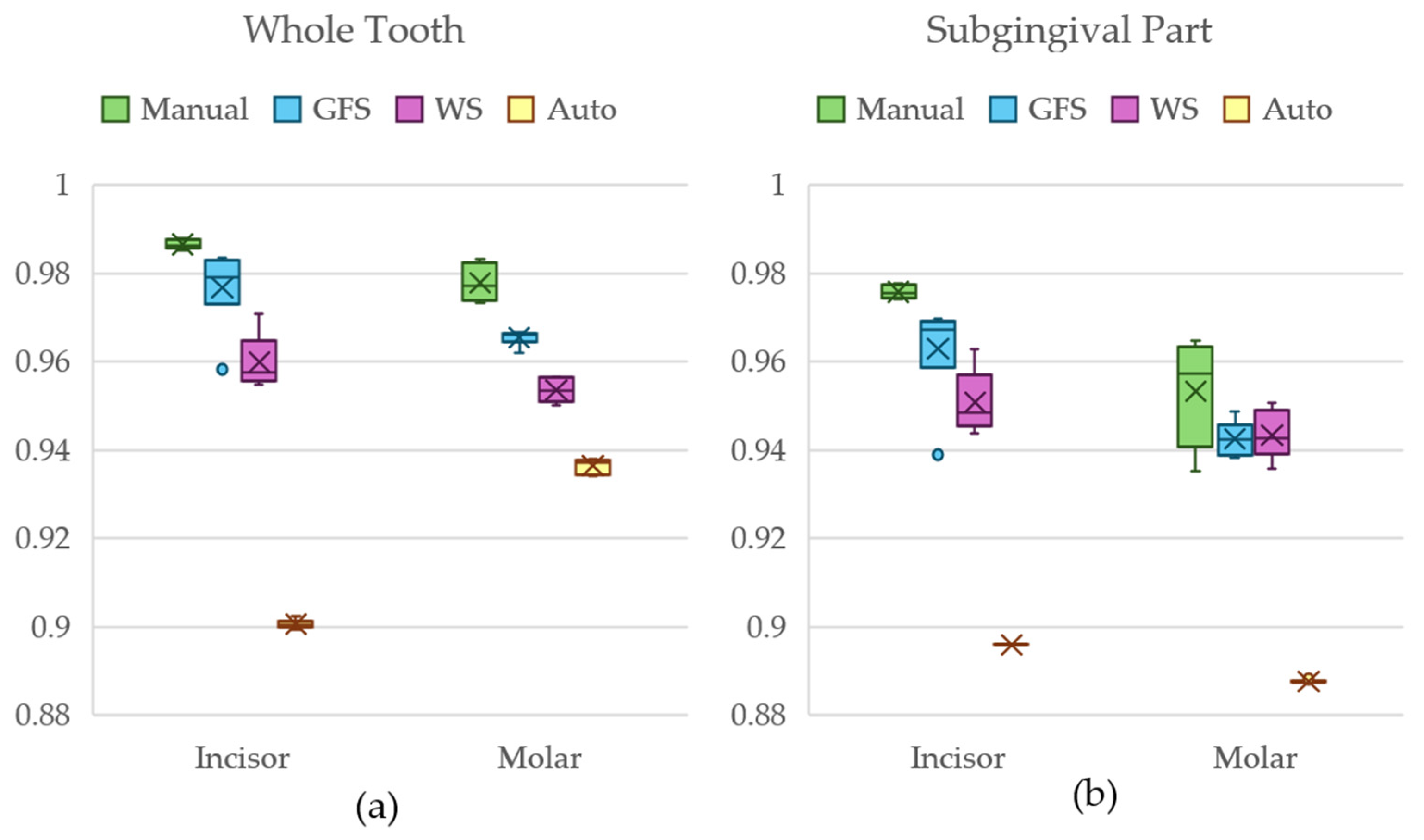
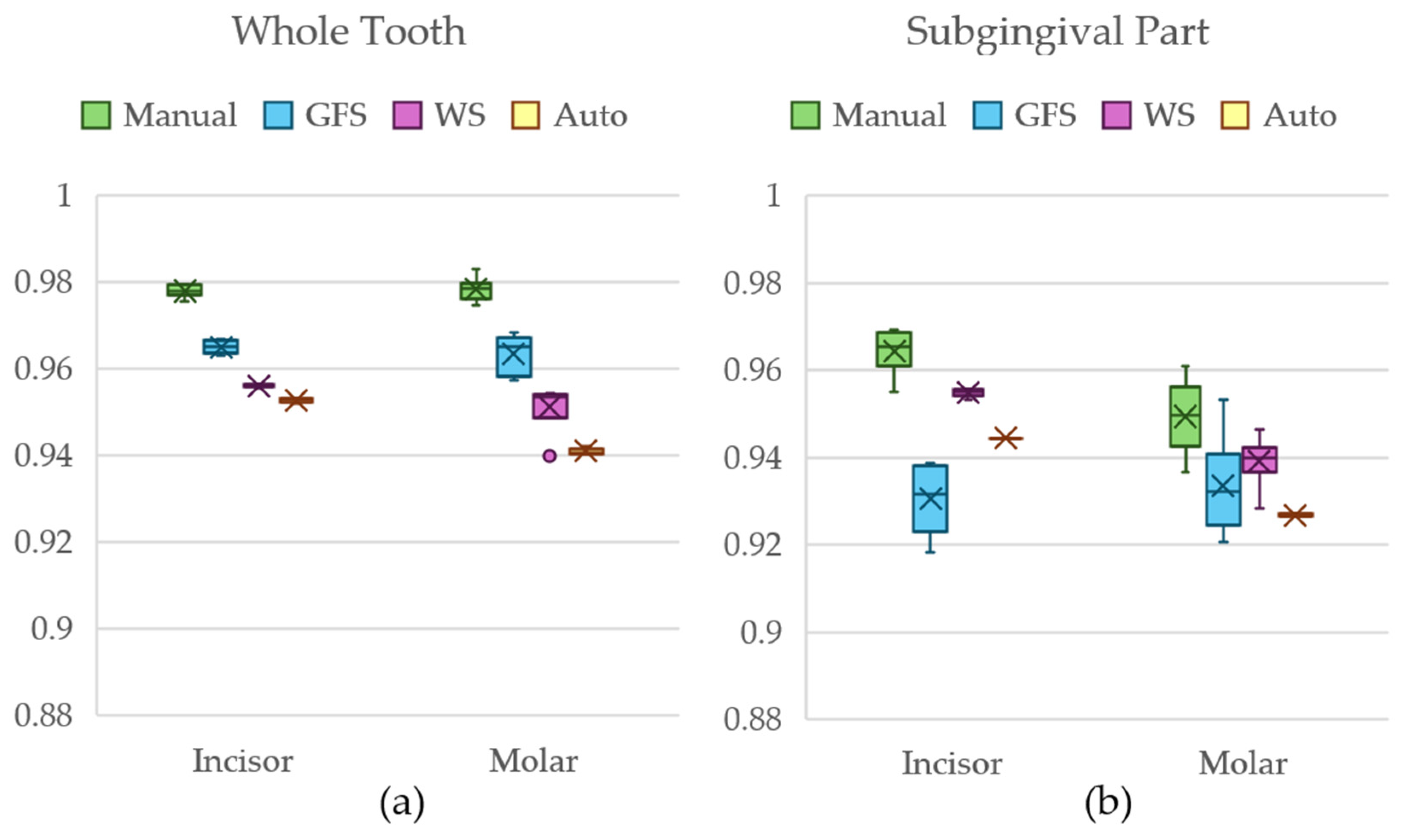
3.2. Statistical Analysis
3.3. Results of Dataset 1
3.4. Results of Dataset 2
3.5. Results of Dataset 3
4. Discussion
4.1. Manual Segmentation Method
4.2. GFS Method
4.3. WS Method
4.4. Automated DentalSegmentator
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CBCT | Cone Beam Computed Tomography |
| GAN | Generative Adversarial Network |
| GFS | Grow From Seeds |
| WS | Watershed |
| DICOM | Digital Imaging and Communications in Medicine |
| ROI | Region of Interest |
References
- Mildenberger, P.; Eichelberg, M.; Martin, E. Introduction to the DICOM standard. Eur. Radiol. 2002, 12, 920–927. [Google Scholar] [CrossRef]
- Kahn, C.E.; Carrino, J.A.; Flynn, M.J.; Peck, D.J.; Horii, S.C. DICOM and Radiology: Past, Present, and Future. J. Am. Coll. Radiol. 2007, 4, 652–657. [Google Scholar] [CrossRef]
- Towers, A.; Field, J.; Stokes, C.; Maddock, S.; Martin, N. A scoping review of the use and application of virtual reality in pre-clinical dental education. Br. Dent. J. 2019, 226, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Bruellmann, D.D.; Tjaden, H.; Schwanecke, U.; Barth, P. An optimized video system for augmented reality in endodontics: A feasibility study. Clin. Oral Investig. 2013, 17, 441–448. [Google Scholar] [CrossRef]
- Wu, C.-H.; Tsai, W.-H.; Chen, Y.-H.; Liu, J.-K.; Sun, Y.-N. Model-Based Orthodontic Assessments for Dental Panoramic Radiographs. IEEE J. Biomed. Health Inform. 2018, 22, 545–551. [Google Scholar] [CrossRef]
- Lai, Y.; Fan, F.; Wu, Q.; Ke, W.; Liao, P.; Deng, Z.; Chen, H.; Zhang, Y. LCANet: Learnable Connected Attention Network for Human Identification Using Dental Images. IEEE Trans. Med. Imaging 2021, 40, 905–915. [Google Scholar] [CrossRef]
- Cui, Z.; Fang, Y.; Mei, L.; Zhang, B.; Yu, B.; Liu, J.; Jiang, C.; Sun, Y.; Ma, L.; Huang, J.; et al. A fully automatic AI system for tooth and alveolar bone segmentation from cone-beam CT images. Nat. Commun. 2022, 13, 2096. [Google Scholar] [CrossRef] [PubMed]
- Jang, T.J.; Kim, K.C.; Cho, H.C.; Seo, J.K. A Fully Automated Method for 3D Individual Tooth Identification and Segmentation in Dental CBCT. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 6562–6568. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Yang, X. Automatic dental root CBCT image segmentation based on CNN and level set method. In Medical Imaging 2019: Image Processing; SPIE: Bellingham, WA, USA, 2019; pp. 668–674. [Google Scholar] [CrossRef]
- Cui, Z.; Li, C.; Wang, W. ToothNet: Automatic Tooth Instance Segmentation and Identification From Cone Beam CT Images. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 16–20 June 2019; pp. 6361–6370. [Google Scholar] [CrossRef]
