PlantSR: Super-Resolution Improves Object Detection in Plant Images
Abstract
1. Introduction
2. Materials and Methods
2.1. PlantSR Dataset
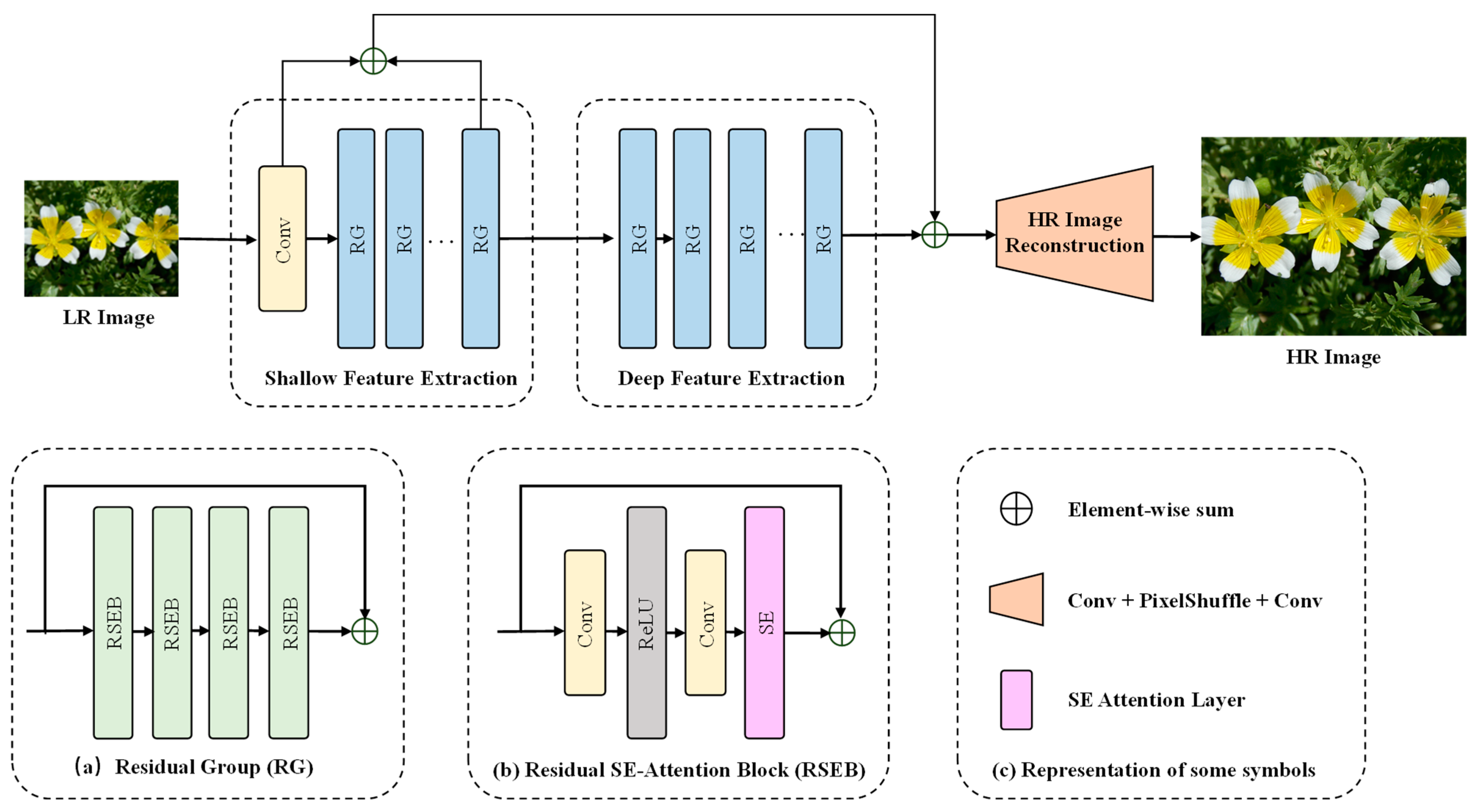
2.2. Architecture of PlantSR Model
2.3. Super-Resolution Effects on Apple Counting Task
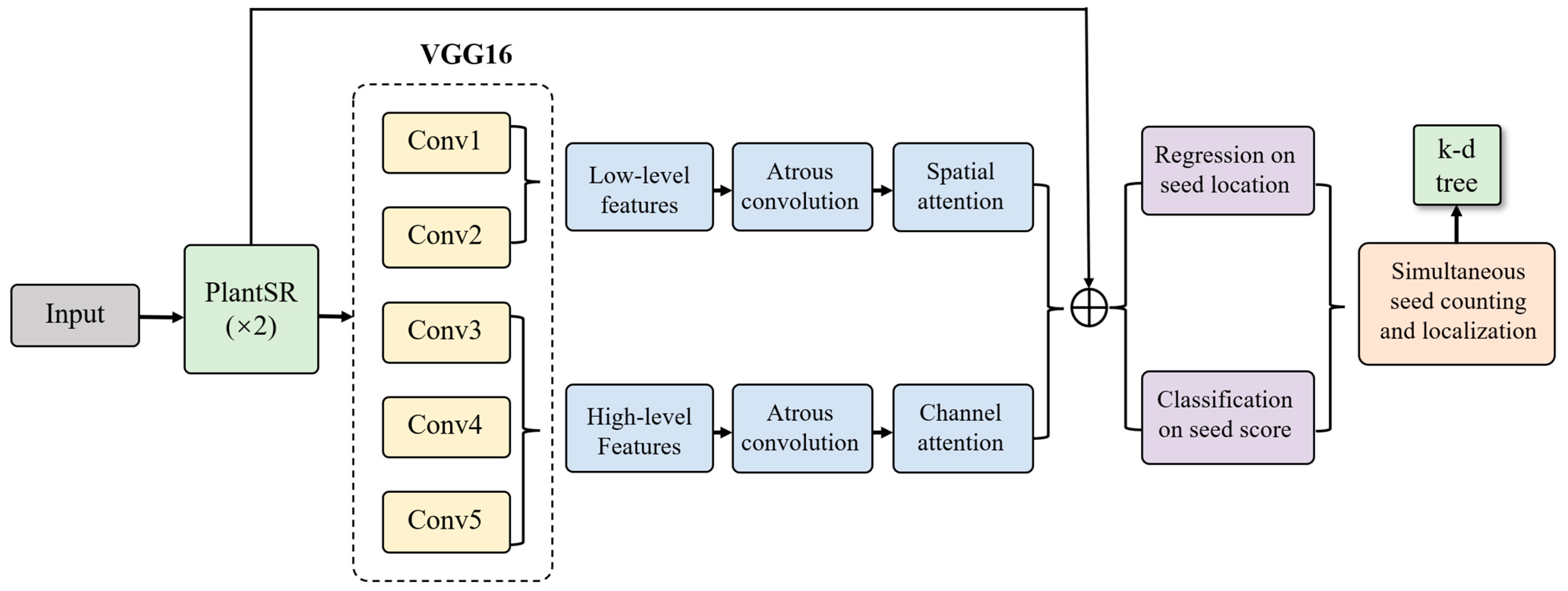
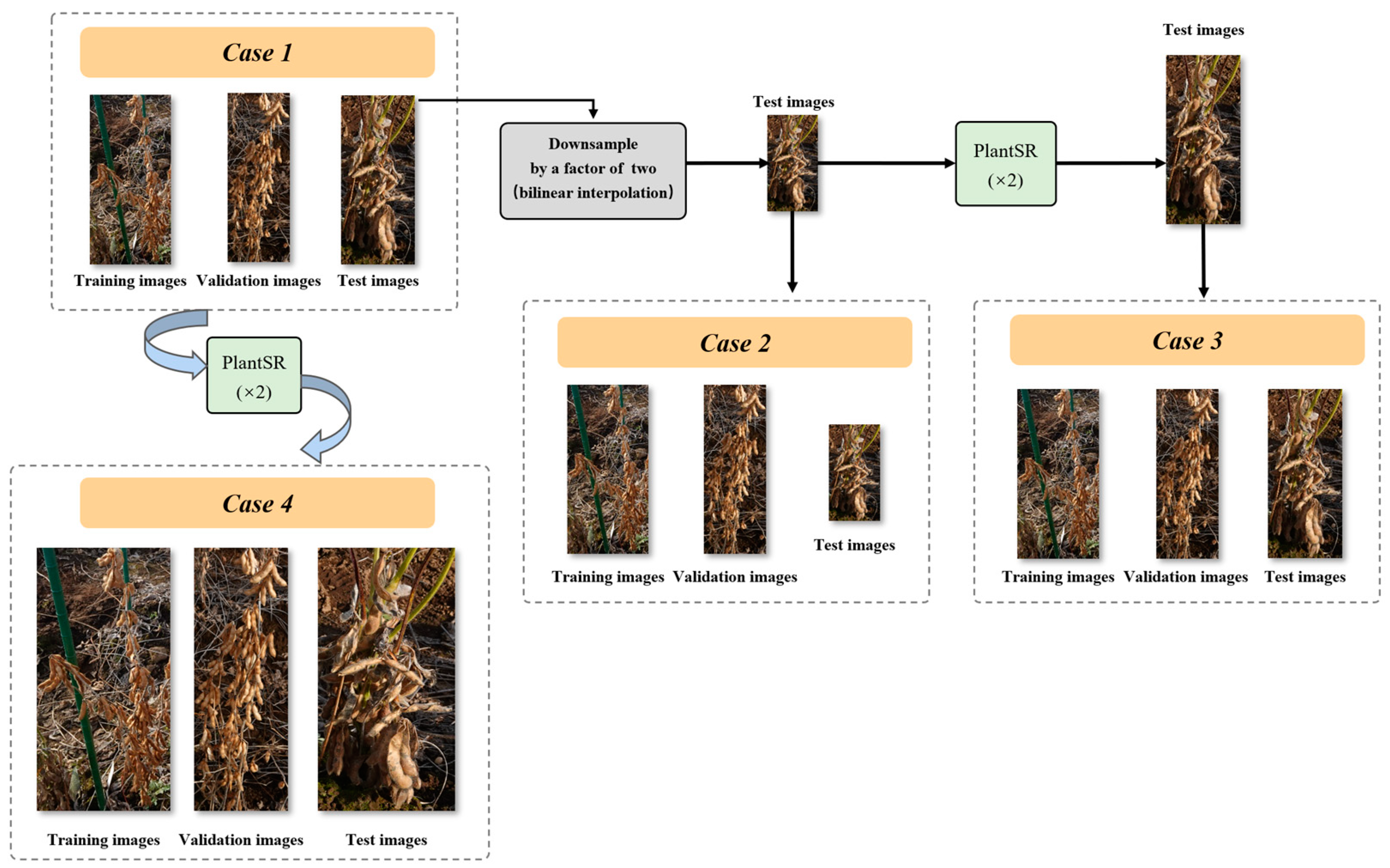
2.4. Super-Resolution Effects on Soybean Seed Counting Task
2.5. Training and Evaluation Settings
2.6. Evaluation Metrics
3. Results
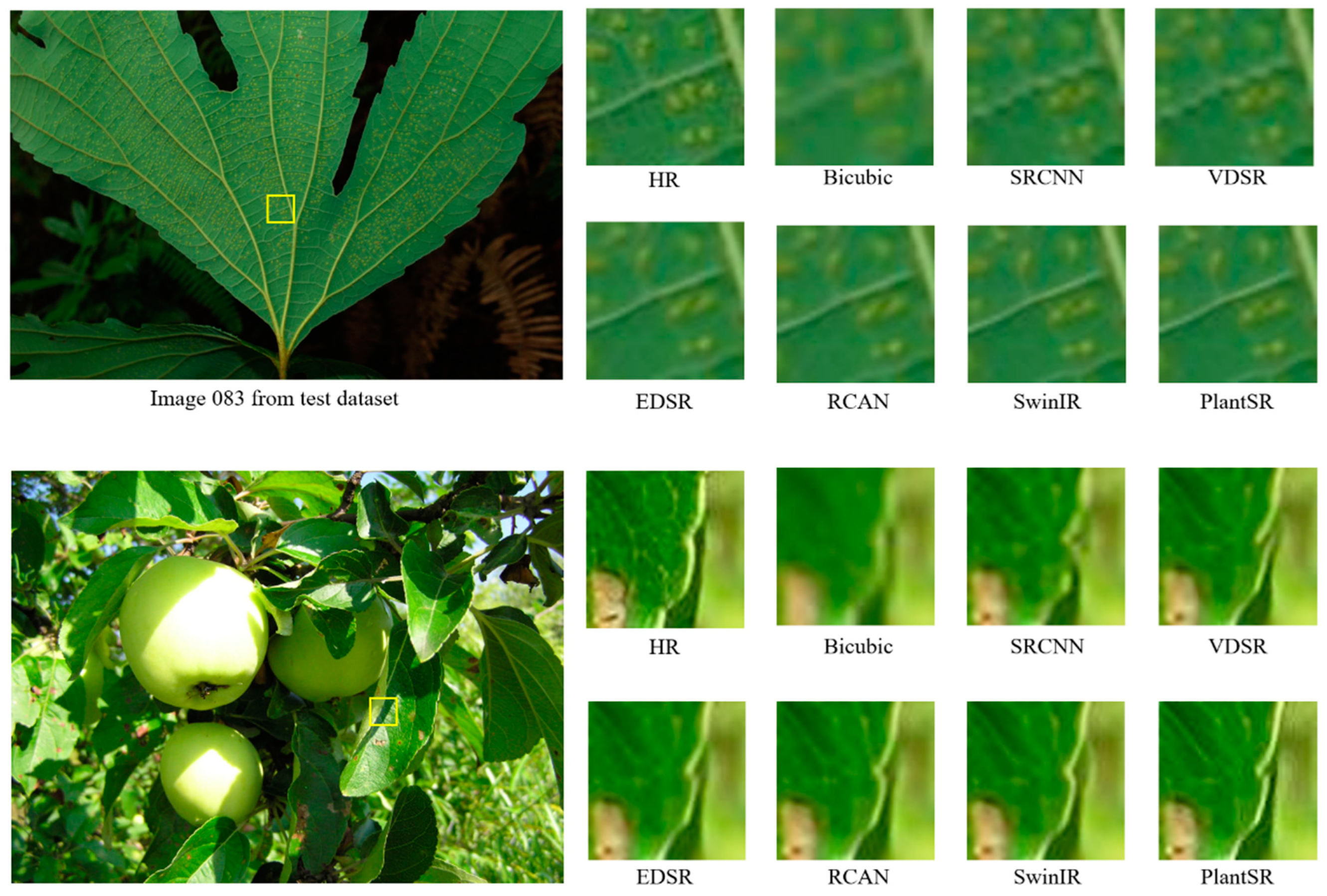
3.1. SR Model Compression
3.2. Super-Resolution Effects on the Apple Counting Task
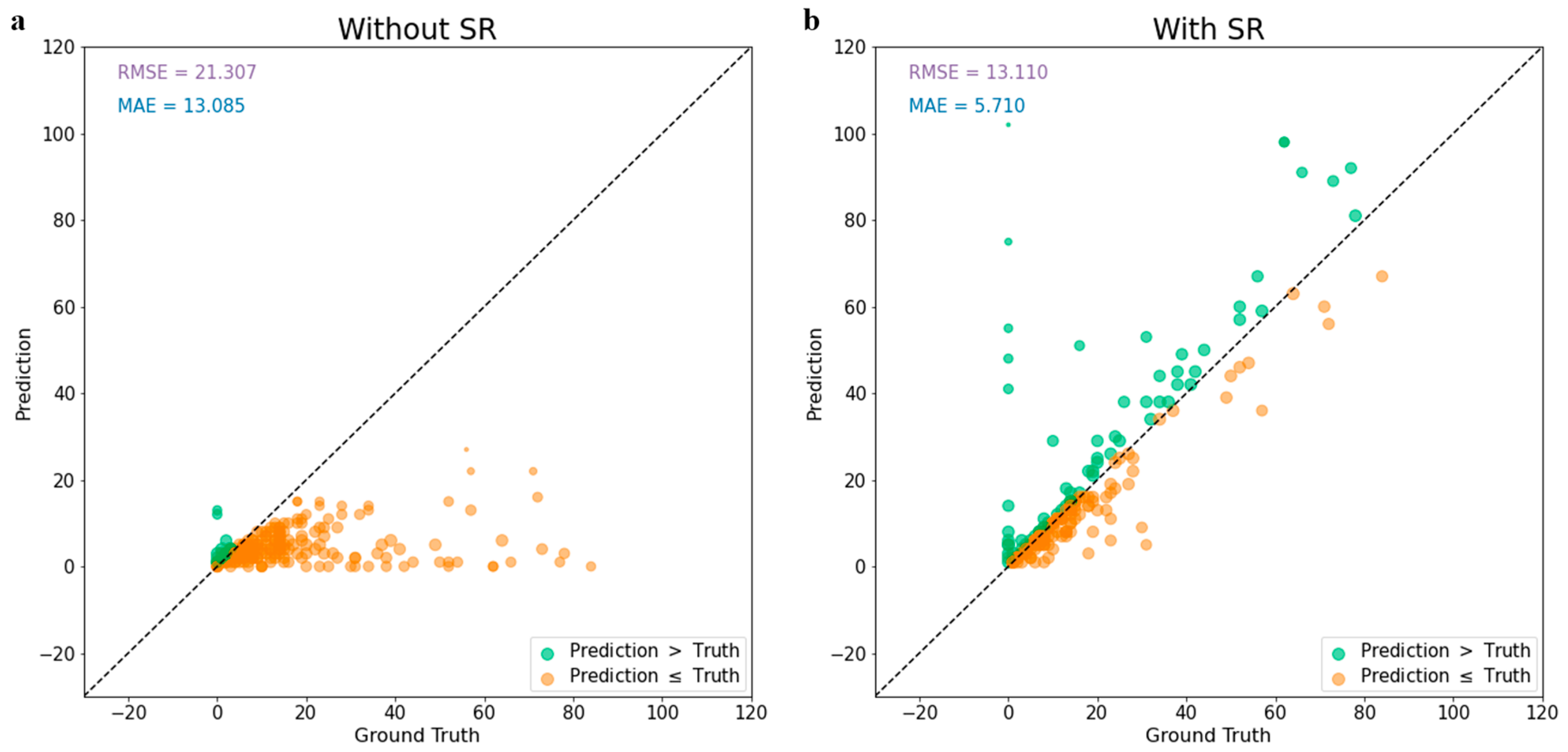
3.3. Super-Resolution Effects on the Soybean Seed Counting Task
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Walter, A.; Liebisch, F.; Hund, A. Plant phenotyping: From bean weighing to image analysis. Plant Methods 2015, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Das Choudhury, S.; Samal, A.; Awada, T. Leveraging image analysis for high-throughput plant phenotyping. Front. Plant Sci. 2019, 10, 508. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jia, J.; Zhang, L.; Khattak, A.M.; Sun, S.; Gao, W.; Wang, M. Soybean seed counting based on pod image using two-column convolution neural network. IEEE Access 2019, 7, 64177–64185. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Wu, T.; Sun, S. Fast Counting Method of Soybean Seeds Based on Density Estimation and VGG-Two. Smart Agric. 2021, 3, 111. [Google Scholar]
