Genome-Wide Identification of Laccase Gene Family from Punica granatum and Functional Analysis towards Potential Involvement in Lignin Biosynthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Determination of Seed Hardness and Lignin Content
2.3. Identification and Physicochemical Properties of PgLAC Family Members
2.4. Bioinformation Analysis of PgLAC Family Members
2.5. Analysis of Cis-Acting Elements and Protein Interaction Networks
2.6. RNA Extraction and Quantitative RT-PCR (qRT-PCR) Analysis of PgLAC Family Members
3. Results
3.1. Comparison of Seed Hardness and Lignin Content during Seed Development Stage
3.2. Identification of PgLAC Gene Family Members
3.3. Bioinformatic Characteristics of PgLAC Gene Family Members
3.4. Motif Distribution and Exon/Intron Analysis of PgLAC Family Members
3.5. Analysis of Cis-Acting Elements in PgLACs Promotors
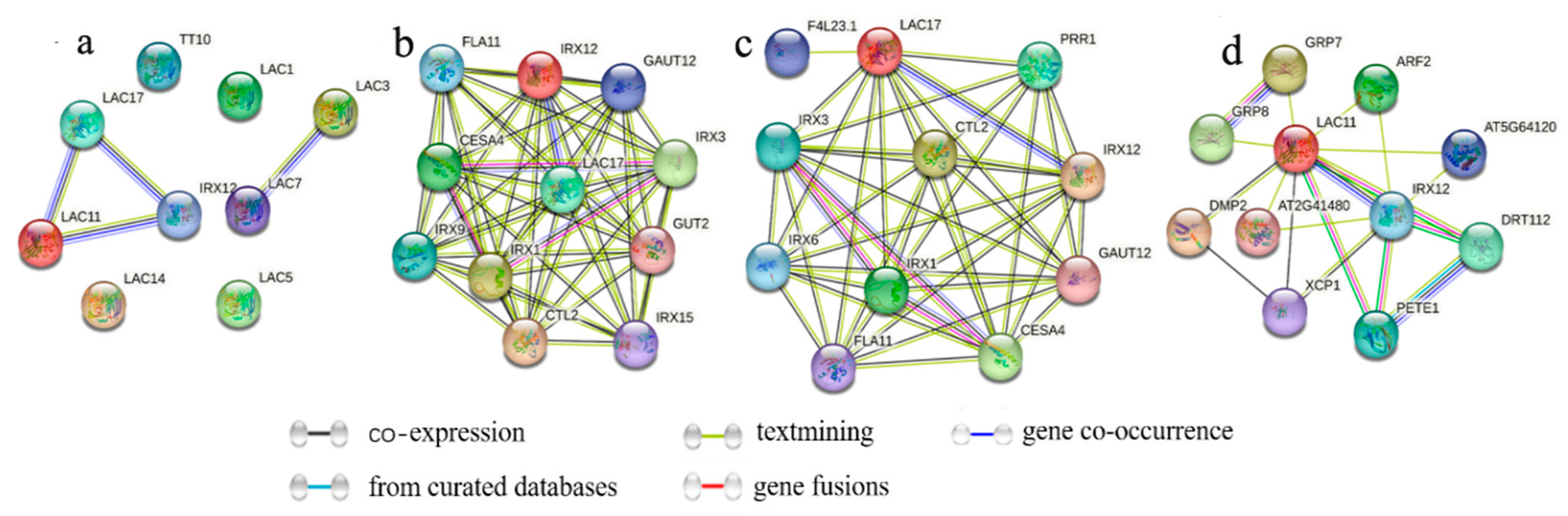
3.6. Analysis of Protein Interaction Networks
3.7. Expression Profiles of PgLACs during the Seed Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yuan, Z.; Fang, Y.; Zhang, T.; Fei, Z.; Han, F.; Liu, C.; Min, L.; Xiao, W.; Zhang, W.; Wu, S.; et al. The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol. J. 2017, 7, 1363–1374. [Google Scholar] [CrossRef] [PubMed]
- Bishayeea, A.; Mandalb, A.; Bhattacharyyac, P.; Bhatiad, D. Pomegranate exerts chemoprevention of experimentally induced mammary tumorigenesis by suppression of cell proliferation and induction of apoptosis. Nutr. Cancer 2016, 68, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Sadeghi, R.; Kokini, J. Pomegranate as a promising opportunity in medicine and nanotechnology. Trends Food Sci. Technol. 2017, 69, 59–73. [Google Scholar] [CrossRef]
