Enhanced CATBraTS for Brain Tumour Semantic Segmentation
Abstract
1. Introduction
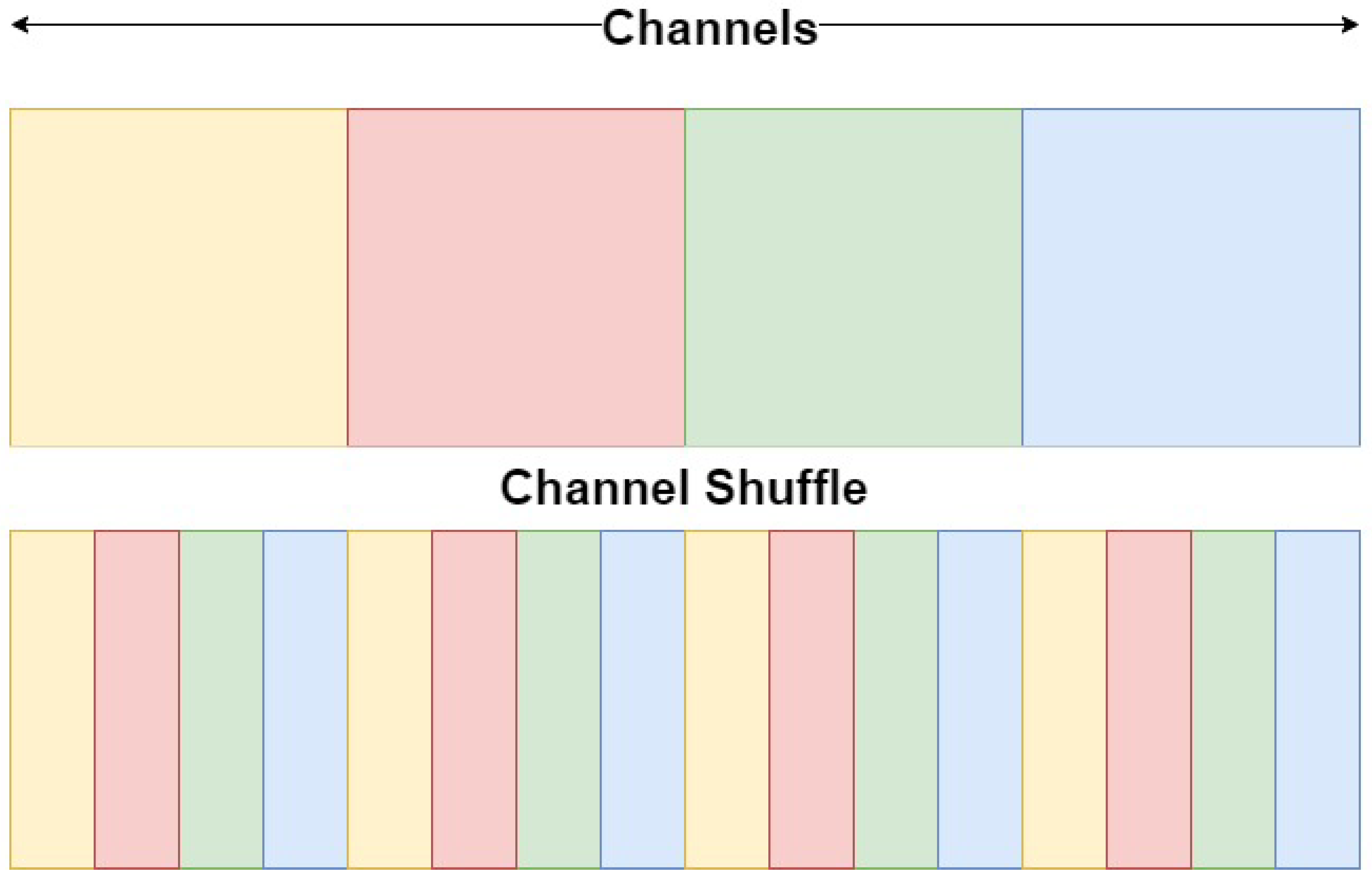
- A novel ViT-CNN model for the automatic segmentation of brain tumours in MRI volumes, which uses channel shuffling and a channel-attention mechanism; this improves both segmentation accuracy and the model’s generalisability on different MRI sequences and multi-modal MRI datasets;
- An original CNN encoding block with a channel-attention module, which can exploit tumour features and optimise the robustness of the segmentation on various brain tumour regions and image artefacts;
- A comprehensive validation on four different datasets, which demonstrates higher segmentation accuracy and generalisability, compared to the current state-of-the-art models.