- Gan, Y.; Xia, Z.; Xiong, J.; Li, G.; Zhao, Q. Tooth and Alveolar Bone Segmentation From Dental Computed Tomography Images. IEEE J. Biomed. Health Inform. 2018, 22, 196–204. [Google Scholar] [CrossRef]
- Wang, L.; Gao, Y.; Shi, F.; Li, G.; Chen, K.-C.; Tang, Z.; Xia, J.J.; Shen, D. Automated segmentation of dental CBCT image with prior-guided sequential random forests. Med. Phys. 2016, 43, 336–346. [Google Scholar] [CrossRef]
- Yau, H.-T.; Yang, T.-J.; Chen, Y.-C. Tooth model reconstruction based upon data fusion for orthodontic treatment simulation. Comput. Biol. Med. 2014, 48, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Chae, O. Individual tooth segmentation from CT images using level set method with shape and intensity prior. Pattern Recognit. 2010, 43, 2406–2417. [Google Scholar] [CrossRef]
- Hosntalab, M.; Zoroofi, R.A.; Tehrani-Fard, A.A.; Shirani, G. Segmentation of teeth in CT volumetric dataset by panoramic projection and variational level set. Int. J. CARS 2008, 3, 257–265. [Google Scholar] [CrossRef]
- Dumont, M.; Prieto, J.C.; Brosset, S.; Cevidanes, L.; Bianchi, J.; Ruellas, A.; Gurgel, M.; Massaro, C.; Del Castillo, A.A.; Ioshida, M.; et al. Patient Specific Classification of Dental Root Canal and Crown Shape. In Shape in Medical Imaging; Reuter, M., Wachinger, C., Lombaert, H., Paniagua, B., Goksel, O., Rekik, I., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 145–153. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Deleat-Besson, R.; Le, C.; Zhang, W.; Turkestani, N.A.; Cevidanes, L.; Bianchi, J.; Ruellas, A.; Gurgel, M.; Massaro, C.; Del Castillo, A.A.; et al. Merging and Annotating Teeth and Roots from Automated Segmentation of Multimodal Images. In Multimodal Learning for Clinical Decision Support; Syeda-Mahmood, T., Li, X., Madabhushi, A., Greenspan, H., Li, Q., Leahy, R., Dong, B., Wang, H., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 81–92. [Google Scholar] [CrossRef]
- Deleat-Besson, R.; Le, C.; Al Turkestani, N.; Zhang, W.; Dumont, M.; Brosset, S.; Prieto, J.C.; Cevidanes, L.; Bianchi, J.; Ruellas, A.; et al. Automatic Segmentation of Dental Root Canal and Merging with Crown Shape. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Mexico City, Mexico, 1–5 November 2021; pp. 2948–2951. [Google Scholar] [CrossRef]
- Wang, Y.; Xia, W.; Yan, Z.; Zhao, L.; Bian, X.; Liu, C.; Qi, Z.; Zhang, S.; Tang, Z. Root canal treatment planning by automatic tooth and root canal segmentation in dental CBCT with deep multi-task feature learning. Med. Image Anal. 2023, 85, 102750. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xia, W.; Dong, J.; Tang, Z.; Zhao, Q. Root Canal Segmentation in CBCT Images by 3D U-Net with Global and Local Combination Loss. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Mexico City, Mexico, 1–5 November 2021; pp. 3097–3100. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016; Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 424–432. [Google Scholar] [CrossRef]