- Kandylis, P.; Kokkinomagoulos, E. Food applications and potential health benefits of pomegranate and its derivatives. Foods 2020, 9, 122. [Google Scholar] [CrossRef]
- Ambigaipalan, P.; de Camargo, A.C.; Shahidi, F. Phenolic compounds of pomegranate byproducts (outer skin. mesocarp. divider membrane) and their antioxidant activities. J. Agric. Food Chem. 2016, 64, 6584–6604. [Google Scholar] [CrossRef]
- Morittu, V.M.; Mastellone, V.; Tundis, R.; Loizzo, M.R.; Tudisco, R.; Figoli, A.; Cassano, A.; Musco, N.; Britti, D.; Infascelli, F.; et al. Antioxidant, biochemical, and in-life effects of Punica granatum L. natural juice vs. clarified juice by polyvinylidene fluoride membrane. Foods 2020, 9, 242. [Google Scholar] [CrossRef]
- Zarei, A.; Zamani, Z.; Fatahi, R.; Mousavi, A.S.; Seyed, A. A mechanical method of determining seed-hardness in pomegranate. J. Crop Improv. 2013, 27, 444–459. [Google Scholar] [CrossRef]
- Xue, H.; Cao, S.; Li, H.; Zhang, J.; Niu, J.; Chen, L.; Zhang, F.; Zhao, D. De novo transcriptome assembly and quantification reveal differentially expressed genes between soft-seed and hard-seed pomegranate (Punica granatum L.). PLoS ONE 2017, 12, e0178809. [Google Scholar] [CrossRef]