- Khaki, S.; Saeed, N.; Pham, H.; Wang, L. WheatNet: A lightweight convolutional neural network for high-throughput image-based wheat head detection and counting. Neurocomputing 2022, 489, 78–89. [Google Scholar] [CrossRef]
- David, E.; Madec, S.; Sadeghi-Tehran, P.; Aasen, H.; Zheng, B.; Liu, S.; Kirchgessner, N.; Ishikawa, G.; Nagasawa, K.; Badhon, M.A.; et al. Global Wheat Head Detection (GWHD) dataset: A large and diverse dataset of high-resolution RGB-labelled images to develop and benchmark wheat head detection methods. Plant Phenomics 2020, 2020, 3521852. [Google Scholar] [CrossRef] [PubMed]
- David, E.; Serouart, M.; Smith, D.; Madec, S.; Velumani, K.; Liu, S.; Wang, X.; Pinto, F.; Shafiee, S.; Tahir, I.S.A.; et al. Global wheat head detection 2021: An improved dataset for benchmarking wheat head detection methods. Plant Phenomics 2021, 2021, 9846158. [Google Scholar] [CrossRef] [PubMed]
- Koziarski, M.; Cyganek, B. Impact of low resolution on image recognition with deep neural networks: An experimental study. Int. J. Appl. Math. Comput. Sci. 2018, 28, 735–744. [Google Scholar] [CrossRef]
- Luke, J.; Joseph, R.; Balaji, M. Impact of image size on accuracy and generalization of convolutional neural networks. Int. J. Res. Anal. Rev. (IJRAR) 2019, 6, 70–80. [Google Scholar]