- Duan, W.; Chen, Y.; Zhang, Q.; Lin, X.; Yang, X. Refined tooth and pulp segmentation using U-Net in CBCT image. Dentomaxillofac. Radiol. 2021, 50, 20200251. [Google Scholar] [CrossRef] [PubMed]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Dot, G.; Chaurasia, A.; Dubois, G.; Savoldelli, C.; Haghighat, S.; Azimian, S.; Taramsari, A.R.; Sivaramakrishnan, G.; Issa, J.; Dubey, A.; et al. DentalSegmentator: Robust open source deep learning-based CT and CBCT image segmentation. J. Dent. 2024, 147, 105130. [Google Scholar] [CrossRef]
- Yepes-Calderon, F.; McComb, J.G. Eliminating the need for manual segmentation to determine size and volume from MRI. A proof of concept on segmenting the lateral ventricles. PLoS ONE 2023, 18, e0285414. [Google Scholar] [CrossRef]
- Jader, G.; Oliveira, L.; Pithon, M. Automatic segmenting teeth in X-ray images: Trends, a novel data set, benchmarking and future perspectives. Expert Syst. Appl. 2018, 107, 15–31. [Google Scholar] [CrossRef]
- Subramanyam, R.B.; Prasad, K.P.; Anuradha, D.B. Different Image Segmentation Techniques for Dental Image Extraction. Int. J. Eng. Res. Appl. 2014, 4, 173–177. [Google Scholar]
- Adams, R.; Bischof, L. Seeded Region Growing. IEEE Trans. Pattern Anal. Mach. Intell. 1994, 16, 641–647. [Google Scholar] [CrossRef]
- Vincent, L.; Soille, P. Watersheds in digital spaces: An efficient algorithm based on immersion simulations. IEEE Trans. Pattern Anal. Mach. Intell. 1991, 13, 583–598. [Google Scholar] [CrossRef]
- Ramesh, K.K.D.; Kumar, G.K.; Swapna, K.; Datta, D.; Rajest, S.S. A Review of Medical Image Segmentation Algorithms. EAI Endorsed Trans. Pervasive Health Technol. 2021, 7, e6. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.-I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef]
- Zheng, Q.; Gao, Y.; Zhou, M.; Li, H.; Lin, J.; Zhang, W.; Chen, X. Semi or fully automatic tooth segmentation in CBCT images: A review. PeerJ Comput. Sci. 2024, 10, e1994. [Google Scholar] [CrossRef]
- Liang, X.; Nguyen, D.; Jiang, S.B. Generalizability issues with deep learning models in medicine and their potential solutions: Illustrated with cone-beam computed tomography (CBCT) to computed tomography (CT) image conversion. Mach. Learn. Sci. Technol. 2020, 2, 015007. [Google Scholar] [CrossRef]
- Verykokou, S.; Ioannidis, C.; Angelopoulos, C. Evaluation of 3D Modeling Workflows Using Dental CBCT Data for Periodontal Regenerative Treatment. J. Pers. Med. 2022, 12, 1355. [Google Scholar] [CrossRef] [PubMed]
- 3D Slicer Image Computing Platform, 3D Slicer. Available online: https://slicer.org/ (accessed on 12 November 2023).