- Zarei, A.; Zamani, Z.; Fatahi, R.; Mousavi, A.; Salami, S.A.; Avila, C.; Canovas, F.M. Differential expression of cell wall related genes in the seeds of soft-and hard-seeded pomegranate genotypes. Sci. Hortic. 2016, 205, 7–16. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Wang, X.; Shen, Z.; Zheng, L. Comprehensive analysis of rice laccase gene (OsLAC) family and ectopic expression of OsLAC10 enhances tolerance to copper stress in Arabidopsis. Int. J. Mol. Sci 2017, 18, 209. [Google Scholar] [CrossRef]
- Turlapati, P.V.; Kim, K.W.; Davin, L.B.; Lewis, N.G. The laccase multigene family in Arabidopsis thaliana: Towards addressing the mystery of their gene function(s). Planta 2011, 233, 439–470. [Google Scholar] [CrossRef] [PubMed]
- Liao, B.; Wang, C.; Li, X.; Man, Y.; Ruan, H.; Zhao, Y. Genome-wide analysis of the Populus trichocarpa laccase gene family and functional identification of PtrLAC23. Front. Plant Sci. 2023, 13, 1063813. [Google Scholar] [CrossRef] [PubMed]
- Qui, K.; Zhou, H.; Pan, H.; Sheng, Y.; Yu, H.; Xie, Q.; Chen, H.; Cai, Y.; Zhang, J.; He, J. Genome-wide identification and functional analysis of the peach (P. persica) laccase gene family reveal members potentially involved in endocarp lignification. Trees 2022, 36, 1477–1496. [Google Scholar] [CrossRef]
- Xu, X.; Zhou, Y.; Wang, B.; Ding, L.; Wang, Y.; Luo, L.; Zhang, Y.; Kong, W. Genome-wide identification and characterization of laccase gene family in Citrus sinensis. Gene 2018, 689, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Li, G.; Ma, C.; Abdullah, M.; Zhang, J.; Zhao, H.; Qing, J.; Cai, Y.; Lin, Y. Comprehensive genome-wide analysis of the pear (Pyrus bretschneideri) laccase gene (PbLAC) family and functional identification of PbLAC1 involved in lignin biosynthesis. PLoS ONE 2019, 14, e0210892. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Xing, Y.; Liu, F.; Zhang, X.; Li, X.; Zhang, J.; Sun, X. The laccase gene family mediate multi-perspective trade-offs during tea plant (Camellia sinensis) development and defense processes. Int. J. Mol. Sci. 2021, 22, 12554. [Google Scholar] [CrossRef]
- Wan, F.; Zhang, L.; Tan, M.; Wang, X.; Wang, G.; Qi, M.; Liu, B.; Gao, J.; Pan, Y.; Wang, Y. Genome-wide identification and characterization of laccase family members in eggplant (Solanum melongena L.). PeerJ 2022, 10, e12922. [Google Scholar] [CrossRef]
- Janusz, G.; Pawlik, A.; Świderska-Burek, U.; Polak, J.; Sulej, J.; Jarosz-Wilkołazka, A.; Paszczyński, A. Laccase properties, physiological functions, and evolution. Int. J. Mol. Sci. 2020, 21, 966. [Google Scholar] [CrossRef]
- Liu, M.; Dong, H.; Wang, M.; Liu, Q. Evolutionary divergence of function and expression of laccase genes in plants. J. Genet. 2020, 99, 23. [Google Scholar] [CrossRef]
- Pourcel, L.; Routaboul, J.M.; Kerhoas, L.; Caboche, M.; Lepiniec, L.; Debeaujon, I. TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 2005, 17, 2966–2980. [Google Scholar] [CrossRef]
- Zaman, F.; Zhang, M.; Liu, Y.; Wang, Z.; Xu, L.; Guo, D.; Luo, Z.; Zhang, Q. DkmiR397 regulates proanthocyanidin biosynthesis via negative modulating DkLAC2 in Chinese PCNA Persimmon. Int. J. Mol. Sci. 2022, 23, 3200. [Google Scholar] [CrossRef]