- Sabottke, C.F.; Spieler, B.M. The effect of image resolution on deep learning in radiography. Radiol. Artif. Intell. 2020, 2, e190015. [Google Scholar] [CrossRef]
- Shermeyer, J.; Van Etten, A. The effects of super-resolution on object detection performance in satellite imagery. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Long Beach, CA, USA, 16–20 June 2019. [Google Scholar]
- Bashir Syed, M.A.; Wang, Y.; Khan, M.; Niu, Y. A comprehensive review of deep learning-based single image super-resolution. PeerJ Comput. Sci. 2021, 7, e621. [Google Scholar] [CrossRef]
- Gendy, G.; He, G.; Sabor, N. Lightweight image super-resolution based on deep learning: State-of-the-art and future directions. Inf. Fusion 2023, 94, 284–310. [Google Scholar] [CrossRef]
- Lepcha, D.C.; Goyal, B.; Dogra, A.; Goyal, V. Image super-resolution: A comprehensive review, recent trends, challenges and applications. Inf. Fusion 2023, 91, 230–260. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, J.; Hoi, S.C.H. Deep learning for image super-resolution: A survey. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 43, 3365–3387. [Google Scholar] [CrossRef]
- Yamamoto, K.; Togami, T.; Yamaguchi, N. Super-Resolution of Plant Disease Images for the Acceleration of Image-based Phenotyping and Vigor Diagnosis in Agriculture. Sensors 2017, 17, 2557. [Google Scholar] [CrossRef] [PubMed]
