Association of Tomato Chlorosis Virus Complicates the Management of Tomato Yellow Leaf Curl Virus in Cultivated Tomato (Solanum lycopersicum) in the Southern United States
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Disease Observation
2.3. Sample Collections
2.4. Virus Cloning and Sequencing
2.5. Virus Quantification
2.6. Statistical Analysis
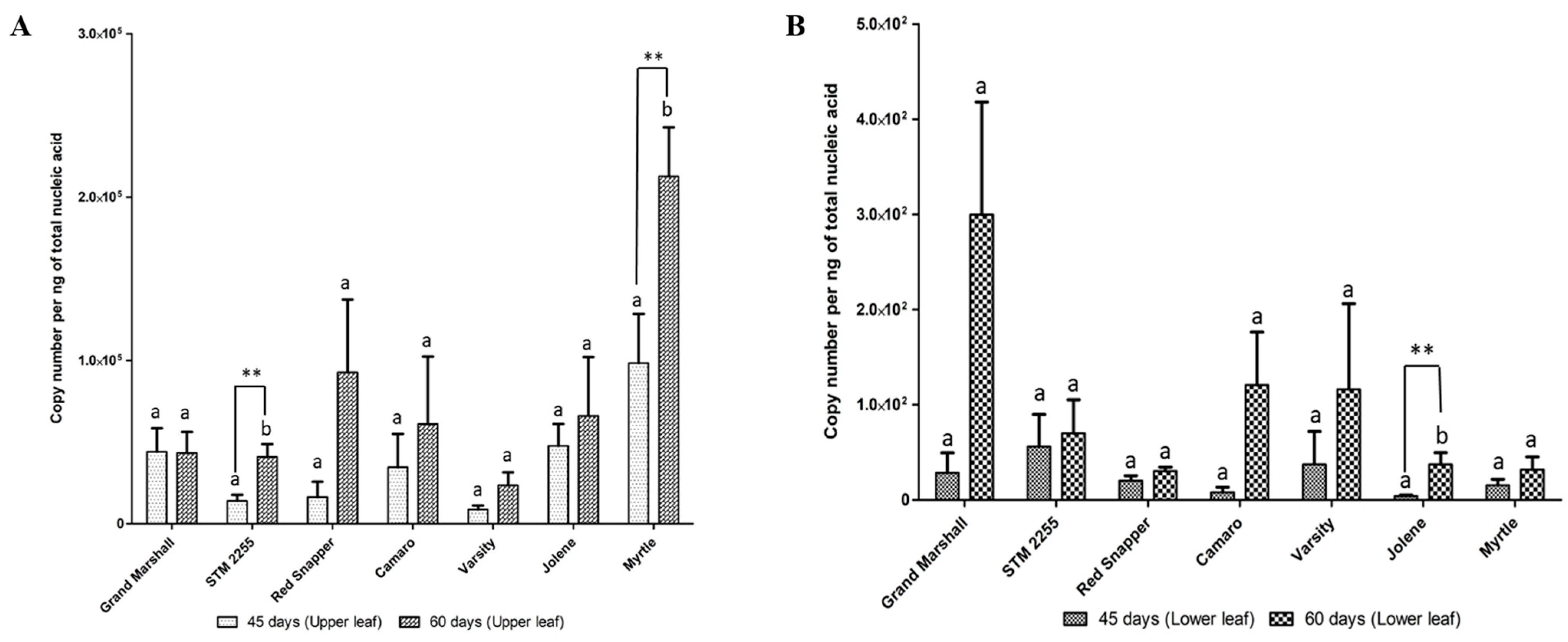
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lefeuvre, P.; Martin, D.P.; Harkins, G.; Lemey, P.; Gray, A.J.A.; Meredith, S.; Lakay, F.; Monjane, A.; Lett, J.-M.; Varsani, A.; et al. The Spread of Tomato Yellow Leaf Curl Virus From The Middle East to The World. PLoS Pathog. 2010, 6, e1001164. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.; Lapidot, M. Effect of Plant Age at Inoculation on Expression of Genetic Resistance to Tomato Yellow Leaf Curl Virus. Arch. Virol. 2008, 153, 171–179. [Google Scholar] [CrossRef]
- Prasad, A.; Sharma, N.; Hari-Gowthem, G.; Muthamilarasan, M.; Prasad, M. Tomato Yellow Leaf Curl Virus: Impact, Challenges, and Management. Trends Plant Sci. 2020, 25, 897–911. [Google Scholar] [CrossRef]
- Zhang, J.; Dong, J.; Xu, Y.; Wu, J. V2 Protein Encoded by Tomato Yellow Leaf Curl China Virus is an RNA Silencing Suppressor. Virus Res. 2012, 163, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chang, Z.; Zhao, S.; Gong, P.; Zhang, M.; Lozano-Durán, R.; Yan, H.; Zhou, X.; Li, F. Functional identification of a novel C7 protein of tomato yellow leaf curl virus. Virology 2023, 585, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Harpaz, I. Periodic, Rather than Continual Acquisition of a New Tomato Virus by Its Vector, the Tobacco Whitefly (Bemisia tabaci Gennadius). Entomol. Exp. Appl. 1964, 7, 155–166. [Google Scholar] [CrossRef]
- Ghanim, M.; Sobol, I.; Ghanim, M.; Czosnek, H. Horizontal Transmission of Begomoviruses between Bemisia tabaci Biotypes. Arthropod-Plant Interact. 2007, 1, 195–204. [Google Scholar] [CrossRef]
- Papayiannis, L.C.; Katis, N.I.; Idris, A.M.; Brown, J.K. Identification of Weed Hosts of Tomato Yellow Leaf Curl Virus in Cyprus. Plant Dis. 2011, 95, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Polston, J.E.; McGovern, R.J.; Brown, L.G. Introduction of Tomato Yellow Leaf Curl Virus in Florida and Implications for the Spread of This and Other Geminiviruses of Tomato. Plant Dis. 1999, 83, 984–988. [Google Scholar] [CrossRef]
