Abstract
The application of gold nanoparticles (AuNPs) is emerging in many fields, raising the need for a systematic investigation on their safety. In particular, for biomedical purposes, a relevant issue are certainly AuNP interactions with biomolecules, among which proteins are the most abundant ones. Elucidating the effects of those interactions on protein structure and on nanoparticle stability is a major task towards understanding their mechanisms at a molecular level. We investigated the interaction of the 3-mercaptopropionic acid coating of AuNPs (MPA-AuNPs) with β2-microglobulin (β2m), which is a paradigmatic amyloidogenic protein. To this aim, we prepared and characterized MPA-AuNPs with an average diameter of 3.6 nm and we employed NMR spectroscopy and fluorescence spectroscopy to probe protein structure perturbations. We found that β2m interacts with MPA-AuNPs through a highly localized patch maintaining its overall native structure with minor conformational changes. The interaction causes the reversible precipitation of clusters that can be easily re-dispersed through brief sonication.
1. Introduction
Proteins play a fundamental role in biological processes. Their activity, indeed, on one hand supports the correct operation of an organism, but on the other, could be responsible for disease onset. Many protein functions are affected by their interaction with other molecules e.g., other proteins, oligonucleotides, hormones, and so on. The fact that the interaction profile can highlight the functions a protein performs implies that understanding the behaviour of a protein at the molecular level is a valuable strategy to get deep insights into the functional role and possibly into the design of new tools to master the protein activity. The spreading application of nanomaterials in different fields such as biomedicine, food, environmental, and material sciences [1,2], has stressed the relevance of understanding at a molecular level protein-nanoparticle interaction, because any contact of nanomaterials with a biological fluid is suddenly followed by the adsorption of proteins [3]. However, the challenging investigation of protein-nanomaterial interface also proved so intriguing that no general trends could be drawn so far. For example, it has been reported that some enzymes, e.g., lysozyme, chymotrypsin, and fibrinogen [4,5], lose their catalytic activity upon interacting with gold nanoparticles (AuNPs), other ones, e.g., pepsin [6], retain their functionality and some other ones, e.g., bovine catalase [7], show a higher stability in harsh conditions in presence of AuNPs. On the other hand, when Aβ peptide, the amyloidogenic fragment from Amyloid Precursor Protein (APP), whose fibril deposition has been related to Alzheimer’s disease onset [8], was incubated with different nanoparticles (NPs), various effects on fibrillogenesis were observed. While titanium oxide NPs promote Aβ aggregation [9], silica NPs leave it unaffected [9], and fullerene even inhibited it [10]. Thus, the interaction between proteins and nanomaterials and the ensuing effects appear to be highly dependent both on the specific protein and on the nanomaterial physico-chemical properties.
Here, we present a NMR and fluorescence-based study of the interaction between 3-mercaptopropionic acid-coated AuNPs (MPA-AuNPs) and β2-microglobulin (β2m).
Gold nanoparticles have been widely used in biomedical research because of their optical features, large surface to volume ratio, gold inertness, ease of production, and surface functionalization [11]. MPA-AuNPs were synthesized through a one-phase direct synthesis that proceeds through three steps [12].
β2m is an amyloidogenic protein responsible for dialysis related amyloidosis (DRA) [13], and it is considered a model for amyloidogenic proteins since it recapitulates the typical features of this class of proteins. β2m is the light chain of class I major histocompatibility complex and in healthy organisms, after its detachment from the heavy chain, it is removed from the blood through the kidneys. In patients that are affected by chronic renal failure, it accumulates in the blood and precipitates into amyloid deposits in correspondence of the joints. In literature, the interaction between β2m and citrate-stabilized AuNPs (Cit-AuNPs) was reported for both the wild-type and D76N variants [14,15]. In both of the cases, a labile interaction was observed that does not affect the overall folding and is mainly located at the N-terminal apical part of the protein structure. Furthermore, it was shown that Cit-AuNPs are able to partially hinder the fibrillogenesis of the most amyloidogenic variant, namely D76N β2m [15].