- Kikinis, R.; Pieper, S.D.; Vosburgh, K.G. 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support. In Intraoperative Imaging and Image-Guided Therapy; Jolesz, F.A., Ed.; Springer: New York, NY, USA, 2014; pp. 277–289. [Google Scholar] [CrossRef]
- Geomagic Wrap|Artec3D. Available online: https://www.artec3d.com/3d-software/geomagic-wrap (accessed on 28 May 2024).
- Morell, G.F.C.; Chen, K.; Flores-Mir, C. 3D reconstruction of lower anterior teeth from CBCT images: Automatic segmentation with manual refinements. Dental Press. J. Orthod. 2023, 28, e232249. [Google Scholar] [CrossRef]
- ITK-SNAP Home. Available online: http://www.itksnap.org/pmwiki/pmwiki.php (accessed on 28 May 2024).
- Luc, P.; Couprie, C.; Chintala, S.; Verbeek, J. Semantic Segmentation using Adversarial Networks. arXiv 2016, arXiv:1611.08408. [Google Scholar] [CrossRef]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. arXiv 2014, arXiv:1406.2661. [Google Scholar] [CrossRef]
- Mirza, M.; Osindero, S. Conditional Generative Adversarial Nets. arXiv 2014, arXiv:1411.1784. [Google Scholar] [CrossRef]
- Makhlouf, A.; Maayah, M.; Abughanam, N.; Catal, C. The use of generative adversarial networks in medical image augmentation. Neural Comput. Appl. 2023, 35, 24055–24068. [Google Scholar] [CrossRef]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised Representation Learning with Deep Convolutional Generative Adversarial Networks. arXiv 2015, arXiv:1511.06434. [Google Scholar] [CrossRef]
- Jeong, J.J.; Tariq, A.; Adejumo, T.; Trivedi, H.; Gichoya, J.W.; Banerjee, I. Systematic Review of Generative Adversarial Networks (GANs) for Medical Image Classification and Segmentation. J. Digit. Imaging 2022, 35, 137–152. [Google Scholar] [CrossRef]
- Tian, S.; Wang, M.; Yuan, F.; Dai, N.; Sun, Y.; Xie, W.; Qin, J. Efficient Computer-Aided Design of Dental Inlay Restoration: A Deep Adversarial Framework. IEEE Trans. Med. Imaging 2021, 40, 2415–2427. [Google Scholar] [CrossRef]
- Tian, S.; Wang, M.; Ma, H.; Huang, P.; Dai, N.; Sun, Y.; Meng, J. Efficient tooth gingival margin line reconstruction via adversarial learning. Biomed. Signal Process. Control 2022, 78, 103954. [Google Scholar] [CrossRef]
- Ashwini, C.S.; Fernandes, S.; Shetty, S.; Pratap, S.; Mesta, S. DentaGAN—Smart Dental Image Segmentation. In Proceedings of the 2024 IEEE International Conference on Distributed Computing, VLSI, Electrical Circuits and Robotics (DISCOVER), Mangalore, India, 18–19 October 2024; pp. 286–291. [Google Scholar] [CrossRef]
- Koike, Y.; Anetai, Y.; Takegawa, H.; Ohira, S.; Nakamura, S.; Tanigawa, N. Deep learning-based metal artifact reduction using cycle-consistent adversarial network for intensity-modulated head and neck radiation therapy treatment planning. Phys. Medica: Eur. J. Med. Phys. 2020, 78, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Park, H.S.; Jeon, K.; Lee, S.-H.; Seo, J.K. Unpaired-Paired Learning for Shading Correction in Cone-Beam Computed Tomography. IEEE Access 2022, 10, 26140–26148. [Google Scholar] [CrossRef]
- Hegazy, M.A.A.; Cho, M.H.; Lee, S.Y. Image denoising by transfer learning of generative adversarial network for dental CT. Biomed. Phys. Eng. Express 2020, 6, 055024. [Google Scholar] [CrossRef]
- Khaleghi, G.; Hosntalab, M.; Sadeghi, M.; Reiazi, R.; Mahdavi, S.R. Metal artifact reduction in computed tomography images based on developed generative adversarial neural network. Inform. Med. Unlocked 2021, 24, 100573. [Google Scholar] [CrossRef]
- Hu, Z.; Jiang, C.; Sun, F.; Zhang, Q.; Ge, Y.; Yang, Y.; Liu, X.; Zheng, H.; Liang, D. Artifact correction in low-dose dental CT imaging using Wasserstein generative adversarial networks. Med. Phys. 2019, 46, 1686–1696. [Google Scholar] [CrossRef]
- Hegazy, M.A.A.; Cho, M.H.; Lee, S.Y. Half-scan artifact correction using generative adversarial network for dental CT. Comput. Biol. Med. 2021, 132, 104313. [Google Scholar] [CrossRef]
- Xun, S.; Li, D.; Zhu, H.; Chen, M.; Wang, J.; Li, J.; Chen, M.; Wu, B.; Zhang, H.; Chai, X.; et al. Generative adversarial networks in medical image segmentation: A review. Comput. Biol. Med. 2022, 140, 105063. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Bi, L.; Kim, J.; Feng, D.; Kumar, A. Controlled Synthesis of Dermoscopic Images via a New Color Labeled Generative Style Transfer Network to Enhance Melanoma Segmentation. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 2591–2594. [Google Scholar] [CrossRef]
- Shen, T.; Gou, C.; Wang, F.-Y.; He, Z.; Chen, W. Learning from adversarial medical images for X-ray breast mass segmentation. Comput. Methods Programs Biomed. 2019, 180, 105012. [Google Scholar] [CrossRef] [PubMed]
- Blue Sky Bio. Available online: https://blueskybio.com/ (accessed on 10 January 2024).