- Momol, M.T.; Simone, G.W.; Dankers, W.; Sprenkel, R.K.; Olson, S.M.; Momol, E.A.; Polston, J.E.; Hiebert, E. First Report of Tomato Yellow Leaf Curl Virus in Tomato in South Georgia. Plant Dis. 1999, 83, 487. [Google Scholar] [CrossRef]
- Pappu, S.S.; Pappu, H.R.; Langston, D.B.; Flanders, J.T.; Riley, D.G.; Diaz-Perez, J.C. Outbreak of Tomato Yellow Leaf Curl Virus (Family Geminiviridae) in Georgia. Plant Dis. 2000, 84, 370. [Google Scholar] [CrossRef]
- Marchant, W.G.; Gautam, S.; Hutton, S.F.; Srinivasan, R. Tomato Yellow Leaf Curl Virus-Resistant and -Susceptible Tomato Genotypes Similarly Impact the Virus Population Genetics. Front. Plant Sci. 2020, 11, 599697. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Chakraborty, P.; Ghosh, A. How Many Begomovirus Copies are Acquired and Inoculated by Its Vector, Whitefly (Bemisia tabaci) During Feeding? PLoS ONE 2021, 16, e0258933. [Google Scholar] [CrossRef] [PubMed]
- Polston, J.E.; Lapidot, M. Management of Tomato yellow leaf curl virus: US and Israel Perspectives. In Tomato Yellow Leaf Curl Virus Disease: Management, Molecular Biology, Breeding for Resistance; Czosnek, H., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 251–262. [Google Scholar]
- Verlaan, M.G.; Szinay, D.; Hutton, S.F.; de Jong, H.; Kormelink, R.; Visser, R.G.; Scott, J.W.; Bai, Y. Chromosomal Rearrangements between Tomato and Solanum chilense Hamper Mapping and Breeding of the TYLCV Resistance Gene Ty-1. Plant J. 2011, 68, 1093–1103. [Google Scholar] [CrossRef] [PubMed]
- El-Sappah, A.H.; Qi, S.; Soaud, S.A.; Huang, Q.; Saleh, A.M.; Abourehab, M.A.S.; Wan, L.; Cheng, G.-T.; Liu, J.; Ihtisham, M.; et al. Natural Resistance of Tomato Plants to Tomato Yellow Leaf Curl Virus. Front. Plant Sci. 2022, 13, 1081549. [Google Scholar] [CrossRef]
- Lapidot, M.; Friedmann, M.; Lachman, O.; Yehezkel, A.; Nahon, S.; Cohen, S.; Pilowsky, M. Comparison of Resistance Level to Tomato Yellow Leaf Curl Virus among Commercial Cultivars and Breeding Lines. Plant Dis. 1997, 81, 1425–1428. [Google Scholar] [CrossRef]
- Kelley, T.W.; Boyhan, G. UGA Extension Bulletin 1312, Commercial Tomato Production Handbook; University of Georgia: Athens, GA, USA, 2017; Available online: https://secure.caes.uga.edu/extension/publications/files/pdf/B%201312_7.PDF (accessed on 17 August 2023).