2. Results
2.1. AuNP Synthesis and Characterization
To synthesize MPA-AuNPs, a reported procedure was employed [12] (Figure 1). Briefly, at the beginning, AuIII is reduced to AuI by the thiols, and then AuI forms with the thiolate polymeric structures ([AuISR]n), and finally gold is further reduced to Au0 by NaBH4, leading to gold clusters that are stabilized by the covalently bound thiolate organic monolayer. When compared to the reference recipe [12], the ligand/Au ratio was changed from 3 to 9 in order to obtain smaller AuNPs.
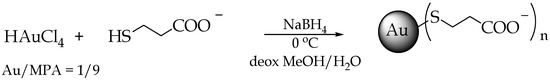
Figure 1.
Scheme representing 3-mercaptopropionic acid-coated AuNPs (MPA-AuNP) synthesis.
From transmission electron microscopy (TEM) images, an average diameter of 3.6 nm was determined (Figure 2a). The small dimensions of these AuNPs are consistent with the weak SPB that is recorded in the UV-Vis spectrum (Figure 2b). MPA-AuNPs can be centrifuged, dried, and dispersed again without any aggregation. To evaluate the NP organic percentage content originating from MPA, thermogravimetric analysis (TGA) analysis was performed (Figure 2c). From TGA results and the NP diameter obtained from TEM, it was possible to estimate the average composition and the molecular weight of MPA-AuNPs (Table 1).
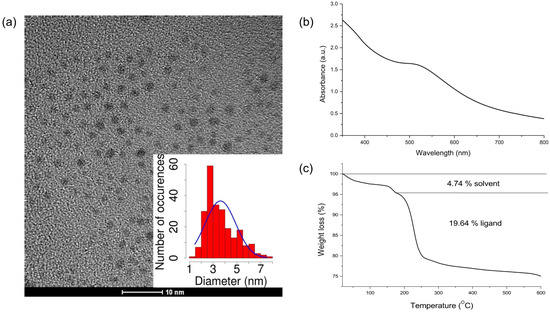
Figure 2.
Characterization of synthesized MPA-AuNPs: (a) transmission electron microscopy (TEM) micrograph of MPA-AuNPs along with the corresponding size histogram; (b) UV-Vis spectrum of MPA-AuNPs; and, (c) thermogravimetric analysis (TGA) analysis of MPA-AuNPs.

Table 1.
Composition of MPA-AuNPs calculated from TEM and TGA.
2.2. Protein-AuNP Interaction
When MPA-AuNPs and β2m were mixed together, and within a few hours a brown precipitate developed on the bottom of the flask. This precipitate could be easily re-dispersed by sonication and dropped onto a TEM grid for imaging. TEM micrographs (Figure 3) showed a well dispersed nanoparticle sample in which the average size of the NP cores was the same as the control (Figure 2a).
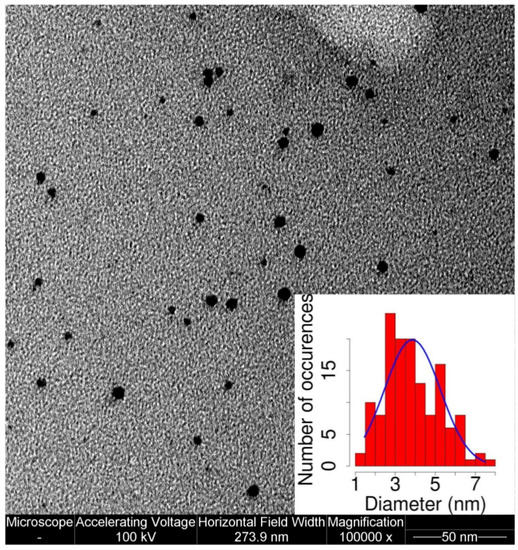
Figure 3.
TEM micrograph of 2.5 µM MPA-AuNPs and 25 µM β2m. The average NP diameter measured in presence of the protein was 3.8 ± 1.3 nm.