- Ledig, C.; Theis, L.; Huszár, F.; Caballero, J.; Cunningham, A.; Acosta, A.; Aitken, A.; Tejani, A.; Totz, J.; Wang, Z.; et al. Photo-realistic single image super-resolution using a generative adversarial network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4681–4690. [Google Scholar]
- Maqsood, M.H.; Mumtaz, R.; Haq, I.U.; Shafi, U.; Zaidi, S.; Hafeez, M. Super resolution generative adversarial network (Srgans) for wheat stripe rust classification. Sensors 2021, 21, 7903. [Google Scholar] [CrossRef] [PubMed]
- Cap, Q.H.; Tani, H.; Kagiwada, S.; Uga, H.; Iyatomi, H. LASSR: Effective super-resolution method for plant disease diagnosis. Comput. Electron. Agric. 2021, 187, 106271. [Google Scholar] [CrossRef]
- Albert, P.; Saadeldin, M.; Narayanan, B.; Fernandez, J.; Mac Namee, B.; Hennessey, D.; O’Connor, N.E.; McGuinness, K. Unsupervised domain adaptation and super resolution on drone images for autonomous dry herbage biomass estimation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 1636–1646. [Google Scholar]
- Martin, D.; Fowlkes, C.; Tal, D.; Malik, J. A database of human segmented natural images and its application to evaluating segmentation algorithms and measuring ecological statistics. In Proceedings of the Eighth IEEE International Conference on Computer Vision, Vancouver, BC, Canada, 7–14 July 2001; Volume 2, pp. 416–423. [Google Scholar]