2. Background
3. Methodology
4. Experiments
4.1. Datasets
4.2. Evaluation
4.3. Implementation Details
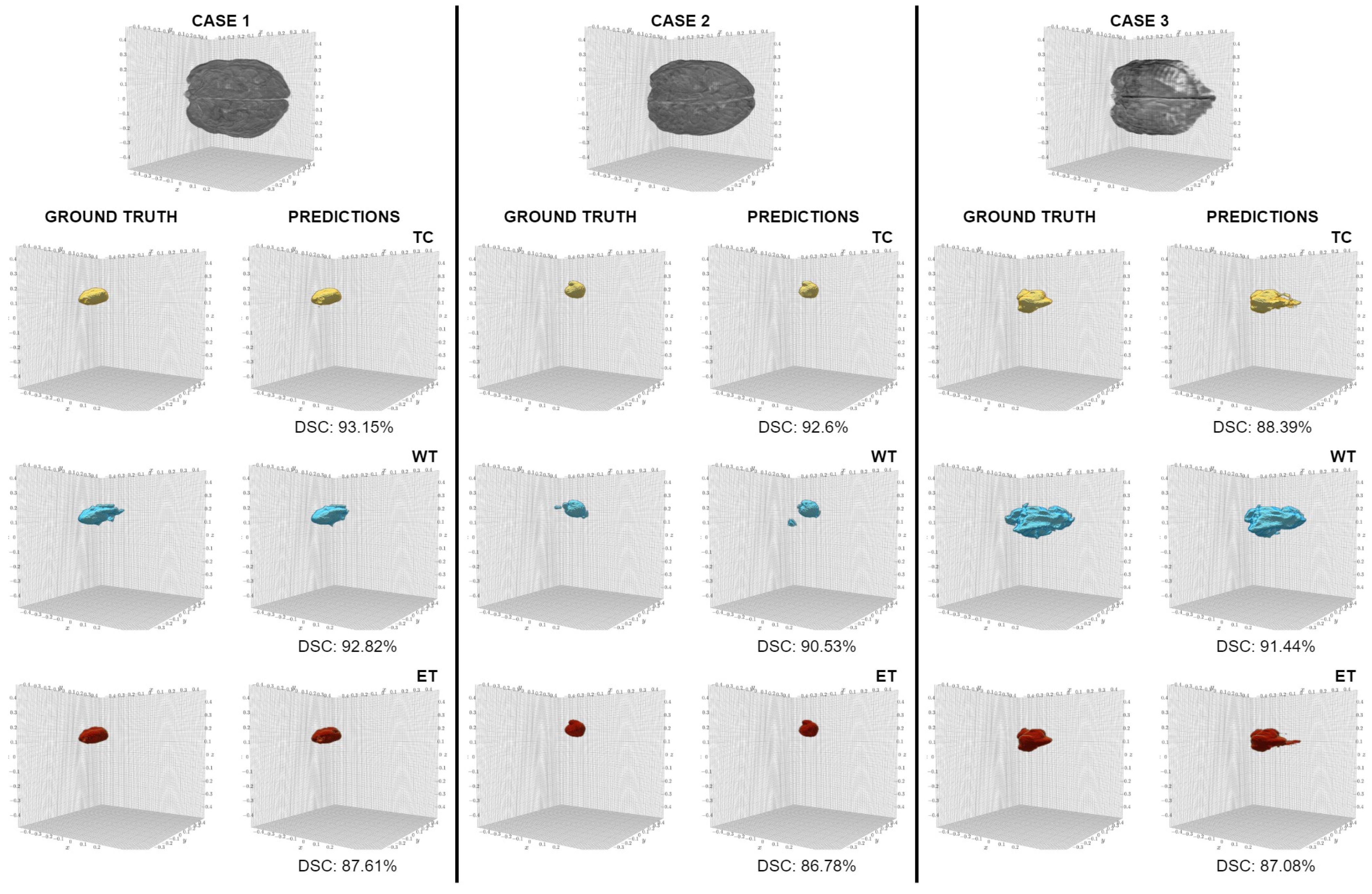
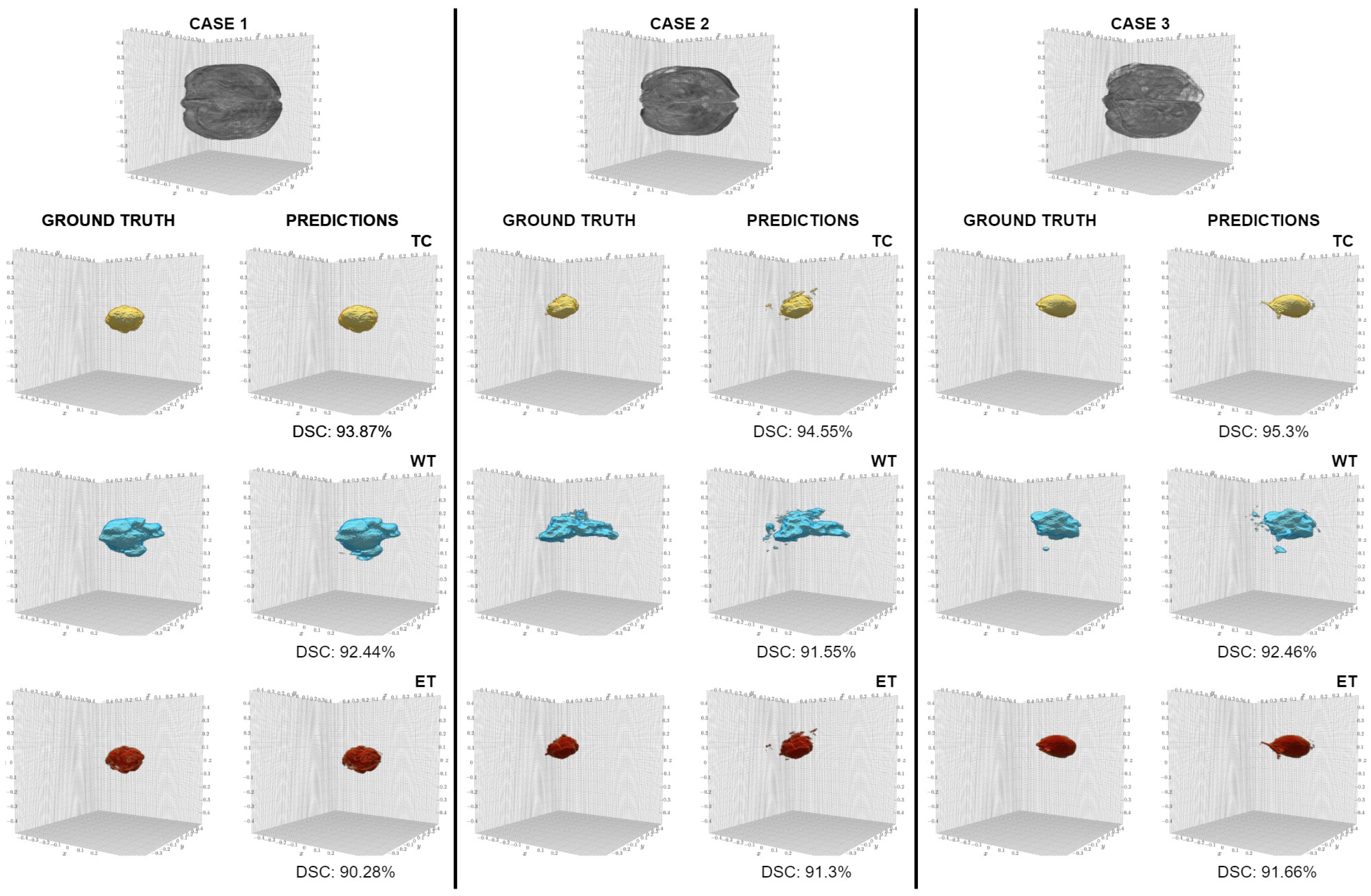
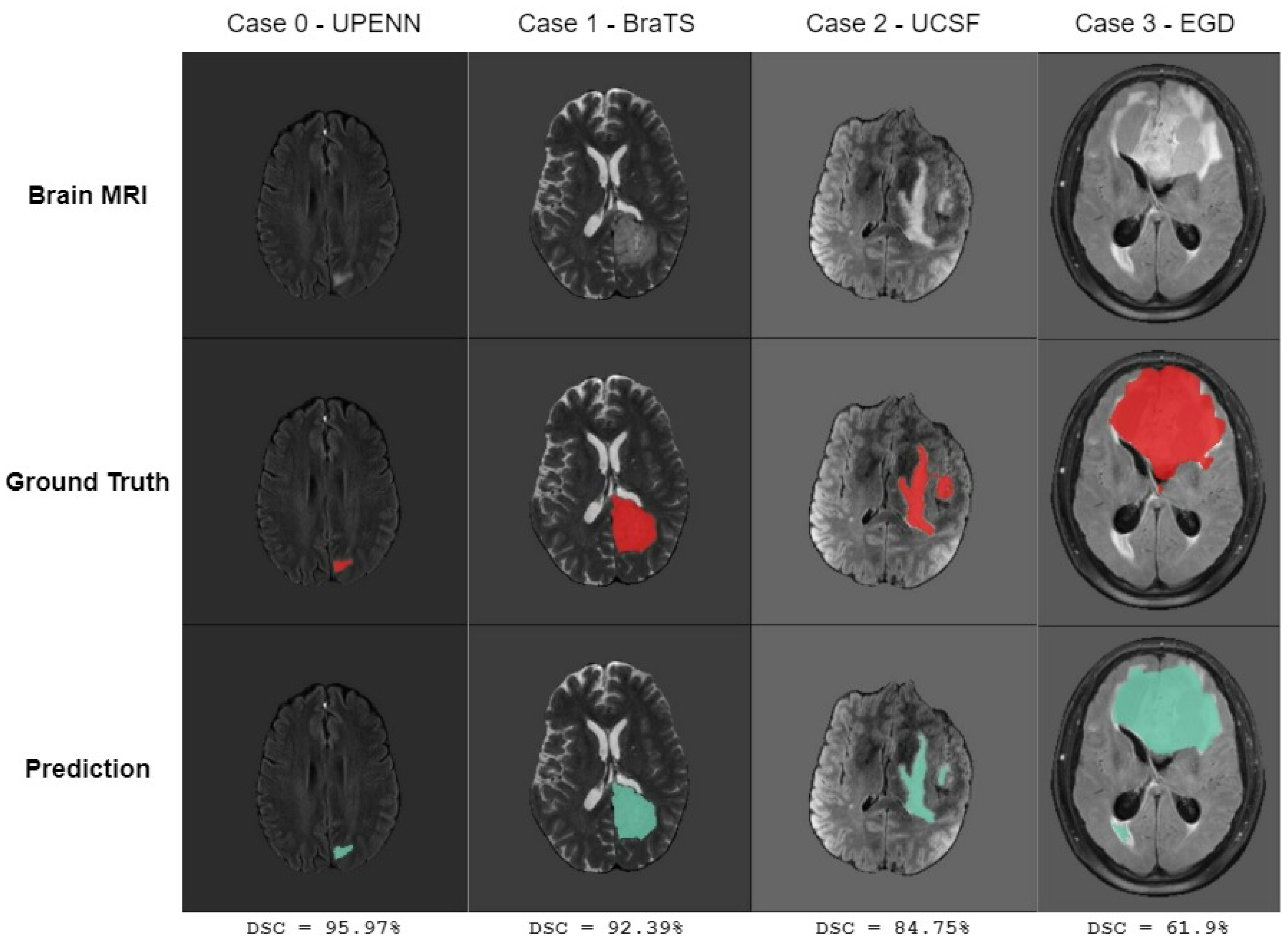
5. Results
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kortz, M.W.; Lillehei, K.O. Insular Cortex; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: http://www.ncbi.nlm.nih.gov/books/NBK570606/ (accessed on 31 July 2024).
- Huisman, T.A. Tumor-like lesions of the brain. Cancer Imaging 2009, 9, S10. [Google Scholar] [CrossRef] [PubMed]
- McFaline-Figueroa, J.R.; Lee, E.Q. Brain Tumors. Am. J. Med. 2018, 131, 874–882. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. Adult Central Nervous System Tumors Treatment (PDQ®)–Patient Version—NCI, 2022. Archive Location: Nciglobal, Ncienterprise. Available online: https://www.cancer.gov/types/brain/patient/adult-brain-treatment-pdq (accessed on 10 April 2024).
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- NHS England. Survival by Cancer Group. 2022. Available online: https://digital.nhs.uk/data-and-information/publications/statistical/cancer-survival-in-england/cancers-diagnosed-2015-to-2019-followed-up-to-2020/survival-by-cancer-group (accessed on 28 October 2024).