By doing simple geometrical considerations, it is possible to estimate the protein adsorption capacity of a single nanoparticle. If the ratio between the volumes is considered, as proposed by Calzolai et al. [16], it is possible to calculate the number of proteins () per nanoparticle applying the following equation:
where corresponds to the sum of the NP radius and the diameter of the protein, is the radius of the nanoparticle, and is the protein radius. However, this equation is suited for spherical proteins, while β2m has an oblate three-dimensional structure with longitudinal and transverse axes of 4.3–3.8 and 2.5–2.0 nm, respectively [17]. When considering the crystallographic cylindrical shape of β2m, the Equation (1) becomes
where is given by the sum of the NP radius and the height of the β2m cylindroid, is the radius of the nanoparticle, and and are the height and the base radius of β2m cylindroid. The NP radius is given by the addition of the alkanethiolate monolayer thickness to the radius of the gold core obtained from TEM. When considering that MPA is 0.55 nm long, approximately, we can assume that the monolayer protected cluster has a diameter of 4.7 nm. Following Equation (2), on the surface of a spherical MPA-AuNP with average diameter of 4.7 nm 36–50 β2m monomers can be accommodated. The maximum packing density factor that is used in this model is referred to spherical proteins. Since the base of β2m cylindroid is a half with respect to its height, substituting symmetric tetramers to cylindroid monomers can improve the geometrical model. This adjustment leads to 9–12 tetramers per NP that means 36–48 monomers. If the ratio between the NP surface area and the cylindroid base is considered, then the number of protein monomers per NP is reduced to 14–22. From all of these considerations, beyond any critical evaluation, the number of β2m molecules that can be adsorbed on a MPA-AuNP goes from 14 to 50, approximately.
Two-dimensional 1H 15N NMR experiments were acquired to ascertain the state of the protein in presence of MPA-AuNPs. Five different protein/NP ratios were examined going from 100 to 10. SOFAST HMQC [18] spectra of β2m alone and in the presence of MPA-AuNPs at protein/NP ratios of 40 and 15 are shown in Figure 4.
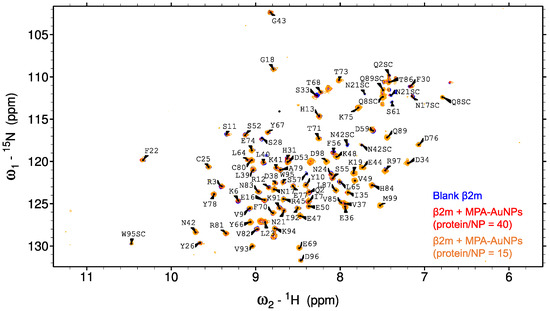
Figure 4.
Superimposition of β2m 1H-15N SOFAST-HMQC spectra in absence of MPA-AuNPs (blue) and in presence of MPA-AuNPs in protein/NP ratio of 40 (red) and of 15 (orange). The label SC indicates side chain NH’s.
By analyzing the spectra intensity, in addition to an overall attenuation reflecting the increase of protein recruitment by the increasing number of available interaction sites on NPs, there was also a preferential intensity decrease (Figure 5a). Moreover, the gradual intensity attenuation was associated with progressive chemical shift variation (Figure 5b). By plotting for each residue, the relative intensity (RI) against the chemical shift perturbation (Δδ), recorded at protein/NP = 25, i.e., the lowest ratio at which most of the peaks were still visible (Figure 5c), it can be seen that there is a quite good correlation between the two variations, i.e., the amino acids that showed the largest chemical shift deviation were usually also characterized by the lowest relative intensity. This suggests that both of the perturbations arose from the same process, namely the exchange interaction with the nanoparticles.
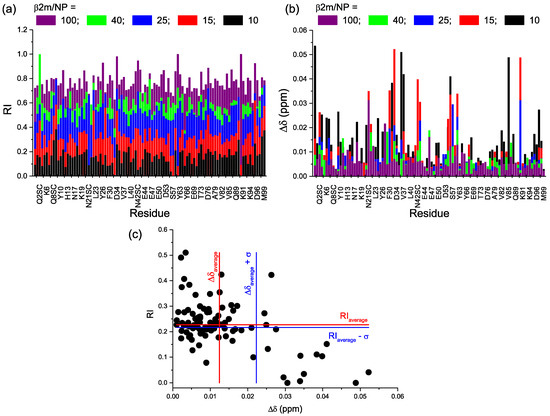
Figure 5.