- Timofte, R.; Agustsson, E.; Gool, L.; Yang, M.H.; Zhang, L.; Lim, B.; Son, S.; Kim, H.; Nah, S.; Lee, K.M.; et al. Ntire 2017 challenge on single image super-resolution: Methods and results. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Honolulu, HI, USA, 21–26 July 2017; pp. 114–125. [Google Scholar]
- Li, Z.; Liu, Y.; Wang, X.; Liu, X.; Zhang, B.; Liu, J. Blueprint separable residual network for efficient image super-resolution. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 833–843. [Google Scholar]
- Zhang, Y.; Tian, Y.; Kong, Y.; Zhong, B.; Fu, Y. Residual dense network for image super-resolution. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 2472–2481. [Google Scholar]
- Zechmeister, H.G.; Grodzińska, K.; Szarek-Łukaszewska, G. Bryophytes. In Trace Metals and Other Contaminants in the Environment; Elsevier: Amsterdam, The Netherlands, 2003; Volume 6, pp. 329–375. [Google Scholar]
- Smith, A.R.; Pryer, K.M.; Schuettpelz, E.; Korall, P.; Schneider, H.; Wolf, P.G. A classification for extant ferns. Taxon 2006, 55, 705–731. [Google Scholar] [CrossRef]
- Hutchinson, K.R.S.; House, H. The Morphology of Gymnosperms; Scientific Publishers: New York, NY, USA, 2015. [Google Scholar]
- Bahadur, B.; Rajam, M.V.; Sahijram, L.; Krishnamurthy, K.V. Angiosperms: An overview. In Plant Biology and Biotechnology: Volume I: Plant Diversity, Organization, Function and Improvement; Springer: New Delhi, India, 2015; pp. 361–383. [Google Scholar]