- Relu® Creator. Available online: https://relu.eu/creator (accessed on 10 January 2024).
- Diagnocat. 2023. Available online: https://diagnocat.com/ (accessed on 10 January 2024).
- Cephalometric Analysis & Teeth Segmentation Service|Cephx. Available online: https://cephx.com/new-front-page/ (accessed on 27 July 2024).
- exocad Ortho, Exocad. Available online: https://exocad.com/our-products/our-products (accessed on 10 January 2024).
- Embodi3D Home Biomedical 3D Printing, embodi3D.Com. Available online: https://www.embodi3d.com (accessed on 10 January 2024).
- Tzeng, L.-T.; Chang, M.-C.; Chang, S.-H.; Huang, C.-C.; Chen, Y.-J.; Jeng, J.-H. Analysis of root canal system of maxillary first and second molars and their correlations by cone beam computed tomography. J. Formos. Med. Assoc. 2020, 119, 968–973. [Google Scholar] [CrossRef] [PubMed]
- SlicerRT. Available online: https://slicerrt.github.io/ (accessed on 15 September 2025).
- Zou, K.H.; Warfield, S.K.; Bharatha, A.; Tempany, C.M.C.; Kaus, M.R.; Haker, S.J.; Wells, W.M.; Jolesz, F.A.; Kikinis, R. Statistical Validation of Image Segmentation Quality Based on a Spatial Overlap Index. Acad. Radiol. 2004, 11, 178–189. [Google Scholar] [CrossRef]
- Wang, H.; Minnema, J.; Batenburg, K.J.; Forouzanfar, T.; Hu, F.J.; Wu, G. Multiclass CBCT Image Segmentation for Orthodontics with Deep Learning. J. Dent. Res. 2021, 100, 943–949. [Google Scholar] [CrossRef]
- Zaiontz, C. Real Statistics Using Excel. Available online: https://real-statistics.com/ (accessed on 16 September 2025).
- Harter, H.L. Tables of Range and Studentized Range. Ann. Math. Stat. 1960, 31, 1122–1147. [Google Scholar] [CrossRef]