(a,b) Bar plots of amide resonance chemical shift perturbations (Δδ) and cross-peak attenuations (RI), respectively. Refer to color legend for the protein/NP ratios; (c) Scatter plot indicating the correlation between chemical shift deviation (Δδ) and relative intensity (RI) attenuation, each point corresponds to an amide resonance. Tick marks and labels are reported every 3 residues, except for the missing and undetected ones, i.e., I1, P5, P14, G29, P32, L54, K58, D59, W60, F62, P72, S88, and P90. The observed side chain (SC) NHs are also included, i.e., two for Q2, two for Q8, one for N17, two for N21, two for N24, two for Q89, and one for W95.
The variation of the resonance position accounts for a change in the chemical environment around a specific amide group and the accompanying signal intensity decrease reflects the line broadening, i.e., transverse relaxation rate increase, from additional exchange and possibly cross-relaxation contributions. These two features are both related to the exchange process between free and NP-contacting states. The occurrence of single resonances whose position reflects the population-weighted chemical shift average of the free and the NP-close forms of the protein implies that the exchange regime is not slow, nor intermediate with respect to the NMR chemical shift scale. The observed pattern is consistent with a nearly fast exchange regime with residual line-broadening contributions that, besides the dissociation rate from the NP adduct, may also reflect more or less transient changes in the local dynamics and the overall rotational tumbling rate that affect both dipolar (DD) and chemical shift anisotropy (CSA) contributions to relaxation, and therefore, the linewidth. The possibility of a progressively slowing exchange rate on increasing the NP concentration that would approach the intermediate exchange regime and stress further the signal attenuation could account for the substantial intensity reductions that were observed at the lowest protein/NP ratios.
To find the residues that proved more affected by the presence of MPA-AuNPs, the amide signals whose RI and Δδ was displaced more than one standard deviation from the average values were identified (Figure 6a). By mapping their positions on the protein three-dimensional structure (Figure 6b), a highly specific and localized region was found to be involved in the interaction. The patch includes two loops, namely BC loop and DE loop, and the spatially close N-terminal tail.
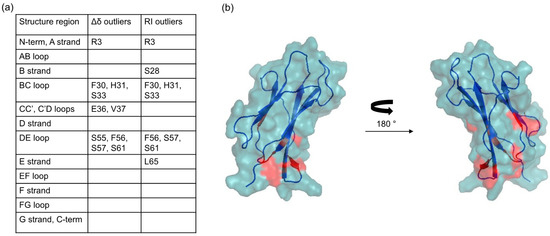
Figure 6.
(a) Synopsis of β2m positions that proved most affected, i.e., displaced more than one standard deviation from the average, by the presence of MPA-AuNP and their secondary structure element location; (b) β2m cartoon highlighting in red the locations of the outlier residues.
This result is consistent with a strong localization of the electrostatic interaction, probably being due to the compactness of the NP electron plasma [14,15]. The character of the interaction surface, as described in Figure 6, can be appreciated when compared to the β2m regions that are involved in the contacts with the heavy chain of type I major histocompatibility complex (MHC-I) [19]. In this complex, β2m establishes the closest contacts with the partner species through a hydrophobic area that extends over strands B (fragment 23–27) and E (fragment 62–66). The interface is also comprised of contacts with hydrophilic stretches at strand A (fragment 8–12), strand D (fragment 51–55) and loop DE (fragment 57–61) that appear less tightly packed in the complex, and are thus more accessible to the solvent. The non-uniform distribution of hydrophilic and hydrophobic patches on β2m surface that is recognized in the quaternary organization of MHC-I is probably relevant for the amyloidogenic propensity of the protein [20]. The regions that are highlighted in Figure 6b, however, appear distinctly different from those involved in the typical hydrophobic-driven interaction that β2m engages in MHC-I. It was not possible to fit the chemical shift perturbation data with a binding isotherm because the signals that showed the highest deviations were also the ones undergoing extensive attenuation up to the complete cancellation in the first titration points.
To further investigate the NP effect on protein conformation, fluorescence experiments were performed. β2m intrinsic fluorescence is mainly due to a buried Trp residue, namely Trp95 [21]. The second Trp residue of the molecule, Trp60, contributes only marginally (20% approximatively) to the overall fluorescence because of its exposure on the protein surface and the consequent quenching effect of the solvent. The β2m intrinsic fluorescence was recorded upon the progressive addition of MPA-AuNPs. After an initial decrease (around 20%), the fluorescence intensity increased and the emission peak shifted (Figure 7a). The initial intensity decrease is likely to be due to the quenching of the limited Trp60 emission contribution to the whole fluorescence. Since Trp60 is exposed on the surface, it is accessible to nanoparticle direct contact. The fluorescence of W60G variant is, indeed, approximately 20% lower than the value of wild-type protein.