- Simko, I. IdeTo: Spreadsheets for Calculation and Analysis of Area under the Disease Progress over Time Data. PhytoFrontier 2021, 1, 244–247. [Google Scholar] [CrossRef]
- Kavalappara, S.R.; Milner, H.; Konakalla, N.C.; Morgan, K.; Sparks, A.N.; McGregor, C.; Culbreath, A.K.; Wintermantel, W.M.; Bag, S. High Throughput Sequencing-Aided Survey Reveals Widespread Mixed Infections of Whitefly-Transmitted Viruses in Cucurbits in Georgia, USA. Viruses 2021, 13, 988. [Google Scholar] [CrossRef]
- European and Mediterranean Plant Protection Organization. PM 7/118 (1) Tomato Chlorosis Virus and Tomato Infectious Chlorosis Virus. EPPO Bull. 2013, 43, 462–470. [CrossRef]
- Legarrea, S.; Barman, A.; Marchant, W.; Diffie, S.; Srinivasan, R. Temporal Effects of a Begomovirus Infection and Host Plant Resistance on the Preference and Development of an Insect Vector, Bemisia tabaci, and Implications for Epidemics. PLoS ONE 2015, 10, e0142114. [Google Scholar] [CrossRef]
- Wintermantel, W.M.; Hladky, L.L. Methods For Detection and Differentiation of Existing and New Crinivirus Species through Multiplex and Degenerate Primer RT-PCR. J. Virol. Methods 2010, 170, 106–114. [Google Scholar] [CrossRef]
- Luckew, A.; Meru, G.; Wang, Y.-Y.; Mwatuwa, R.; Paret, M.; Carvalho, R.; Kalischuk, M.; Ribeiro da Silva, A.L.B.; Candian, J.; Dutta, B.; et al. Field Evaluation of Cucurbita Germplasm for Resistance to Whiteflies and Whitefly-transmitted Viruses. HortScience 2022, 57, 337–344. [Google Scholar] [CrossRef]
- Orfanidou, C.G.; Pappi, P.G.; Efthimiou, K.E.; Katis, N.I.; Maliogka, V.I. Transmission of Tomato Chlorosis Virus (ToCV) by Bemisia tabaci Biotype Q and Evaluation of Four Weed Species as Viral Sources. Plant Dis. 2016, 100, 2043–2049. [Google Scholar] [CrossRef]
- Kavalappara, S.R.; Riley, D.G.; Cremonez, P.S.G.; Perier, J.D.; Bag, S. Wild Radish (Raphanus raphanistrum L.) Is a Potential Reservoir Host of Cucurbit Chlorotic Yellows Virus. Viruses 2022, 14, 593. [Google Scholar] [CrossRef] [PubMed]
- Thesnim, P.; Jangra, S.; Kumar, M.; Ghosh, A. Effect of Silencing Bemisia tabaci TLR3 and TOB1 on Fitness and Begomovirus Transmission. Front. Plant Sci. 2023, 14, 1136262. [Google Scholar] [CrossRef]