- Liedke, G.S.; Spin-Neto, R.; Vizzotto, M.B.; da Silveira, P.F.; Wenzel, A.; da Silveira, H.E.D. Diagnostic accuracy of cone beam computed tomography sections with various thicknesses for detecting misfit between the tooth and restoration in metal-restored teeth. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2015, 120, e131–e137. [Google Scholar] [CrossRef]
- Alqahtani, A.S.; Habib, S.R.; Ali, M.; Alshahrani, A.S.; Alotaibi, N.M.; Alahaidib, F.A. Maxillary anterior teeth dimension and relative width proportion in a Saudi subpopulation. J. Taibah Univ. Med. Sci. 2021, 16, 209–216. [Google Scholar] [CrossRef]
- Nogueira-Reis, F.; Morgan, N.; Suryani, I.R.; Tabchoury, C.P.M.; Jacobs, R. Full virtual patient generated by artificial intelligence-driven integrated segmentation of craniomaxillofacial structures from CBCT images. J. Dent. 2024, 141, 104829. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, E.; Leite, A.; Alqahtani, K.A.; Smolders, A.; Van Gerven, A.; Willems, H.; Jacobs, R. A novel deep learning system for multi-class tooth segmentation and classification on cone beam computed tomography. A validation study. J. Dent. 2021, 115, 103865. [Google Scholar] [CrossRef] [PubMed]







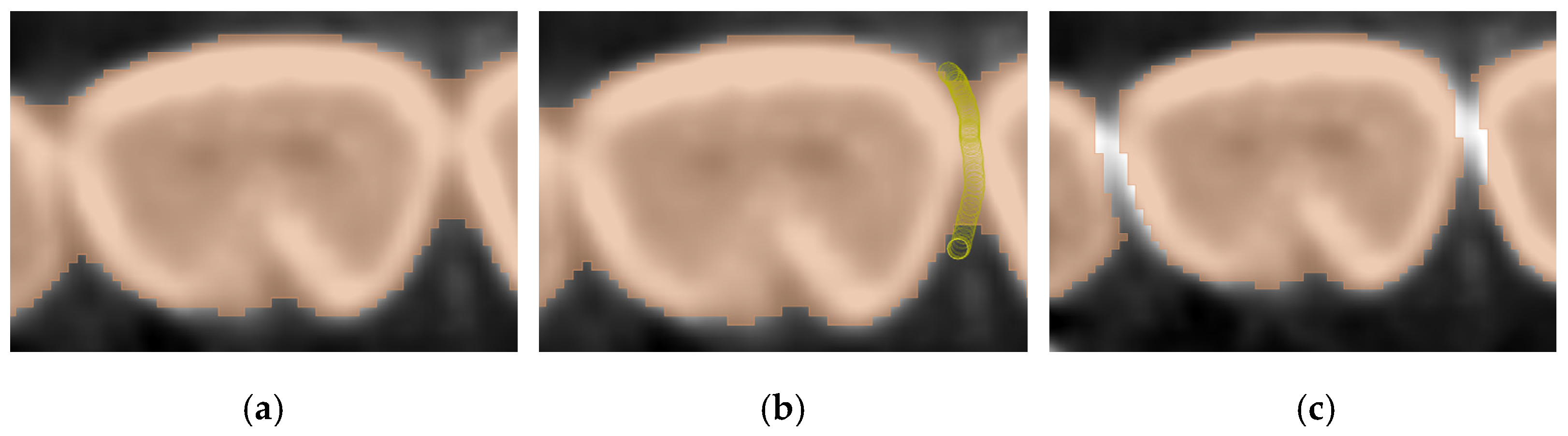
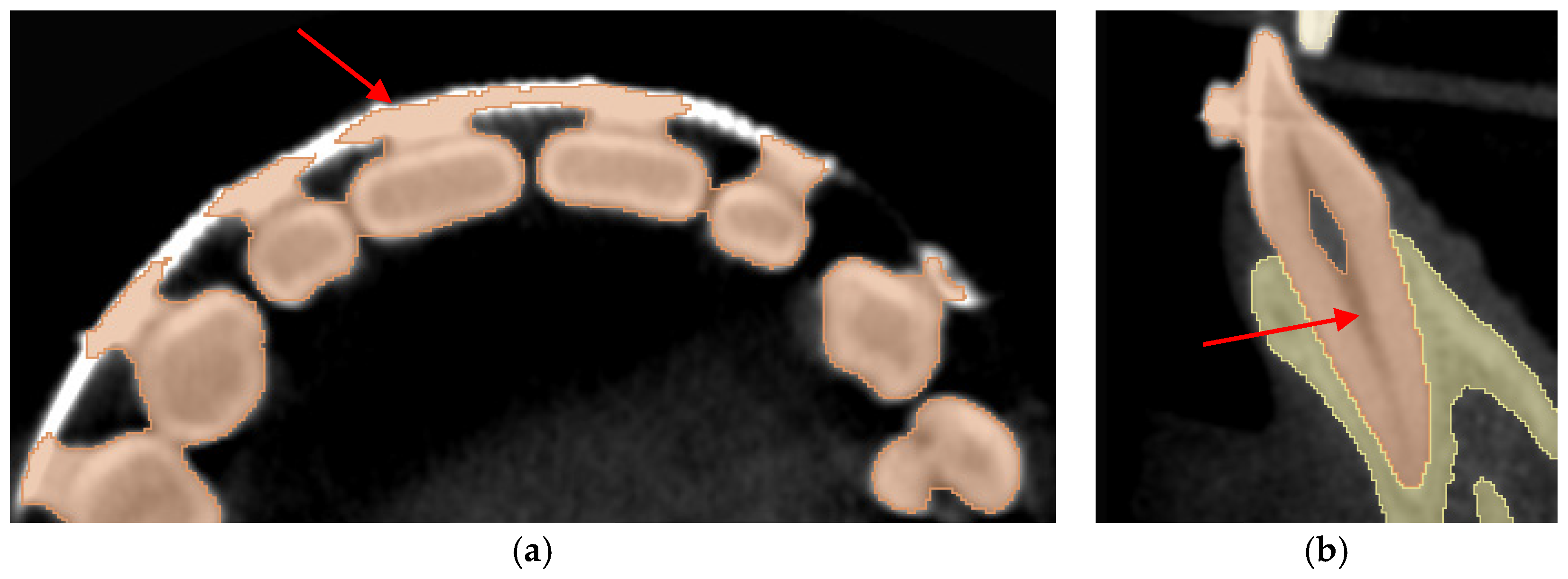
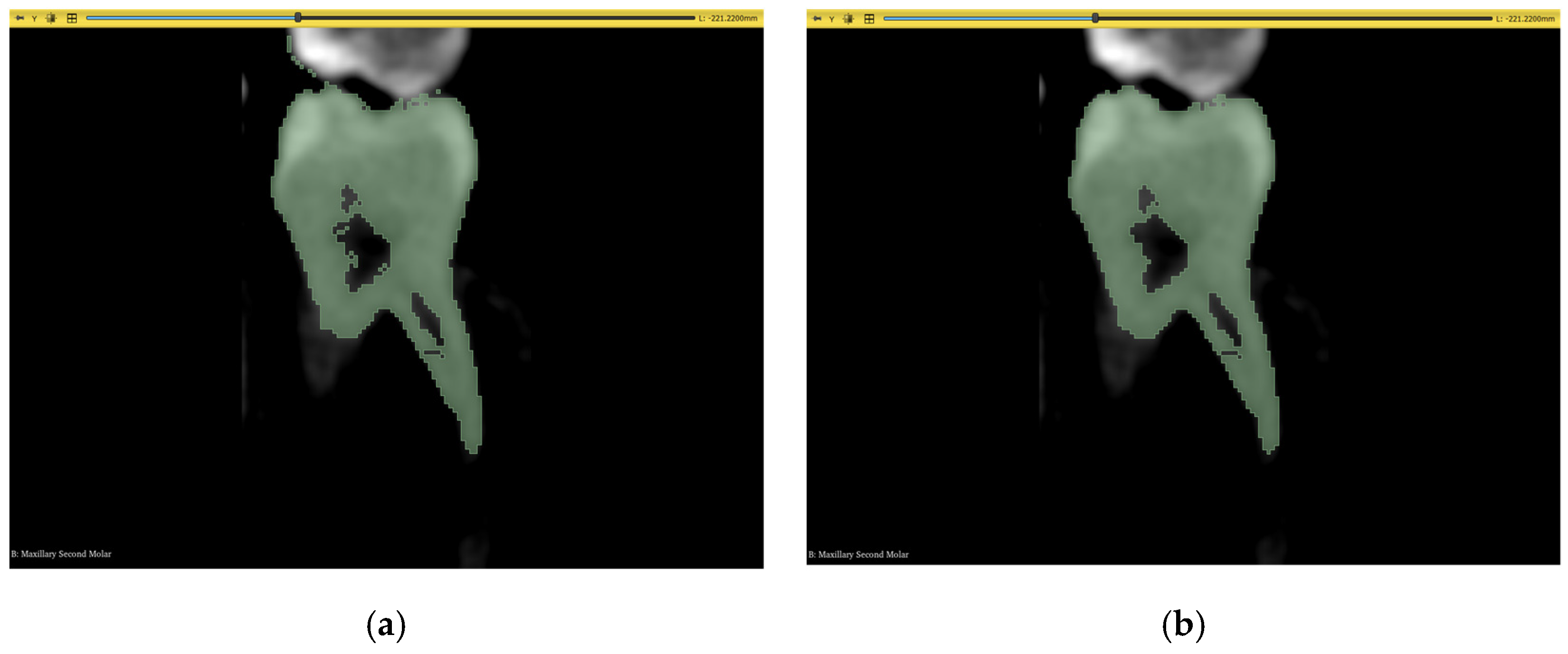
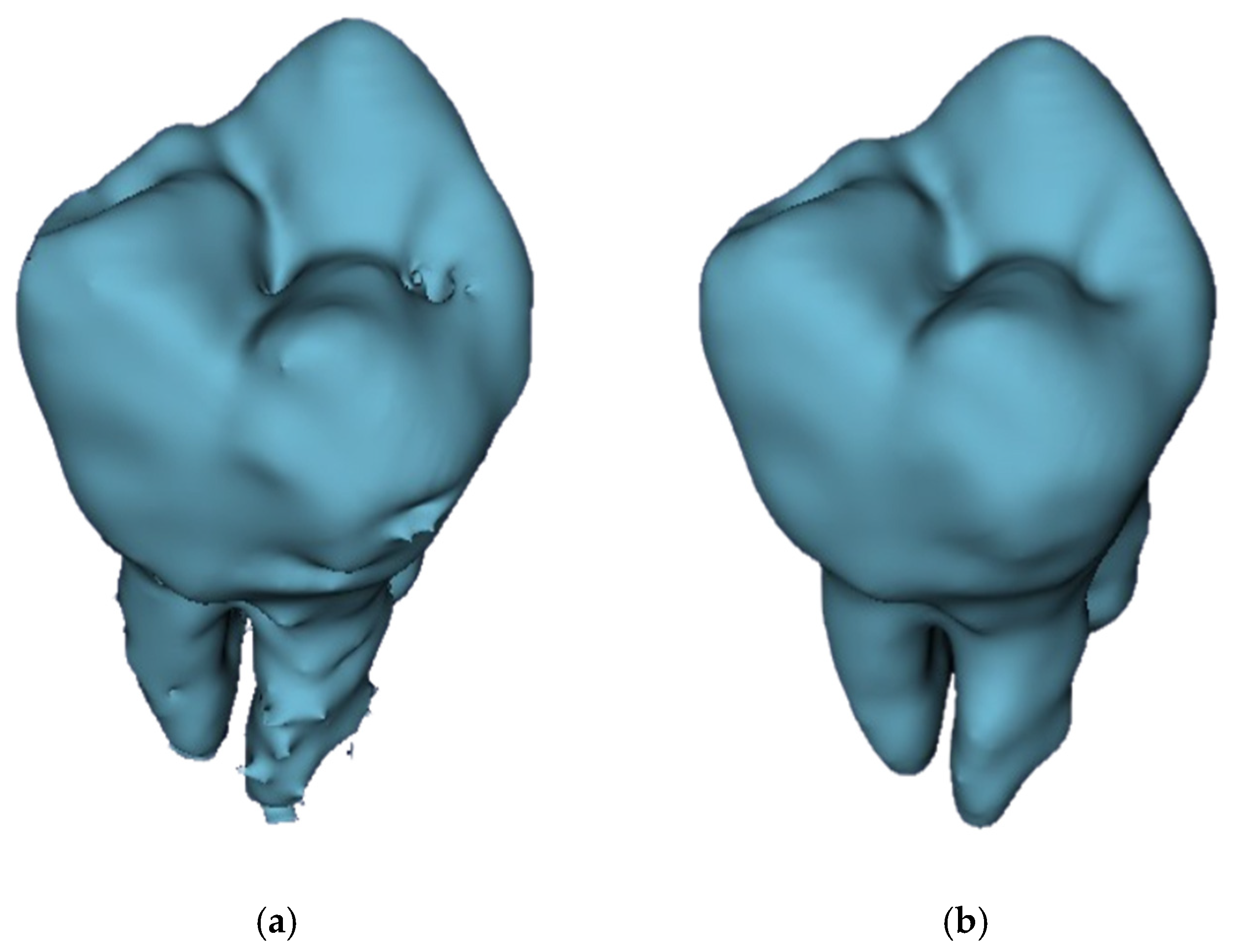




| Datasets | Manual | GFS | WS | Auto |
|---|---|---|---|---|
| Dataset 1 | 7 Incisor (including 1 benchmark) 7 Molars (including 1 benchmark) | 6 Incisors 6 Molars | 6 Incisors 6 Molars | 6 Incisors 6 Molars |
| Dataset 2 | 7 Incisor (including 1 benchmark) 7 Molars (including 1 benchmark) | 6 Incisors 6 Molars | 6 Incisors 6 Molars | 6 Incisors 6 Molars |
| Dataset 3 | 7 Incisor (including 1 benchmark) 7 Molars (including 1 benchmark) | 6 Incisors 6 Molars | 6 Incisors 6 Molars | 6 Incisors 6 Molars |