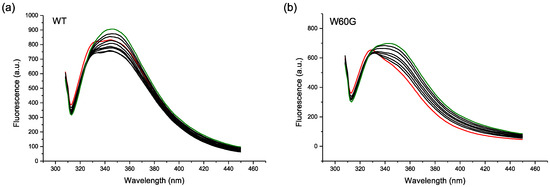
Figure 7.
(a,b) Fluorescence quenching of 0.5 µM wild-type (WT) and W60G β2m, respectively, with MPA-AuNP concentrations ranging from 0 to 50 nM. The control sample spectrum is shown in red, while the last titration point in green. Each titration point corresponds to a NP increase of 5 or 7 nM, approximately, for the experiments done with WT or W60G, respectively. For WT, the first two additions caused an intensity decrease and the following ones a progressive increase. For W60G, all of the consecutive additions led to progressive intensity increase.
When considering only the data prior to the shift of the fluorescence emission frequency that certainly pertain to the natively folded β2m species, the bimolecular quenching constant (kq) values, calculated from the apparent Stern-Volmer constant (Table 2), assuming a fluorescence lifetime of 1–10 ns for the indole chromophore [22], were higher than the collisional rate limit, i.e., 2 × 1010 M−1·s−1. Hence, it can be concluded that the quenching of the external tryptophan is not purely collisional.

Table 2.
Parameters obtained from fluorescence quenching data fitted with Stern-Volmer equation [23]. Only the initial points of the titration in which the fluorescence decreases increasing the titrant concentration were used for the Stern-Volmer fitting.
After Trp60 complete quenching, the effect of the actual NP interaction becomes evident. Accordingly, the protein conformation perturbation induced by MPA-AuNPs leads to a Trp95 fluorescence intensity increase and an emission spectrum shift. This interpretation is supported by the result observed with W60G β2m variant that exhibited only the intensity increase and the shift of the emission band (Figure 7b).
3. Discussion
The interaction of β2m and its variants with AuNPs has been thoroughly investigated by our group [14,15]. We found that the actual interaction between the citrate-coated surface of AuNPs and β2m is essentially of electrostatic nature, although the overall protein charge should be around zero or slightly negative. The protein regions that are involved in the preferential contacts with AuNPs possess a local dipole or polarity distribution that must favor pairing with the NP surface, irrespective of the overall net charge and dipole moment. An overall weak interaction was observed that involved the N-terminal apical part of the protein, in particular, Q2 and R3 in the N-terminus, and K58 and D59 in the DE loop, in good agreement the simulations results [14,15]. In addition, we could also identify other close interaction sites, namely S55 and F56 in DE loop, and residues Y26, S28, G29, F30, and S33 of the adjacent BC loop. Additional residues appeared selectively perturbed when the very amyloidogenic variant D76N β2m was assayed [15]. These additional involvements did not map the protein-NP interface, but rather the protein-protein association equilibria. The citrate coated AuNPs that were employed had average diameters of 7.5 nm at most and we could even test the in vitro amyloidogenesis inhibition of those NP preparations [15]. Larger AuNPs, as checked with thiol-coated ones (unpublished results), do not support β2m stability towards unfolding as the protein precipitates out of solution shortly after preparation. In general, large NPs had been previously shown to rather accelerate β2m fibrillogenesis [24]. For AuNPs, this effect stems from the well-established affinity increase of proteins for gold surfaces with small curvature, i.e., large sphere diameters, that enhances the contacts, thereby destabilizing the protein folding [25].
Now, we have shown that the same regions of β2m, which establish contacts with citrate-coated AuNPs, which support the conformational stability of the protein and even inhibit fibrillogenesis in vitro, remain involved in the interaction also when the citrate is replaced by 3-mercaptopropionate. With this coating, however, the NP dimensions are sensibly reduced. With an average diameter of 3.6 nm, MPA-AuNPs become much more easy to handle for any preparation, keep the protein stable in solution while engaging an efficient fast-exchange mild interaction, and can be exploited to reach larger NP/protein concentration ratios that may be necessary to exploit the efficiency and the mildness of the interaction in vivo.