- Anco, D.J.; Rouse, L.; Lucas, L.; Parks, F.; Mellinger, H.C.; Adkins, S.; Kousik, C.S.; Roberts, P.D.; Stansly, P.A.; Ha, M.; et al. Spatial and Temporal Physiognomies of Whitefly and Tomato Yellow Leaf Curl Virus Epidemics in Southwestern Florida Tomato Fields. Phytopathology 2020, 110, 130–145. [Google Scholar] [CrossRef]
- Riley, D.G.; Srinivasan, R. Integrated Management of Tomato Yellow Leaf Curl Virus and Its Whitefly Vector in Tomato. J. Econ. Entomol. 2019, 112, 1526–1540. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Tao, X.; Li, D.; Yang, X.; Zhou, X. Ty-5 Confers Broad-Spectrum Resistance to Geminiviruses. Viruses 2022, 14, 1804. [Google Scholar] [CrossRef]
- Verlaan, M.G.; Hutton, S.F.; Ibrahem, R.M.; Kormelink, R.; Visser, R.G.F.; Scott, J.W.; Edwards, J.D.; Bai, Y. The Tomato Yellow Leaf Curl Virus Resistance Genes Ty-1 and Ty-3 Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases. PLoS Genet. 2013, 9, e1003399. [Google Scholar] [CrossRef] [PubMed]
- Prasanna, H.C.; Kashyap, S.P.; Krishna, R.; Sinha, D.P.; Reddy, S.; Malathi, V.G. Marker Assisted Selection of Ty-2 and Ty-3 Carrying Tomato Lines and Their Implications in Breeding Tomato Leaf Curl Disease Resistant Hybrids. Euphytica 2015, 204, 407–418. [Google Scholar] [CrossRef]
- Ohnishi, J.; Yamaguchi, H.; Saito, A. Analysis of the Mild Strain of Tomato Yellow Leaf Curl Virus, Which Overcomes Ty-2 Gene-mediated Resistance in Tomato Line H24. Arch. Virol. 2016, 161, 2207–2217. [Google Scholar] [CrossRef]
- Voorburg, C.M.; Yan, Z.; Bergua-Vidal, M.; Wolters, A.A.; Bai, Y.; Kormelink, R. Ty-1, a Universal Resistance Gene against Geminiviruses that Is Compromised by Co-replication of a Betasatellite. Mol. Plant Pathol. 2020, 21, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Wintermantel, W.M.; Wisler, G.C. Vector Specificity, Host Range, and Genetic Diversity of Tomato Chlorosis Virus. Plant Dis. 2006, 90, 814–819. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Navas-Castillo, J. Tomato Chlorosis Virus, an Emergent Plant Virus Still Expanding Its Geographical and Host Ranges. Mol. Plant. Pathol. 2019, 20, 1307–1320. [Google Scholar] [CrossRef]
- Sundaraj, S.; Srinivasan, R.; Webster, C.G.; Adkins, S.; Perry, K.; Riley, D. First Report of Tomato Chlorosis Virus Infecting Tomato in Georgia. Plant Dis. 2011, 95, 881. [Google Scholar] [CrossRef]