- Hanna, T.P.; King, W.D.; Thibodeau, S.; Jalink, M.; Paulin, G.A.; Harvey-Jones, E.; O’Sullivan, D.E.; Booth, C.M.; Sullivan, R.; Aggarwal, A. Mortality due to cancer treatment delay: Systematic review and meta-analysis. BMJ 2020, 371, m4087. [Google Scholar] [CrossRef]
- McDonald, R.J.; Schwartz, K.M.; Eckel, L.J.; Diehn, F.E.; Hunt, C.H.; Bartholmai, B.J.; Erickson, B.J.; Kallmes, D.F. The Effects of Changes in Utilization and Technological Advancements of Cross-Sectional Imaging on Radiologist Workload. Acad. Radiol. 2015, 22, 1191–1198. [Google Scholar] [CrossRef] [PubMed]
- Villarini, B.; Asaturyan, H.; Kurugol, S.; Afacan, O.; Bell, J.D.; Thomas, E.L. 3D Deep Learning for Anatomical Structure Segmentation in Multiple Imaging Modalities. In Proceedings of the 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), Virtual Conference, 7–9 June 2021; pp. 166–171, ISSN 2372-9198. [Google Scholar] [CrossRef]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef]
- Albawi, S.; Mohammed, T.A.; Al-Zawi, S. Understanding of a convolutional neural network. In Proceedings of the 2017 International Conference on Engineering and Technology (ICET), Antalya, Turkey, 21–23 August 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Li, Q.; Cai, W.; Wang, X.; Zhou, Y.; Feng, D.D.; Chen, M. Medical image classification with convolutional neural network. In Proceedings of the 2014 13th International Conference on Control Automation Robotics & Vision (ICARCV), Singapore, 10–12 December 2014; pp. 844–848. [Google Scholar] [CrossRef]
- Tabrizchi, H.; Parvizpour, S.; Razmara, J. An Improved VGG Model for Skin Cancer Detection. Neural Process. Lett. 2023, 55, 3715–3732. [Google Scholar] [CrossRef]
- Cen, L.P.; Ji, J.; Lin, J.W.; Ju, S.T.; Lin, H.J.; Li, T.P.; Wang, Y.; Yang, J.F.; Liu, Y.F.; Tan, S.; et al. Automatic detection of 39 fundus diseases and conditions in retinal photographs using deep neural networks. Nat. Commun. 2021, 12, 4828. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. arXiv 2017, arXiv:1706.03762. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2021, arXiv:2010.11929. Available online: http://arxiv.org/abs/2010.11929 (accessed on 24 January 2024).