4. Materials and Methods
4.1. MPA-AuNP Synthesis and Characterization
All the reagents used in the synthesis were purchased from Sigma Aldrich (St. Louis, MO, USA) To a solution of HAuCl4·3H2O (49.2 mg, 0.125 mmol) in deoxygenated methanol (5 mL) cooled at 0 °C and purged with nitrogen, three equivalents (65.3 µL) of 3-mercaptopropionic acid, dissolved in deoxygenated water (5 mL), were added under stirring. Upon the addition of the alkanethiol, the solution colour changed from yellow to cloudy white. After two hours of stirring, a freshly prepared cooled aqueous solution of NaBH4 (1.25 mmol, 47.3 mg, in 2 mL of water) was dripped inside the gold/thiol solution. One hour later, the brown solution that was obtained was purified by centrifugation (5000 rpm, 15 min at 25 °C), repeating the removal of the supernatant and the dispersion in methanol five times.
For the UV-Vis characterization, a V-750 spectrophotometer (Jasco, Oklahoma City, OK, USA) was used and the spectra were recorded at 20 °C, from 400 nm to 800 nm with a data pitch of 0.2 nm, a scan rate of 200 nm/min and a bandwidth of 2 nm. To prepare the samples for the TEM imaging, a small amount of the nanoparticle solution was dropped on a TEM grid and left for 5 min. Filter paper was used to remove the excess of the solution. TEM images were acquired with EM 208 microscope (Philips, Amsterdam, The Netherlands). The size distribution was calculated by measuring a minimum of 200 particles using ImageJ software (National Institutes of Health, Bethesda, MD, USA). The average number of gold atoms per NP (NAu) was calculated from the following equation:
where d is the nanoparticle diameter expressed in nm, ρ is the density for face-centered cubic gold (19.3 g/cm3) and MAu stands for the atomic weight of gold (197 g/mol).
NAu = πρd3/6MAu = 30.89602d3,
The number of ligands that are bound to the gold core was estimated from thermogravimetric analysis (TGA, collected with TGA 8000, Perkin Elmer, Waltham, MA, USA). This experiment gives the percentage of weight loss during a temperature ramp that is addressed exclusively to the burning of the organic component, following solvent removal. Applying the following equation, it is possible to calculate the monolayer composition:
where NL is the number of ligands, W% is the percentage of weight loss due to organic ligands burning and Mthiolate is the molecular weight of the thiolate molecule. TGA analysis was performed with a SDT Q600 instrument (TA instruments, New Castle, DE, USA) under N2 at a heating rate of 10 °C/min going from 0 to 600 °C.
NL = (NAu·MAu·W%)/((1 − W%) Mthiolate),
4.2. NMR Experiments
NMR experiments were performed on uniformly 15N-labeled β2m wild-type dissolved in HEPES 50 mM pH 7, and diluted to 25 µM with different concentrations of MPA-AuNPs or 2 mM MPA for the control sample. Proteins were expressed and purified, as previously described [14,15]. D2O (5%) was added to each sample for lock purposes. 15N-1H SOFAST-HMQC experiments were collected on the Bruker Avance spectrometer (Bruker, Billerica, MA, USA) (-) at the Udine University Biophysics Laboratory, operating at 500 MHz (1H). Experiments were run at 298 K over spectral widths of 30 ppm (15N, t1) and 14 ppm (1H, t2) with 128 and 1024 points, respectively. For each t1 dimension point, 800 or 1600 scans were accumulated. The data were processed with Topspin 2.1 and were analyzed with Sparky. The β2m assignment was based on the file that was deposited on the Biological Magnetic Resonance Data Bank (Accession Code: 17165). Chemical shift perturbations were calculated as Δδ (ppm) = [(ΔδH)2 + (ΔδN/6.5)2]1/2, where ΔδH and ΔδN are the chemical shift variations for 1H and 15N, respectively [26,27]. The relative intensities (RI) correspond to the ratio between the signal intensity in presence of NPs and in absence of NPs.