- Li, J.; Wang, J.-C.; Ding, T.-B.; Chu, D. Synergistic Effects of a Tomato Chlorosis Virus and Tomato Yellow Leaf Curl Virus Mixed Infection on Host Tomato Plants and the Whitefly Vector. Front. Plant Sci. 2021, 12, 672400. [Google Scholar] [CrossRef] [PubMed]
- Ontiveros, I.; López-Moya, J.J.; Díaz-Pendón, J.A. Coinfection of Tomato Plants with Tomato Yellow Leaf Curl Virus and Tomato Chlorosis Virus Affects the Interaction with Host and Whiteflies. Phytopathology 2022, 112, 944–952. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.-Y.-H.; Liao, J.-Y.; Fajar, A.; Chen, J.-B.; Wei, Y.; Zhang, Z.-H.; Zhang, Z.; Zheng, L.-M.; Tan, X.-Q.; Zhou, X.-G.; et al. Co-infection of TYLCV and ToCV Increases Cathepsin B and Promotes ToCV Transmission by Bemisia tabaci MED. Front. Microbiol. 2023, 14, 1107038. [Google Scholar] [CrossRef]
- García-Cano, E.; Resende, R.O.; Fernández-Muñoz, R.; Moriones, E. Synergistic Interaction between Tomato Chlorosis Virus and Tomato Spotted Wilt Virus Results in Breakdown of Resistance in Tomato. Phytopathology 2006, 96, 1263–1269. [Google Scholar] [CrossRef]
- Butterbach, P.; Verlaan, M.G.; Dullemans, A.; Lohuis, D.; Visser, R.G.F.; Bai, Y.; Kormelink, R. Tomato Yellow Leaf Curl Virus Resistance by Ty-1 Involves Increased Cytosine Methylation of Viral Genomes and Is Compromised by Cucumber Mosaic Virus Infection. Proc. Natl. Acad. Sci. USA 2014, 111, 12942–12947. [Google Scholar] [CrossRef] [PubMed]
- Fortes, I.M.; Fernández-Muñoz, R.; Moriones, E. The Crinivirus Tomato Chlorosis Virus Compromises the Control of Tomato Yellow Leaf Curl Virus in Tomato Plants by the Ty-1 Gene. Phytopathology, 2023; Online ahead of print. [Google Scholar] [CrossRef]

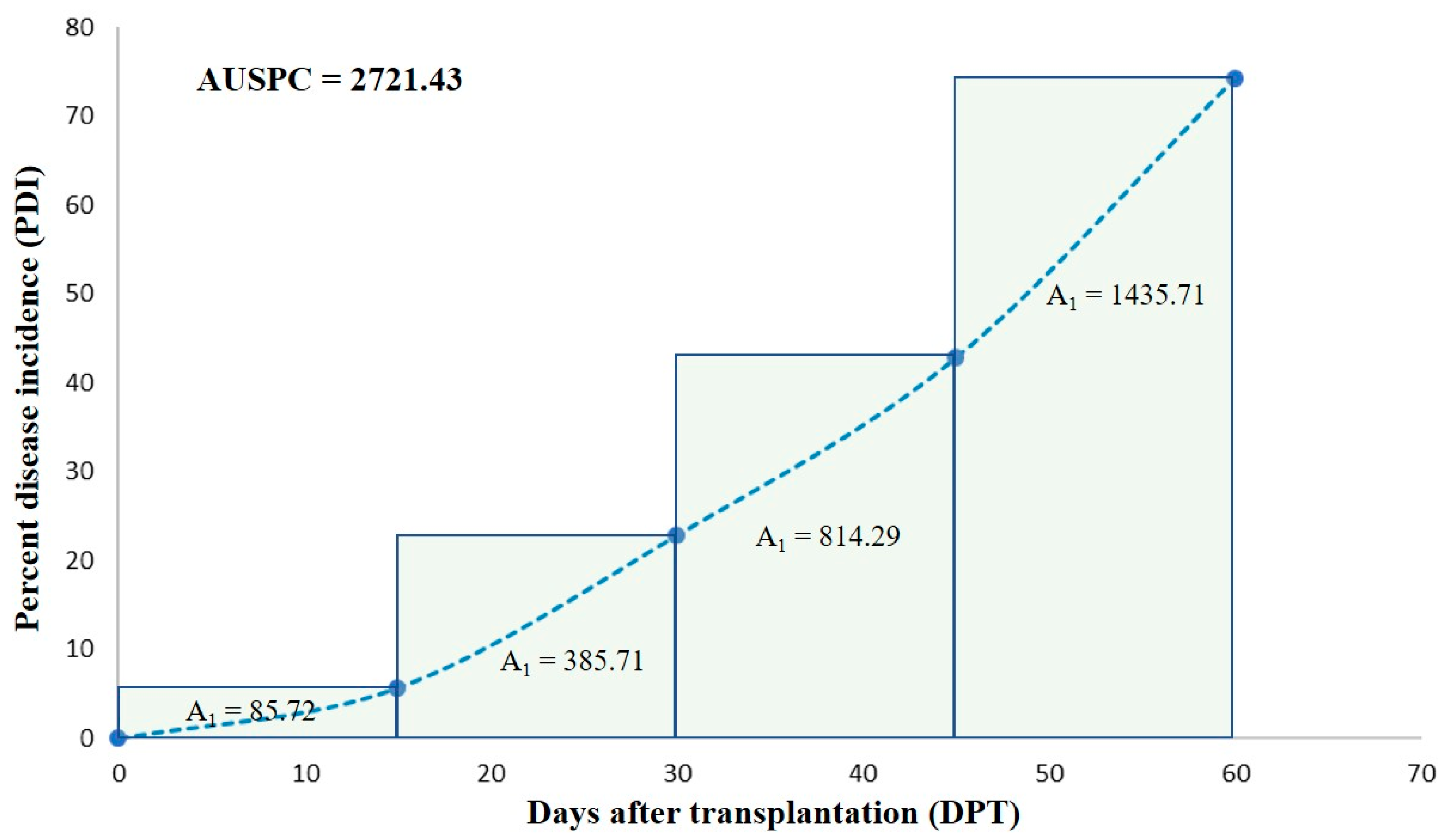

| Sl. No. | Tomato Cultivar | Source Used ≠ | Ty-Gene | AUSPC (Mean) € | Standard Error | Multiple Mean Comparison © |
|---|---|---|---|---|---|---|
| 1 | Grand Marshall | Sakata Seeds, Morgan Hill, CA, USA | Ty3 and Ty6 | 1593.75 | 242.03 | a |
| 2 | STM 2255 | Sakata Seeds, Morgan Hill, CA, USA | Ty3 and Ty6 | 2018.75 | 184.67 | a |
| 3 | Red Snapper | Sakata Seeds, Morgan Hill, CA, USA | Ty3 and Ty6 | 1868.75 | 149.09 | a |
| 4 | Camaro | Sakata Seeds, Morgan Hill, CA, USA | Ty3 and Ty6 | 1850.00 | 200.78 | a |
| 5 | Varsity | Syngenta Vegetable Seeds, Greensboro, NC, USA | Ty1 | 2231.25 | 187.47 | a |
| 6 | Jolene | Bejo Seeds, Inc. Oceano, CA, USA | Ty3 and Ty6 | 1700.00 | 195.52 | a |