- van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in medical imaging—“How-to” guide and critical reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. Available online: http://arxiv.org/abs/1505.04597 (accessed on 22 March 2024).
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. arXiv 2015, arXiv:1411.4038. Available online: http://arxiv.org/abs/1411.4038 (accessed on 8 November 2024).
- Myronenko, A. 3D MRI Brain Tumor Segmentation Using Autoencoder Regularization. arXiv 2018, arXiv:1810.11654. Available online: http://arxiv.org/abs/1810.11654 (accessed on 25 May 2024).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. Available online: http://arxiv.org/abs/1512.03385 (accessed on 9 July 2024).
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. arXiv 2019, arXiv:1811.02629. Available online: http://arxiv.org/abs/1811.02629 (accessed on 8 October 2024).
- Ying, X. An Overview of Overfitting and Its Solutions. J. Phys. Conf. Ser. 2019, 1168, 022022. [Google Scholar] [CrossRef]
- Hatamizadeh, A.; Nath, V.; Tang, Y.; Yang, D.; Roth, H.; Xu, D. Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images. arXiv 2022, arXiv:2201.01266. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer Using Shifted Windows. arXiv 2021, arXiv:2103.14030. Available online: http://arxiv.org/abs/2103.14030 (accessed on 12 August 2024).
- Isensee, F.; Petersen, J.; Klein, A.; Zimmerer, D.; Jaeger, P.F.; Kohl, S.; Wasserthal, J.; Koehler, G.; Norajitra, T.; Wirkert, S.; et al. nnU-Net: Self-adapting Framework for U-Net-Based Medical Image Segmentation. arXiv 2018, arXiv:1809.10486. Available online: http://arxiv.org/abs/1809.10486 (accessed on 7 October 2024).
- Isensee, F.; Jäger, P.F.; Full, P.M.; Vollmuth, P.; Maier-Hein, K.H. nnU-Net for Brain Tumor Segmentation. arXiv 2020, arXiv:2011.00848. Available online: http://arxiv.org/abs/2011.00848 (accessed on 19 September 2024).
- Wang, W.; Chen, C.; Ding, M.; Li, J.; Yu, H.; Zha, S. TransBTS: Multimodal Brain Tumor Segmentation Using Transformer. arXiv 2021, arXiv:2103.04430. Available online: http://arxiv.org/abs/2103.04430 (accessed on 10 September 2024).
- Goyal, B.; Dogra, A.; Agrawal, S.; Sohi, B. Noise Issues Prevailing in Various Types of Medical Images. Biomed. Pharmacol. J. 2018, 11, 1227–1237. [Google Scholar] [CrossRef]
- El Badaoui, R.; Coll, E.B.; Psarrou, A.; Villarini, B. 3D CATBraTS: Channel Attention Transformer for Brain Tumour Semantic Segmentation. In Proceedings of the 2023 IEEE 36th International Symposium on Computer-Based Medical Systems (CBMS), L’Aquila, Italy, 22–24 June 2023; pp. 489–494, ISSN 2372-9198. [Google Scholar] [CrossRef]
- Lin, M.; Chen, Q.; Yan, S. Network in Network. arXiv 2014, arXiv:1312.4400. Available online: http://arxiv.org/abs/1312.4400 (accessed on 9 October 2024).