4.3. TEM Imaging of Stained Samples
To prepare samples for the imaging, a small amount of the nanoparticle-β2m solution was dropped on a TEM grid and left for 5 min. Filter paper was used to remove the excess of the solution. The solution was stained with 2% uranyl acetate solution in water for 2 min.
4.4. Fluorescence Experiments
β2m intrinsic fluorescence was recorded in absence of AuNPs and after the progressive addition of small amounts of nanoparticles using a Cary Eclipse Fluorescence Spectrophotometer (Agilent, Santa Clara, CA, USA). For the measurement, fluorescence semi-micro cuvettes were used (5 mm × 5 mm). The samples were excited at 295 nm and the emission was recorded from 300 to 450 nm, using 5 nm slit for both excitation and emission. Each spectrum was the average of 5 consecutive measurements, and three individual experiments were repeated for each sample. The initial quenching data were fitted with the linear Stern-Volmer equation [23]:
where and are the fluorescence intensities of the protein in absence and in presence of Cit-AuNPs, respectively, and the Stern-Volmer constant, KSV, is the product of the diffusion-limited bimolecular quenching constant, kq, and the fluorophore fluorescence lifetime, τ0.
Acknowledgments
This work received financial support from PRIN project No. 2012A7LMS3. We acknowledge the New York University Abu Dhabi for access to the Core Technology Platform. We also thank Makek A. for the assistance.
Author Contributions
C.C. and G.E. conceived and designed the experiments; C.C. and P.B. performed the experiments; C.C. and G.E. analyzed the data; S.G., P.P.M. and V.B. expressed and purified the proteins; C.C. and G.E. wrote the draft. All authors read, corrected and discussed the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- De, M.; Ghosh, P.S.; Rotello, V.M. Applications of nanoparticles in biology. Adv. Mater. 2008, 20, 4225–4241. [Google Scholar] [CrossRef]
- Liu, W.-T. Nanoparticles and their biological and environmental applications. J. Biosci. Bioeng. 2006, 102, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lynch, I.; Dawson, K.A. Protein-nanoparticle interactions. Nano Today 2008, 3, 40–47. [Google Scholar] [CrossRef]
- Gagner, J.E.; Lopez, M.D.; Dordick, J.S.; Siegel, R.W. Effect of gold nanoparticle morphology on adsorbed protein structure and function. Biomaterials 2011, 32, 7241–7252. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.J.; Liang, M.; Monteiro, M.; Toth, I.; Minchin, R.F. Nanoparticle-induced unfolding of fibrinogen promotes Mac-1 receptor activation and inflammation. Nat. Nanotechnol. 2011, 6, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Gole, A.; Dash, C.; Ramakrishnan, V.; Sainkar, S.R.; Mandale, A.B.; Rao, M.; Sastry, M. Pepsin-gold colloid conjugates: Preparation, characterization, and enzymatic activity. Langmuir 2001, 17, 1674–1679. [Google Scholar] [CrossRef]
- Bailes, J.; Gazi, S.; Ivanova, R.; Soloviev, M. Effect of gold nanoparticle conjugation on the activity and stability of functional proteins. In Nanoparticles in Biology and Medicine; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2012; pp. 89–99. ISBN 978-1-61779-952-5. [Google Scholar]
- Murphy, M.P.; LeVine, H. Alzheimer’s disease and the β-amyloid peptide. J. Alzheimers Dis. 2010, 19, 311. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Sun, X.; Yu, Y.; Hu, J.; Zhao, L.; Liu, Q.; Zhao, Y.; Li, Y. TiO2 nanoparticles promote β-amyloid fibrillation in vitro. Biochem. Biophys. Res. Commun. 2008, 373, 315–318. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Lee, M. Fullerene inhibits β-amyloid peptide aggregation. Biochem. Biophys. Res. Commun. 2003, 303, 576–579. [Google Scholar] [CrossRef]