- POWO. Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. 2024. Available online: http://www.plantsoftheworldonline.org/ (accessed on 11 November 2023).
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Zhao, H.; Gallo, O.; Frosio, I.; Kautz, J. Loss functions for image restoration with neural networks. IEEE Trans. Comput. Imaging 2016, 3, 47–57. [Google Scholar] [CrossRef]
- Wang, C.Y.; Bochkovskiy, A.; Liao, H.Y.M. YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 7464–7475. [Google Scholar]
- Song, Q.; Wang, C.; Jiang, Z.; Wang, Y.; Tai, Y.; Wang, C.; Li, J.; Huang, F.; Wu, Y. Rethinking counting and localization in crowds: A purely point-based framework. In Proceedings of the IEEE International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 3365–3374. [Google Scholar]
- Zhao, J.; Kaga, A.; Yamada, T.; Komatsu, K.; Hirata, K.; Kikuchi, A.; Hirafuji, M.; Ninomiya, S.; Guo, W. Improved field-based soybean seed counting and localization with feature level considered. Plant Phenomics 2023, 5, 0026. [Google Scholar] [CrossRef]
- Pharr, M.; Jakob, W.; Humphreys, G. Physically Based Rendering: From Theory to Implementation; Morgan Kaufmann: Burlington, MA, USA, 2016. [Google Scholar]
- P2PNet-Soy Project. Available online: https://github.com/UTokyo-FieldPhenomics-Lab/P2PNet-Soy (accessed on 21 November 2023).
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Loy, C.C.; He, K.; Tang, X. Image super-resolution using deep convolutional networks. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 38, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, J.K.; Lee, K.M. Accurate image super-resolution using very deep convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 1646–1654. [Google Scholar]
- Lim, B.; Son, S.; Kim, H.; Nah, S.; Lee, K.M. Enhanced deep residual networks for single image super-resolution. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Honolulu, HI, USA, 21–26 July 2017; pp. 136–144. [Google Scholar]
- Zhang, Y.; Li, K.; Li, K.; Wang, L.; Zhong, B.; Fu, Y. Image super-resolution using very deep residual channel attention networks. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 286–301. [Google Scholar]
- Liang, J.; Cao, J.; Sun, G.; Zhang, K.; Gool, L.V.; Timofte, R. Swinir: Image restoration using swin transformer. In Proceedings of the IEEE International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 1833–1844. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B.; Wei, Y. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Advances in Neural Information Processing Systems; 2017; Volume 30, Available online: https://proceedings.neurips.cc/paper_files/paper/2017/hash/3f5ee243547dee91fbd053c1c4a845aa-Abstract.html (accessed on 13 January 2024).