| Total Models | 42 models | 36 models | 36 models | 36 models |
| 150 models | ||||
| Dataset | Incisor | Molar | ||
|---|---|---|---|---|
| Window | Level | Window | Level | |
| Dataset 1 | 1600 | 1400 | 3000 | 1600 |
| Dataset 2 | 2300 | 1800 | 2100 | 1500 |
| Dataset 3 | 4500 | 2800 | 3500 | 2400 |
| Dataset | Incisor | Molar | ||
|---|---|---|---|---|
| Min | Max | Min | Max | |
| Dataset 1 | 1000 | 7820 (Max) | 600 | 7827 (Max) |
| Dataset 2 | 750 | 3608 (Max) | 600 | 2549 (Max) |
| Dataset 3 | 800 | 4200 | 9300 | 5300 |
| Group 1 | Group 2 | Tooth Anatomy | q-Stat | Significantly Different |
|---|---|---|---|---|
| Manual | GFS | Incisor | 2.194 | No |
| Molar | 2.078 | No | ||
| Manual | WS | Incisor | 4.041 | Yes |
| Molar | 4.157 | Yes | ||
| Manual | Auto | Incisor | 6.235 | Yes |
| Molar | 6.235 | Yes |
| Group 1 | Group 2 | Tooth Anatomy | q-Stat | Significantly Different |
|---|---|---|---|---|
| Manual | GFS | Incisor | 2.425 | No |
| Molar | 1.612 | No | ||
| Manual | WS | Incisor | 3.81 | Yes |
| Molar | 1.501 | No | ||
| Manual | Auto | Incisor | 6.235 | Yes |
| Molar | 5.196 | Yes |
| Group 1 | Group 2 | Tooth Anatomy | q-Stat | Significantly Different |
|---|---|---|---|---|
| Manual | GFS | Incisor | 2.078 | No |
| Molar | 2.078 | No | ||
| Manual | WS | Incisor | 4.157 | Yes |
| Molar | 4.503 | Yes | ||
| Manual | Auto | Incisor | 6.235 | Yes |
| Molar | 5.889 | Yes |
| Group 1 | Group 2 | Tooth Anatomy | q-Stat | Significantly Different |
|---|---|---|---|---|
| Manual | GFS | Incisor | 5.889 | Yes |
| Molar | 3.233 | No | ||
| Manual | WS | Incisor | 1.386 | No |
| Molar | 3.349 | No | ||
| Manual | Auto | Incisor | 3.811 | Yes |
| Molar | 5.889 | Yes |
| Group 1 | Group 2 | Tooth Anatomy | q-Stat | Significantly Different |