- Giljohann, D.A.; Seferos, D.S.; Daniel, W.L.; Massich, M.D.; Patel, P.C.; Mirkin, C.A. Gold nanoparticles for biology and medicine. Angew. Chem. Int. Ed. 2010, 49, 3280–3294. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, L.; Cai, W. Size-tunable synthesis of monodisperse water-soluble gold nanoparticles with high X-ray attenuation. Chem. Eur. J. 2010, 16, 1459–1463. [Google Scholar] [CrossRef] [PubMed]
- Gejyo, F.; Yamada, T.; Odani, S.; Nakagawa, Y.; Arakawa, M.; Kunitomo, T.; Kataoka, H.; Suzuki, M.; Hirasawa, Y.; Shirahama, T.; et al. A new form of amyloid protein associated with chronic hemodialysis was identified as β2-microglobulin. Biochem. Biophys. Res. Commun. 1985, 129, 701–706. [Google Scholar] [CrossRef]
- Brancolini, G.; Corazza, A.; Vuano, M.; Fogolari, F.; Mimmi, M.C.; Bellotti, V.; Stoppini, M.; Corni, S.; Esposito, G. Probing the influence of citrate-capped gold nanoparticles on an amyloidogenic protein. ACS Nano 2015, 9, 2600–2613. [Google Scholar] [CrossRef] [PubMed]
- Cantarutti, C.; Raimondi, S.; Brancolini, G.; Corazza, A.; Giorgetti, S.; Ballico, M.; Zanini, S.; Palmisano, G.; Bertoncin, P.; Marchese, L.; et al. Citrate-stabilized gold nanoparticles hinder fibrillogenesis of a pathological variant of β2-microglobulin. Nanoscale 2017, 9, 3941–3951. [Google Scholar] [CrossRef] [PubMed]
- Calzolai, L.; Franchini, F.; Gilliland, D.; Rossi, F. Protein−Nanoparticle interaction: Identification of the Ubiquitin−Gold nanoparticle interaction site. Nano Lett. 2010, 10, 3101–3105. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.W.; Reeke, G.N. Three-dimensional structure of β2-microglobulin. Proc. Natl. Acad. Sci. USA 1985, 82, 4225–4229. [Google Scholar] [CrossRef] [PubMed]
- Schanda, P.; Kupče, Ē.; Brutscher, B. SOFAST-HMQC experiments for recording two-dimensional deteronuclear correlation spectra of proteins within a few seconds. J. Biomol. NMR 2005, 33, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Bjorkman, P.J.; Saper, M.A.; Samaroui, B.; Bennet, W.S.; Strominger, J.L.; Wiley, D.C. Structure of the human class I histocompatibility antigen, HLA-A2. Nature 1987, 329, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Esposito, G.; Michelutti, R.; Verdone, G.; Viglino, P.; Hernandez, H.; Robinson, C.V.; Amoresano, A.; Dal Piaz, F.; Monti, M.; Pucci, P.; et al. Removal of the N-terminal hexapeptide from human β2-microglobulin facilitates protein aggregation and fibril formation. Protein Sci. 2000, 9, 831–845. [Google Scholar] [CrossRef] [PubMed]
- Kihara, M.; Chatani, E.; Iwata, K.; Yamamoto, K.; Matsuura, T.; Nakagawa, A.; Naiki, H.; Goto, Y. Conformation of amyloid fibrils of β2-microglobulin probed by tryptophan mutagenesis. J. Biol. Chem. 2006, 281, 31061–31069. [Google Scholar] [CrossRef] [PubMed]
- Van de Weert, M.; Stella, L. Fluorescence quenching and ligand binding: A critical discussion of a popular methodology. J. Mol. Struct. 2011, 998, 144–150. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer: New York, NY, USA, 2006. [Google Scholar]
- Linse, S.; Cabaleiro-Lago, C.; Xue, W.-F.; Lynch, I.; Lindman, S.; Thulin, E.; Radford, S.E.; Dawson, K.A. Nucleation of protein fibrillation by nanoparticles. Proc. Natl. Acad. Sci. USA 2007, 104, 8691–8696. [Google Scholar] [CrossRef] [PubMed]
- Lacerda, S.H.D.P.; Park, J.J.; Meuse, C.; Pristinski, D.; Becker, M.L.; Karim, A.; Douglas, J.F. Interaction of gold nanoparticles with common human blood proteins. ACS Nano 2010, 4, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Mulder, F.A.A.; Schipper, D.; Bott, R.; Boelens, R. Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol. 1999, 292, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Williamson, M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 73, 1–16. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).