- Song, Z.; Qiu, D.; Zhao, X.; Lin, D.; Hui, Y. Channel attention generative adversarial network for super-resolution of glioma magnetic resonance image. Comput. Methods Programs Biomed. 2023, 229, 107255. [Google Scholar] [CrossRef]
- Calabrese, E.; Villanueva-Meyer, J.E.; Rudie, J.D.; Rauschecker, A.M.; Baid, U.; Bakas, S.; Cha, S.; Mongan, J.T.; Hess, C.P. The University of California San Francisco Preoperative Diffuse Glioma MRI Dataset. Radiol. Artif. Intell. 2022, 4, e220058. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.; Villanueva-Meyer, J.; Rudie, J.; Rauschecker, A.; Baid, U.; Bakas, S.; Cha, S.; Mongan, J.; Hess, C. The University of California San Francisco Preoperative Diffuse Glioma MRI (UCSF-PDGM). arXiv 2023, arXiv:2109.00356. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- Bakas, S.; Sako, C.; Akbari, H.; Bilello, M.; Sotiras, A.; Shukla, G.; Rudie, J.D.; Flores Santamaria, N.; Fathi Kazerooni, A.; Pati, S.; et al. Multi-Parametric Magnetic Resonance Imaging (mpMRI) Scans for De Novo Glioblastoma (GBM) Patients from the University of Pennsylvania Health System (UPENN-GBM). 2021. Available online: https://www.cancerimagingarchive.net/collection/upenn-gbm/ (accessed on 23 December 2023). [CrossRef]
- Bakas, S.; Sako, C.; Akbari, H.; Bilello, M.; Sotiras, A.; Shukla, G.; Rudie, J.D.; Santamaría, N.F.; Kazerooni, A.F.; Pati, S.; et al. The University of Pennsylvania glioblastoma (UPenn-GBM) cohort: Advanced MRI, clinical, genomics, & radiomics. Sci. Data 2022, 9, 453. [Google Scholar] [CrossRef] [PubMed]
- van der Voort, S.R.; Incekara, F.; Wijnenga, M.M.J.; Kapsas, G.; Gahrmann, R.; Schouten, J.W.; Dubbink, H.J.; Vincent, A.J.P.E.; van den Bent, M.J.; French, P.J.; et al. The Erasmus Glioma Database (EGD): Structural MRI scans, WHO 2016 subtypes, and segmentations of 774 patients with glioma. Data Brief 2021, 37, 107191. [Google Scholar] [CrossRef]
- Marcus, D.S.; Olsen, T.R.; Ramaratnam, M.; Buckner, R.L. The extensible neuroimaging archive toolkit. Neuroinformatics 2007, 5, 11–33. [Google Scholar] [CrossRef] [PubMed]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef] [PubMed]
- Baid, U.; Ghodasara, S.; Mohan, S.; Bilello, M.; Calabrese, E.; Colak, E.; Farahani, K.; Kalpathy-Cramer, J.; Kitamura, F.C.; Pati, S.; et al. The RSNA-ASNR-MICCAI BraTS 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification. arXiv 2021, arXiv:2107.02314. Available online: http://arxiv.org/abs/2107.02314 (accessed on 12 April 2024).
- Cardoso, M.J.; Li, W.; Brown, R.; Ma, N.; Kerfoot, E.; Wang, Y.; Murrey, B.; Myronenko, A.; Zhao, C.; Yang, D.; et al. MONAI: An Open-Source Framework for Deep Learning in Healthcare. arXiv 2022, arXiv:2211.02701. Available online: http://arxiv.org/abs/2211.02701 (accessed on 12 February 2024).
- Loshchilov, I.; Hutter, F. SGDR: Stochastic Gradient Descent with Warm Restarts. arXiv 2017, arXiv:1608.03983. Available online: http://arxiv.org/abs/1608.03983 (accessed on 4 June 2024).
- Loshchilov, I.; Hutter, F. Decoupled Weight Decay Regularization. arXiv 2019, arXiv:1711.05101. Available online: http://arxiv.org/abs/1711.05101 (accessed on 4 June 2024).