| 7 | Myrtle | Bayer Crop Science, Creve Coeur, MO, USA | none | 2431.25 | 79.30 | a |
| 8 | SkyWay 687 £ | Enza Zaden, Enkhuizen, Netherlands | Ty3 and Ty6 | 225.0 | 159.10 | a |
| 9 | HM 8148 £ | HM Clause, Halls, NY | Ty3 and Ty6 | 112.50 | 64.95 | a |
| 10 | Saybrook £ | Bayer Crop Science, USA | none | ND * | ND * | a |
| Primer Name | Used for | Sequence 5′–3′ | Tm (°C) | Amplicon Size (bp) | Reference |
|---|---|---|---|---|---|
| ToCV-258F | qPCR | GTCTGTTCCGGCTGATTACAAGT | 60 | 74 | [21] |
| ToCV-331R | qPCR | AATTGAAACCCAAAGAGGAACAAA | |||
| TYLC-C2-For | qPCR | GCAGTGATGAGTTCCCCTGT | 65 | 102 | [22] |
| TYLC-C2-Rev | qPCR | CCAATAAGGCGTAAGCGTGT | |||
| ToCV-172 (F) | PCR | GCTTCCGAAACTCCGTCTTG | 60 | 439 | [21] |
| ToCV-610 (R) | PCR | TGTCGAAAGTACCGCCACC | |||
| ToCV-RdRP | PCR | GCACCCTGATTGGTTCTAAAC | 60 | 643 | [23] |
| Solanaceae reverse primer | PCR | GTGTTBGAYAACCAWGTGTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, M.; Kavalappara, S.R.; McAvoy, T.; Hutton, S.; Simmons, A.M.; Bag, S. Association of Tomato Chlorosis Virus Complicates the Management of Tomato Yellow Leaf Curl Virus in Cultivated Tomato (Solanum lycopersicum) in the Southern United States. Horticulturae 2023, 9, 948. https://doi.org/10.3390/horticulturae9080948
Kumar M, Kavalappara SR, McAvoy T, Hutton S, Simmons AM, Bag S. Association of Tomato Chlorosis Virus Complicates the Management of Tomato Yellow Leaf Curl Virus in Cultivated Tomato (Solanum lycopersicum) in the Southern United States. Horticulturae. 2023; 9(8):948. https://doi.org/10.3390/horticulturae9080948
Chicago/Turabian StyleKumar, Manish, Saritha Raman Kavalappara, Theodore McAvoy, Samuel Hutton, Alvin M. Simmons, and Sudeep Bag. 2023. "Association of Tomato Chlorosis Virus Complicates the Management of Tomato Yellow Leaf Curl Virus in Cultivated Tomato (Solanum lycopersicum) in the Southern United States" Horticulturae 9, no. 8: 948. https://doi.org/10.3390/horticulturae9080948
APA StyleKumar, M., Kavalappara, S. R., McAvoy, T., Hutton, S., Simmons, A. M., & Bag, S. (2023). Association of Tomato Chlorosis Virus Complicates the Management of Tomato Yellow Leaf Curl Virus in Cultivated Tomato (Solanum lycopersicum) in the Southern United States. Horticulturae, 9(8), 948. https://doi.org/10.3390/horticulturae9080948