|---|---|---|---|---|
| Manual | GFS | Incisor | 3.811 | Yes |
| Molar | 2.309 | No | ||
| Manual | WS | Incisor | 2.425 | No |
| Molar | 3.406 | No | ||
| Manual | Auto | Incisor | 6.235 | Yes |
| Molar | 6.062 | Yes |
| Group 1 | Group 2 | Tooth Anatomy | q-Stat | Significantly Different |
|---|---|---|---|---|
| Manual | GFS | Incisor | 6.004 | Yes |
| Molar | 3.406 | No | ||
| Manual | WS | Incisor | 1.617 | No |
| Molar | 1.617 | No | ||
| Manual | Auto | Incisor | 3.926 | Yes |
| Molar | 4.907 | Yes |
| Group 1 | Tooth Anatomy | Average Time (Min) |
|---|---|---|
| Manual | Incisor | 19 |
| Molar | 45 | |
| GFS | Incisor | 10 |
| Molar | 20 | |
| WS | Incisor | 8 |
| Molar | 18 | |
| Auto | Incisor | 6 |
| Molar | 7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abo El Ela, Y.; Badran, M. Development of Semi-Automatic Dental Image Segmentation Workflows with Root Canal Recognition for Faster Ground Tooth Acquisition. J. Imaging 2025, 11, 340. https://doi.org/10.3390/jimaging11100340
Abo El Ela Y, Badran M. Development of Semi-Automatic Dental Image Segmentation Workflows with Root Canal Recognition for Faster Ground Tooth Acquisition. Journal of Imaging. 2025; 11(10):340. https://doi.org/10.3390/jimaging11100340
Chicago/Turabian StyleAbo El Ela, Yousef, and Mohamed Badran. 2025. "Development of Semi-Automatic Dental Image Segmentation Workflows with Root Canal Recognition for Faster Ground Tooth Acquisition" Journal of Imaging 11, no. 10: 340. https://doi.org/10.3390/jimaging11100340
APA StyleAbo El Ela, Y., & Badran, M. (2025). Development of Semi-Automatic Dental Image Segmentation Workflows with Root Canal Recognition for Faster Ground Tooth Acquisition. Journal of Imaging, 11(10), 340. https://doi.org/10.3390/jimaging11100340