- Bi, J.; Zhu, Z.; Meng, Q. Transformer in computer vision. In Proceedings of the 2021 IEEE International Conference on Computer Science, Electronic Information Engineering and Intelligent Control Technology (CEI), Fuzhou, China, 24–26 September 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 178–188. [Google Scholar]
- Csurka, G. Domain adaptation for visual applications: A comprehensive survey. arXiv 2017, arXiv:1702.05374. [Google Scholar]
- Figshare. Dataset. PlantSR Dataset. Available online: https://doi.org/10.6084/m9.figshare.24648150.v1 (accessed on 13 January 2024).
- Figshare. Dataset. HR_Soybean. Available online: https://doi.org/10.6084/m9.figshare.24994253.v1 (accessed on 28 November 2023).


| Method | Scale | PSNR | SSIM | Params (M) | FPS | Input Size (Pixels) |
|---|---|---|---|---|---|---|
| Bicubic | ×2 | 38.23 | 0.9617 | / | / | 64 |
| SRCNN | 39.42 | 0.9667 | 0.069 | 507.0 | ||
| VDSR | 40.31 | 0.9716 | 0.667 | 231.3 | ||
| EDSR | 40.26 | 0.9706 | 1.370 | 123.7 | ||
| RCAN | 40.15 | 0.9708 | 15.445 | 6.6 | ||
| SwinIR | 40.34 | 0.9715 | 16.619 | 0.8 | ||
| PlantSR (Ours) | 40.36 | 0.9716 | 1.397 | 37.1 | ||
| Bicubic | ×3 | 33.56 | 0.9128 | / | / | 63 |
| SRCNN | 34.24 | 0.9190 | 0.069 | 475.4 | ||
| VDSR | 34.79 | 0.9255 | 0.667 | 235.3 | ||
| EDSR | 35.20 | 0.9273 | 1.554 | 71.9 | ||
| RCAN | 35.24 | 0.9290 | 15.629 | 6.6 | ||
| SwinIR | 35.23 | 0.9290 | 16.803 | 0.8 | ||
| PlantSR (Ours) | 35.26 | 0.9291 | 5.760 | 18.1 | ||
| Bicubic | ×4 | 32.13 | 0.8844 | / | / | 64 |
| SRCNN | 32.64 | 0.8927 | 0.069 | 473.2 | ||
| VDSR | 33.53 | 0.9031 | 0.667 | 236.6 | ||
| EDSR | 33.76 | 0.9063 | 1.518 | 57.5 | ||
| RCAN | 33.78 | 0.9071 | 15.888 | 6.5 | ||
| SwinIR | 33.83 | 0.9075 | 16.766 | 0.7 | ||
| PlantSR (Ours) | 33.85 | 0.9077 | 13.531 | 9.8 |
| Case | Preprocessing | MAE | RMSE |
|---|---|---|---|
| Case 1 | No | 19.16 | 21.49 |
| Case 2 | Downsample test images | 59.23 | 61.47 |
| Case 3 | Downsample test images and then upscale them using a PlantSR (×2) model | 19.82 | 22.91 |
| Case 4 | Upscale all the images | 15.09 | 22.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, T.; Yu, Q.; Zhong, Y.; Shao, M. PlantSR: Super-Resolution Improves Object Detection in Plant Images. J. Imaging 2024, 10, 137. https://doi.org/10.3390/jimaging10060137
Jiang T, Yu Q, Zhong Y, Shao M. PlantSR: Super-Resolution Improves Object Detection in Plant Images. Journal of Imaging. 2024; 10(6):137. https://doi.org/10.3390/jimaging10060137
Chicago/Turabian StyleJiang, Tianyou, Qun Yu, Yang Zhong, and Mingshun Shao. 2024. "PlantSR: Super-Resolution Improves Object Detection in Plant Images" Journal of Imaging 10, no. 6: 137. https://doi.org/10.3390/jimaging10060137
APA StyleJiang, T., Yu, Q., Zhong, Y., & Shao, M. (2024). PlantSR: Super-Resolution Improves Object Detection in Plant Images. Journal of Imaging, 10(6), 137. https://doi.org/10.3390/jimaging10060137