- Berthet, S.; Demont-Caulet, N.; Pollet, B.; Bidzinski, P.; Cézard, L.; Bris, P.L.; Borrega, N.; Hervé, J.; Blondet, E.; Balzergue, S.; et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 2011, 23, 1124–1137. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Nakashima, J.; Chen, F.; Yin, Y.; Fu, C.; Yun, J.; Shao, H.; Wang, X.; Wang, Z.; Dixon, R.A.; et al. LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell 2013, 25, 3976–3987. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bouchabke-Coussa, O.; Lebris, P.; Antelme, S.; Soulhat, C.; Gineau, E.; Dalmais, M.; Bendahmane, A.; Morin, H.; Mouille, G.; et al. LACCASE5 is required for lignification of the Brachypodium distachyon culm. Plant Physiol. 2015, 168, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, S.; Yu, Y.; Luo, Y.; Liu, Q.; Ju, C.; Zhang, Y.; Qu, L.; Lucas, W.J.; Wang, X.; et al. MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol. J. 2014, 12, 1132–1142. [Google Scholar] [CrossRef]
- Wang, X.; Zhuo, C.; Xiao, X.; Wang, X.; Docampo-Palacios, M.; Chen, F.; Dixon, R.A. Substrate specificity of LACCASE8 facilitates polymerization of caffeyl alcohol for c-lignin biosynthesis in the seed coat of cleome hassleriana. Plant Cell 2020, 32, 3825–3845. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wang, Q.; Li, G.; Zheng, K.; Zhu, X.; Ma, J.; Wang, D.; Tang, K.; Feng, X.; Leng, J.; Yu, H.; et al. The soybean laccase gene family: Evolution and possible roles in plant defense and stem strength selection. Genes 2019, 10, 701. [Google Scholar] [CrossRef]