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. ShuffleNet: An Extremely Efficient Convolutional Neural Network for Mobile Devices. arXiv 2017, arXiv:1707.01083. Available online: http://arxiv.org/abs/1707.01083 (accessed on 12 October 2024).



| Network | |||||||
|---|---|---|---|---|---|---|---|
| Metric | Region | SegResNet | UNETR | Swin UNetR | 3D CATBraTS |
E-CATBraTS (GRP = 2) |
E-CATBraTS (GRP = 4) |
| DSC | Mean | 0.673 ± 0.031 | 0.757 ± 0.045 | 0.749 ± 0.039 | 0.735 ± 0.038 | 0.761 ± 0.035 | 0.795 ± 0.034 |
| TC | 0.651 ± 0.073 | 0.694 ± 0.067 | 0.667 ± 0.048 | 0.667 ± 0.064 | 0.680 ± 0.044 | 0.722 ± 0.0589 | |
| WT | 0.779 ± 0.027 | 0.833 ± 0.026 | 0.834 ± 0.033 | 0.802 ± 0.018 | 0.851 ± 0.023 | 0.884 ± 0.013 | |
| ET | 0.588 ± 0.040 | 0.744 ± 0.054 | 0.748 ± 0.048 | 0.737 ± 0.045 | 0.753 ± 0.05 | 0.778 ± 0.043 | |
| Jaccard | Mean | 0.551 ± 0.028 | 0.653 ± 0.050 | 0.645 ± 0.041 | 0.624 ± 0.038 | 0.659 ± 0.036 | 0.697 ± 0.039 |
| TC | 0.54 ± 0.074 | 0.586 ± 0.073 | 0.55 ± 0.043 | 0.552 ± 0.064 | 0.568 ± 0.037 | 0.612 ± 0.065 | |
| WT | 0.659 ± 0.031 | 0.735 ± 0.032 | 0.746 ± 0.04 | 0.69 ± 0.022 | 0.761 ± 0.0305 | 0.803 ± 0.018 | |
| ET | 0.455 ± 0.037 | 0.639 ± 0.056 | 0.641 ± 0.05 | 0.628 ± 0.044 | 0.649 ± 0.052 | 0.675 ± 0.046 | |
| Hausdorff | Mean | 25.685 ± 4.238 | 25.755 ± 2.600 | 21.968 ± 5.591 | 25.783 ± 5.542 | 18.323 ± 5.883 | 13.918 ± 2.482 |
| TC | 22.404 ± 7.582 | 23.152 ± 4.408 | 23.909 ± 4.513 | 24.951 ± 5.816 | 20.768 ± 5.929 | 16.553 ± 2.684 | |
| WT | 36.426 ± 5.929 | 33.393 ± 5.179 | 20.8 ± 7.744 | 30.48 ± 6.31 | 18.333 ± 6.156 | 15.748 ± 4.457 | |
| ET | 18.226 ± 5.422 | 20.719 ± 5.665 | 21.194 ± 5.384 | 21.918 ± 6.532 | 15.866 ± 5.994 | 9.453 ± 2.238 | |
| Network | |||||||
|---|---|---|---|---|---|---|---|
| Metric | Region | SegResNet | UNETR | Swin UNetR | 3D CATBraTS | E-CATBraTS (GRP = 2) | E-CATBraTS (GRP = 4) |
| DSC | Mean | 0.751 ± 0.009 | 0.856 ± 0.017 | 0.857 ± 0.019 | 0.824 ± 0.014 | 0.871 ± 0.014 | 0.850 ± 0.009 |
| TC | 0.789 ± 0.015 | 0.823 ± 0.024 | 0.811 ± 0.027 | 0.782 ± 0.021 | 0.857 ± 0.02 | 0.802 ± 0.015 | |
| WT | 0.826 ± 0.008 | 0.891 ± 0.012 | 0.905 ± 0.013 | 0.854 ± 0.009 | 0.9 ± 0.012 | 0.909 ± 0.009 | |
| ET | 0.637 ± 0.02 | 0.854 ± 0.02 | 0.855 ± 0.02 | 0.837 ± 0.019 | 0.856 ± 0.015 | 0.839 ± 0.012 | |
| Jaccard | Mean | 0.623 ± 0.01 | 0.765 ± 0.025 | 0.769 ± 0.028 | 0.719 ± 0.019 | 0.790 ± 0.021 | 0.759 ± 0.012 |
| TC | 0.672 ± 0.02 | 0.718 ± 0.034 | 0.705 ± 0.041 | 0.665 ± 0.028 | 0.774 ± 0.028 | 0.693 ± 0.021 | |
| WT | 0.713 ± 0.01 | 0.813 ± 0.017 | 0.835 ± 0.019 | 0.753 ± 0.012 | 0.828 ± 0.016 | 0.84 ± 0.014 | |
| ET | 0.485 ± 0.019 | 0.763 ± 0.03 | 0.767 ± 0.03 | 0.739 ± 0.026 | 0.766 ± 0.021 | 0.744 ± 0.015 | |
| Hausdorff | Mean | 13.649 ± 1.452 | 11.128 ± 4.426 | 7.320 ± 1.726 | 15.407 ± 7.867 | 7.093 ± 5.05 | 7.905 ± 2.143 |
| TC | 10.367 ± 1.794 | 11.872 ± 3.647 | 9.496 ± 1.36 | 13.768 ± 7.784 | 7.776 ± 6.171 | 10.727 ± 1.614 | |
| WT | 19.904 ± 5.558 | 17.012 ± 11.923 | 7.366 ± 6.142 | 24.189 ± 11.635 | 9.273 ± 5.196 | 7.406 ± 4.835 | |
| ET | 10.677 ± 1.793 | 4.501 ± 0.795 | 5.097 ± 1.154 | 8.263 ± 5.56 | 4.23 ± 5.711 | 5.583 ± 0.773 | |
| Network | |||||
|---|---|---|---|---|---|
| Metric | Region | SegResNet | Swin UNetR | 3D CATBraTS | E-CATBraTS |
| DSC | WT | 0.738 ± 0.022 | 0.774 ± 0.026 | 0.732 ± 0.032 | 0.768 ± 0.024 |