- Li, R.; Zhao, Y.; Sun, Z.; Wu, Z.; Wang, H.; Fu, C.; Zhao, H.; He, F. Genome-Wide identification of switchgrass laccases involved in lignin biosynthesis and heavy-metal responses. Int. J. Mol. Sci. 2022, 23, 6530. [Google Scholar] [CrossRef]
- Li, C.; Li, D.; Zhou, H.; Li, J.; Lu, S. Analysis of the laccase gene family and miR397-/miR408-mediated posttranscriptional regulation in Salvia miltiorrhiza. PeerJ 2019, 7, e7605. [Google Scholar] [CrossRef]
- Wang, J.; Feng, J.; Jia, W.; Fan, P.; Bao, H.; Li, S.; Li, Y. Genome-Wide identification of sorghum bicolor laccases reveals potential targets for lignin modification. Front. Plant Sci. 2017, 8, 714. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Gao, H.; Wang, S.; Wu, W.; Tong, R.; Wang, S.; Li, M.; Jian, Z.; Wan, R.; Hu, Q.; et al. Exogenous arginine treatment maintains the appearance and nutraceutical properties of hard-and soft-seed pomegranates in cold storage. Front. Nutr. 2022, 9, 828946. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Li, H.; Wu, Z.; Yao, W.; Zhao, P.; Cao, D.; Yu, H.; Li, K.; Poudel, K.; Zhao, D.; et al. The pomegranate (Punica granatum L.) draft genome dissects genetic divergence between soft-and hard-seeded cultivars. Plant Biotechnol. J. 2019, 18, 955–968. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Cao, D.; Li, H.; Xue, H.; Chen, L.; Liu, B.; Cao, S. Quantitative proteomics of pomegranate varieties with contrasting seed hardness during seed development stages. Tree Genet. Genomes 2018, 14, 14. [Google Scholar] [CrossRef]
- Giuliano, G.; Pichersky, E.; Malik, V.S.; Timko, M.P.; Scolnik, P.A.; Cashmore, A.R. An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc. Natl. Acad. Sci. USA 1988, 85, 7089–7093. [Google Scholar] [CrossRef]