| Jaccard | WT | 0.625 ± 0.024 | 0.661 ± 0.031 | 0.616 ± 0.034 | 0.658 ± 0.032 |
| Hausdorff | WT | 41.790 ± 8.125 | 38.313 ± 8.024 | 44.999 ± 7.836 | 34.255± 8.341 |
| Network | |||||
|---|---|---|---|---|---|
| Metric | Region | SegResNet | Swin UNetR | 3D CATBraTS | E-CATBraTS |
| DSC | Mean | 0.724 ± 0.059 | 0.751 ± 0.047 | 0.809 ± 0.033 | 0.770 ± 0.080 |
| TC | 0.737 ± 0.084 | 0.682 ± 0.084 | 0.784 ± 0.045 | 0.726 ± 0.102 | |
| WT | 0.818 ± 0.016 | 0.823 ± 0.021 | 0.851 ± 0.03 | 0.802 ± 0.032 | |
| ET | 0.616 ± 0.087 | 0.748 ± 0.055 | 0.792 ± 0.03 | 0.781 ± 0.154 | |
| Jaccard | Mean | 0.606 ± 0.054 | 0.648 ± 0.053 | 0.716 ± 0.04 | 0.658 ± 0.072 |
| TC | 0.630 ± 0.077 | 0.576 ± 0.093 | 0.690 ± 0.054 | 0.613 ± 0.098 | |
| WT | 0.709 ± 0.021 | 0.726 ± 0.025 | 0.766 ± 0.034 | 0.696 ± 0.043 | |
| ET | 0.480 ± 0.073 | 0.643 ± 0.059 | 0.692 ± 0.037 | 0.666 ± 0.132 | |
| Hausdorff | Mean | 18.372 ± 9.995 | 31.620 ± 4.594 | 10.034 ± 2.136 | 13.627 ± 2.284 |
| TC | 12.671 ± 11.545 | 23.667 ± 6.574 | 9.474 ± 2.286 | 9.723 ± 1.651 | |
| WT | 29.155 ± 7.718 | 50.733 ± 5.304 | 13.709 ± 4.411 | 24.559 ± 6.921 | |
| ET | 13.291 ± 11.915 | 20.461 ± 7.383 | 6.919 ± 2.164 | 6.598 ± 1.783 | |
| Network | |||||
|---|---|---|---|---|---|
| Metric | Datasets | E-CATBraTS | No Attention | No Shuffle |
No Shuffle No Attention |
| DSC | UCSF | 0.795 ± 0.034 | 0.745 ± 0.033 | 0.762 ± 0.031 | 0.719 ± 0.041 |
| UPENN | 0.871 ± 0.014 | 0.854 ± 0.010 | 0.859 ± 0.021 | 0.836 ± 0.02 | |
| EGD | 0.768 ± 0.024 | 0.753 ± 0.037 | 0.755 ± 0.028 | 0.740 ± 0.034 | |
| BraTS 2021 | 0.770 ± 0.080 | 0.724 ± 0.047 | 0.707 ± 0.037 | 0.709 ± 0.052 | |
| Jaccard | UCSF | 0.697 ± 0.039 | 0.640 ± 0.031 | 0.657 ± 0.032 | 0.605 ± 0.044 |
| UPENN | 0.790 ± 0.021 | 0.765 ± 0.013 | 0.775 ± 0.030 | 0.741 ± 0.03 | |
| EGD | 0.658 ± 0.032 | 0.639 ± 0.034 | 0.639 ± 0.032 | 0.625 ± 0.035 | |
| BraTS 2021 | 0.658 ± 0.072 | 0.615 ± 0.051 | 0.596 ± 0.035 | 0.603 ± 0.053 | |
| Hausdorff | UCSF | 13.918 ± 2.482 | 17.461 ± 2.600 | 25.196 ± 3.243 | 33.820 ± 4.635 |
| UPENN | 7.093 ± 5.05 | 8.769 ± 2.250 | 6.609 ± 1.898 | 6.711 ± 1.241 | |
| EGD | 34.255 ± 8.341 | 40.145 ± 8.628 | 39.773 ± 7.702 | 46.015 ± 8.462 | |
| BraTS 2021 | 13.627 ± 2.284 | 23.249 ± 3.504 | 18.341 ± 2.524 | 14.272 ± 2.964 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Badaoui, R.; Bonmati Coll, E.; Psarrou, A.; Asaturyan, H.A.; Villarini, B. Enhanced CATBraTS for Brain Tumour Semantic Segmentation. J. Imaging 2025, 11, 8. https://doi.org/10.3390/jimaging11010008
El Badaoui R, Bonmati Coll E, Psarrou A, Asaturyan HA, Villarini B. Enhanced CATBraTS for Brain Tumour Semantic Segmentation. Journal of Imaging. 2025; 11(1):8. https://doi.org/10.3390/jimaging11010008
Chicago/Turabian StyleEl Badaoui, Rim, Ester Bonmati Coll, Alexandra Psarrou, Hykoush A. Asaturyan, and Barbara Villarini. 2025. "Enhanced CATBraTS for Brain Tumour Semantic Segmentation" Journal of Imaging 11, no. 1: 8. https://doi.org/10.3390/jimaging11010008
APA StyleEl Badaoui, R., Bonmati Coll, E., Psarrou, A., Asaturyan, H. A., & Villarini, B. (2025). Enhanced CATBraTS for Brain Tumour Semantic Segmentation. Journal of Imaging, 11(1), 8. https://doi.org/10.3390/jimaging11010008