- Kobayashi, K.; Obayashi, T.; Masuda, T. Role of the G-box element in regulation of chlorophyll biosynthesis in Arabidopsis roots. Plant Signal. Behav. 2012, 7, 922–926. [Google Scholar] [CrossRef]
- Liu, C.; Yu, H.; Rao, X.; Li, L.; Dixon, R.A. Abscisic acid regulates secondary cell-wall formation and lignin deposition in Arabidopsis thaliana through phosphorylation of NST1. Proc. Natl. Acad. Sci. USA 2021, 118, 23. [Google Scholar] [CrossRef]
- Jenkins, P.A.; Shepherd, K.R. Seasonal changes in levels of indole-acetic acid and abscisic acid in stem tissues of Pinus radiata. N. Z. J. Bot. 1974, 4, 511–519. [Google Scholar]
- Wodzicki, T.J.; Wodzicki, A.B. Seasonal abscisic acid accumulation in stem cambial region of Pinus silvestris, and its contribution to the hypothesis of a late-wood control system in conifers. Physiol. Plant. 1980, 48, 443–447. [Google Scholar] [CrossRef]
- Cui, M.; Wang, W.; Guo, F.; Fan, X.; Guan, L.; Zheng, T.; Zhu, X.; Jia, H.; Fang, J.; Wang, C.; et al. Characterization and temporal-spatial expression analysis of LEC1 gene in the development of seedless berries in grape induced by gibberellin. Plant Growth Regul. 2020, 90, 585–596. [Google Scholar] [CrossRef]
- Cheng, C.; Xu, X.; Singer, S.D.; Li, J.; Zhang, H.; Gao, M.; Wang, L.; Song, J.; Wang, X. Effect of GA3 treatment on seed development and seed related gene expression in grape. PLoS ONE 2013, 8, e80044. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xuan, X.; Su, Z.; Wang, W.; Abdelrahman, M.; Jiu, S.; Zhang, X.; Liu, Z.; Wang, X.; Wang, C.; et al. Identification of miRNAs-mediated seed and stone-hardening regulatory networks and their signal pathway of GA-induced seedless berries in grapevine (V. vinifera L.). BMC Plant Biol. 2021, 21, 442. [Google Scholar] [CrossRef] [PubMed]
- Lelievre, J.M.; Oliveira, L.O.; Nielsen, N.C. 5′-CATGCAT-3′ elements modulate the expression of glycinin genes. Plant Physiol. 1992, 98, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, G.; Martin, N.; Golovko, A.; Sundström, J.F.; Rask, L.; Ezcurra, I. The RY/Sph element mediates transcriptional repression of maturation genes from late maturation to early seedling growth. New Phytol. 2009, 184, 552–565. [Google Scholar] [CrossRef]

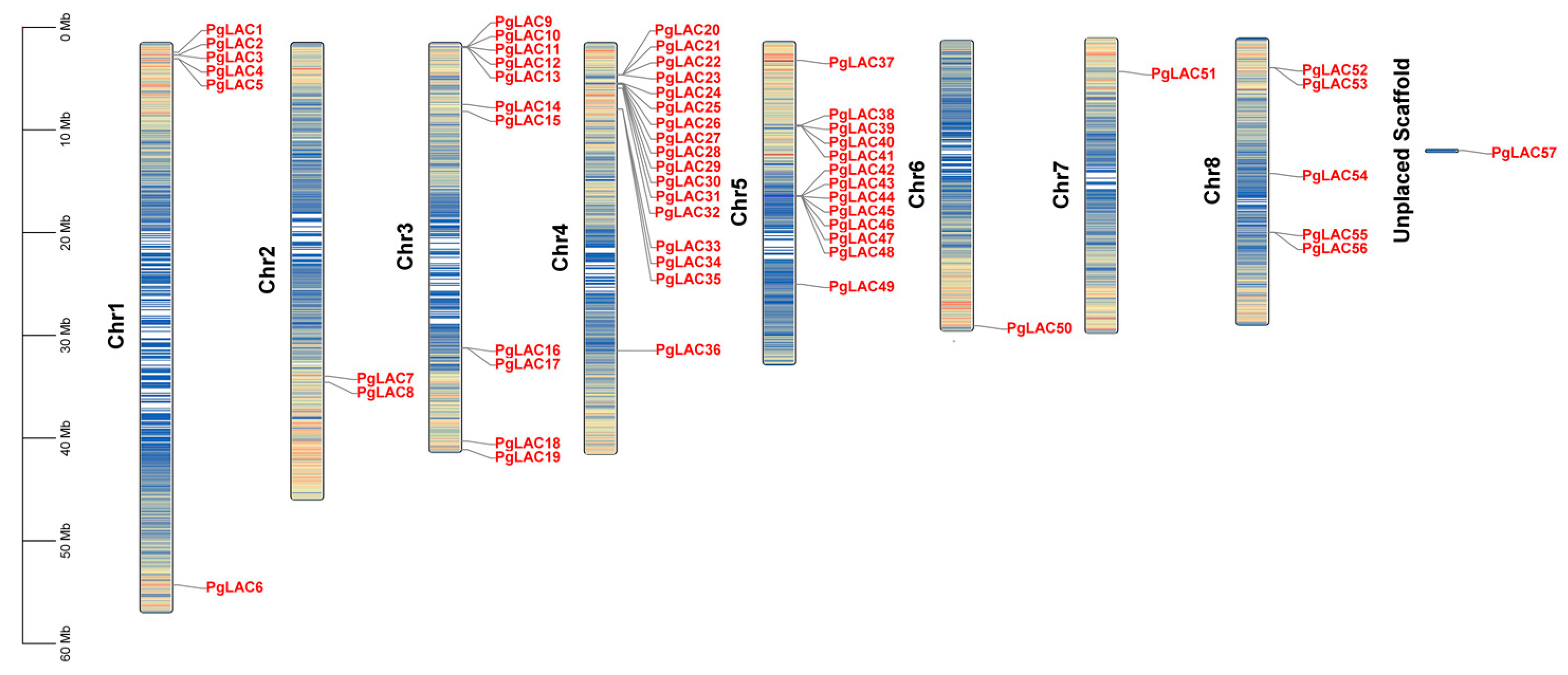

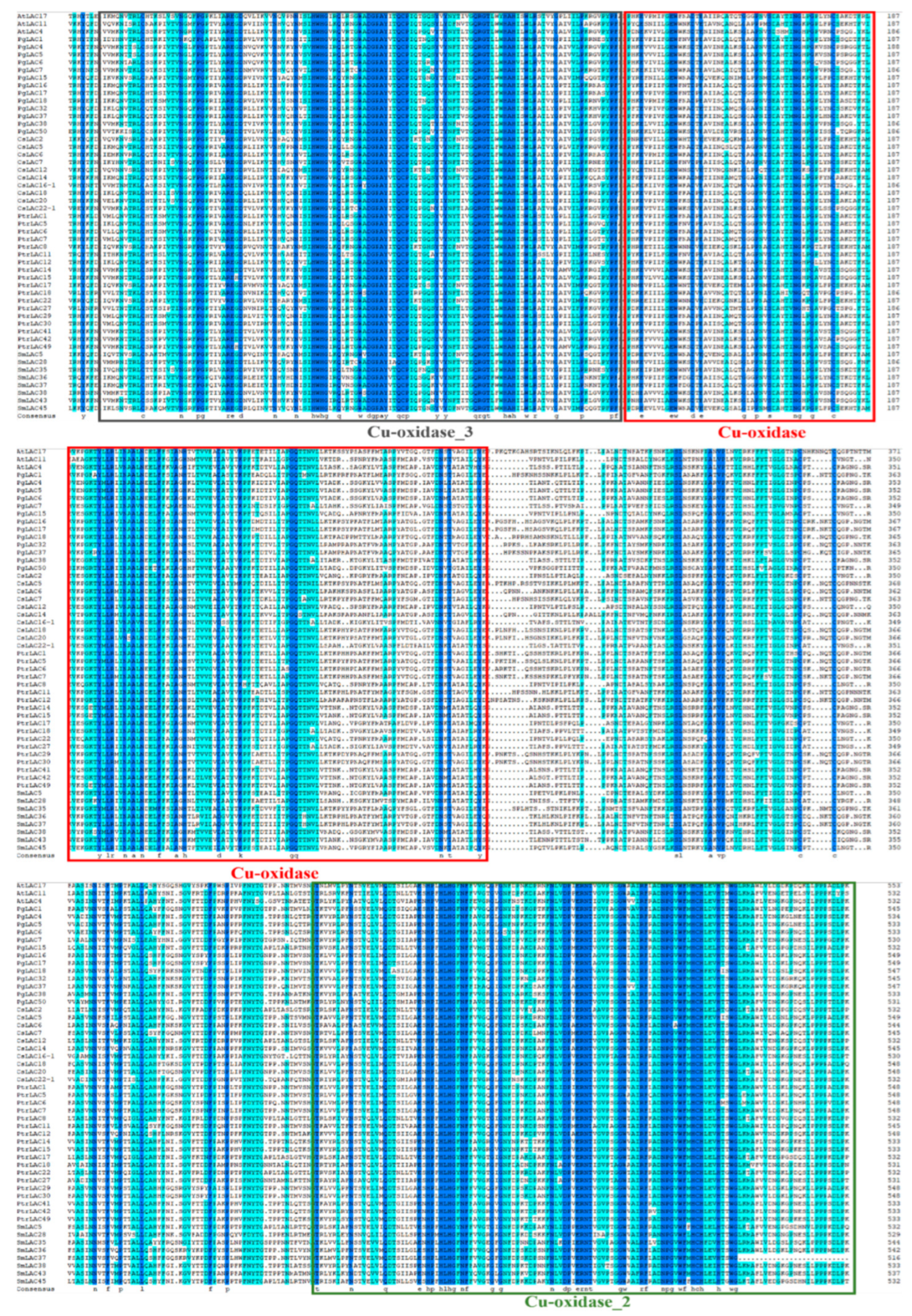
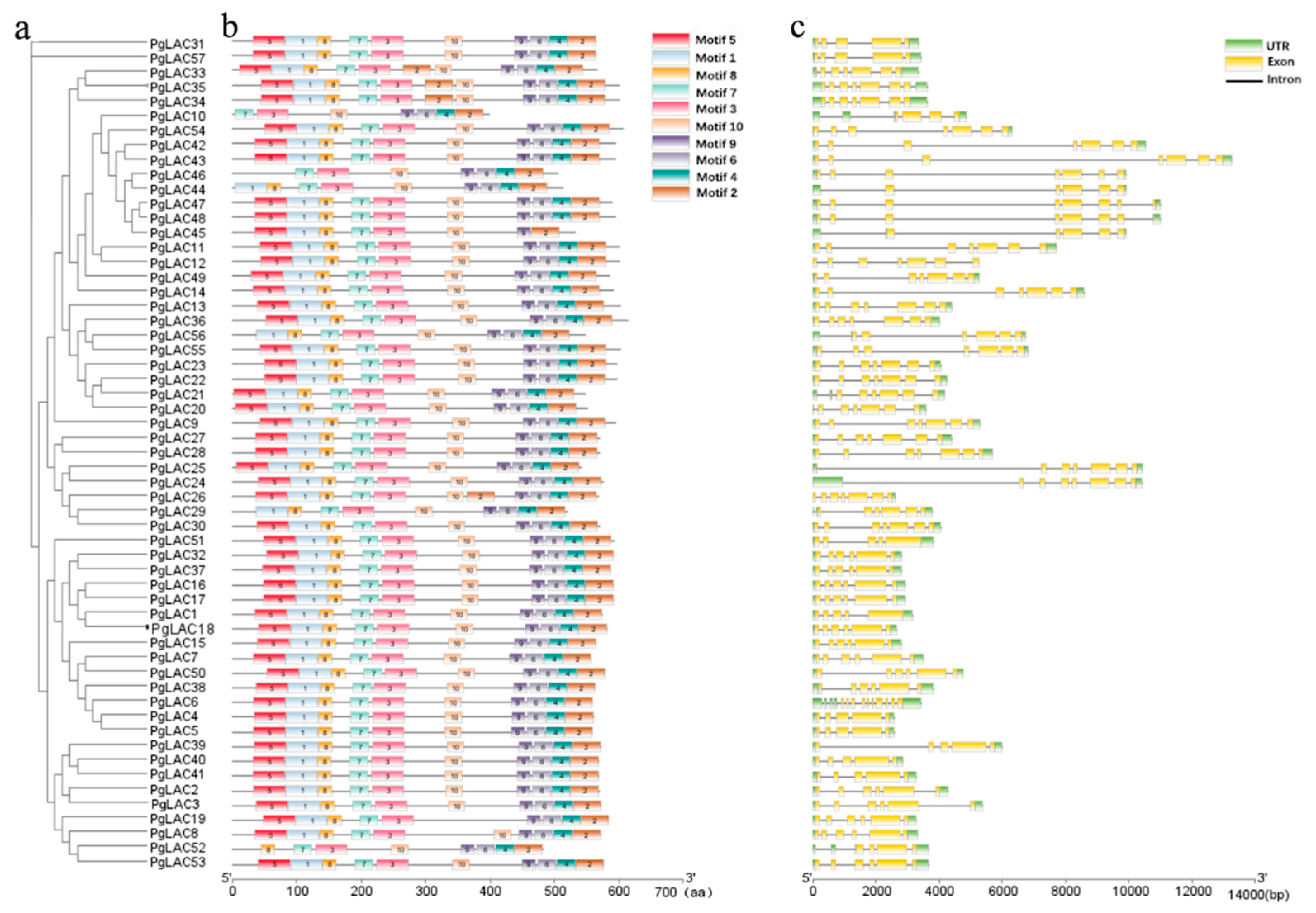
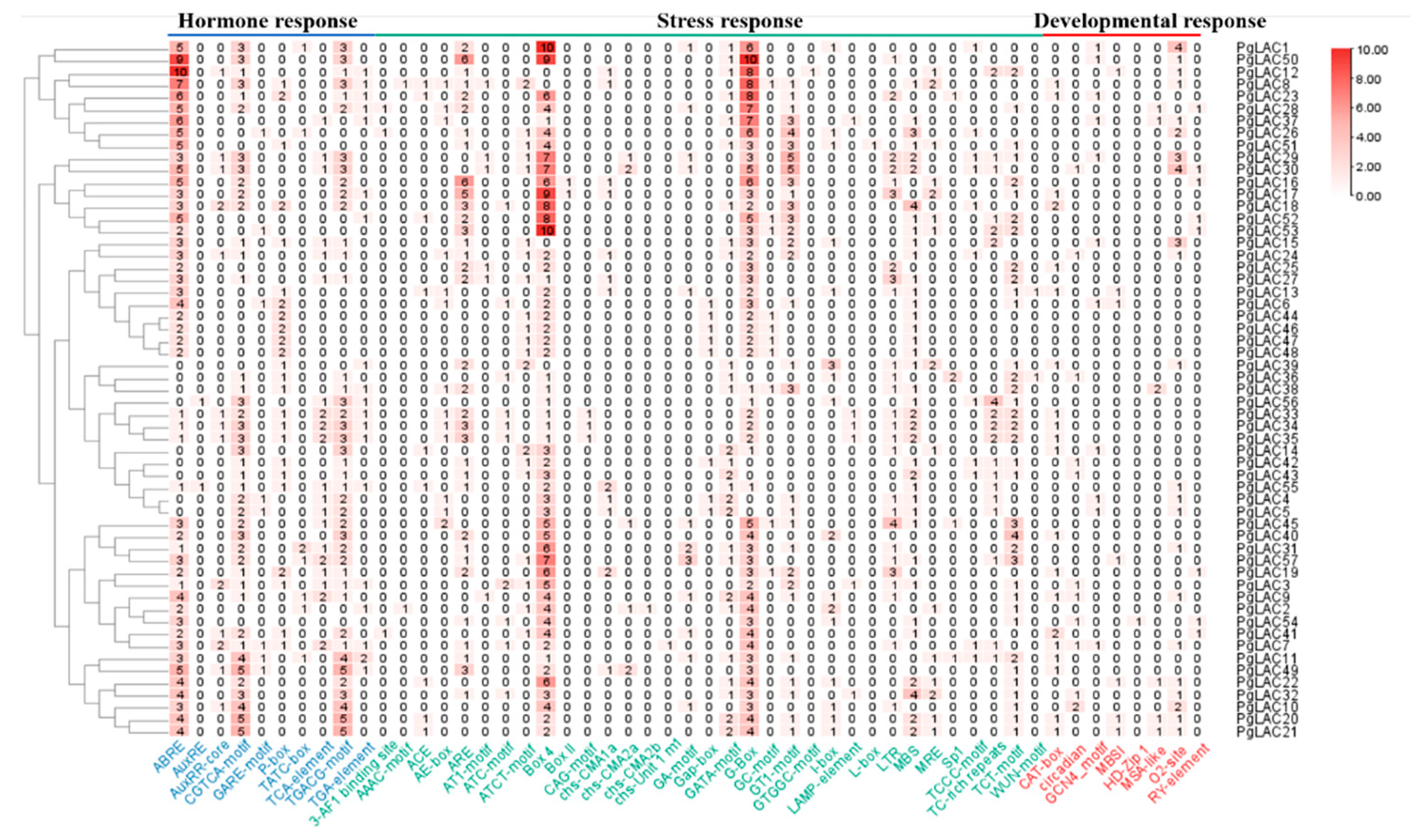

| Gene Name | Gene ID | Protein ID | Gene Length (bp) | Chromosome Position | Amino Acid (aa) | pI | MW (kDa) | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| PgLAC1 | LOC116200081 | XP_031386621.1 | 1811 | Chr1 | 573 | 9.59 | 63.46 | Chloroplast |
| PgLAC2 | LOC116192628 | XP_031377084.1 | 1822 | Chr1 | 568 | 8.22 | 62.61 | Vacuolar membrane |
| PgLAC3 | LOC116193231 | XP_031377897.1 | 1811 | Chr1 | 572 | 8.84 | 62.79 | Vacuolar membrane |
| PgLAC4 | LOC116193760 | XP_031378369.1 | 1796 | Chr1 | 560 | 9.48 | 61.62 | Chloroplast |
| PgLAC5 | LOC116193760 | XP_031378376.1 | 1793 | Chr1 | 559 | 9.48 | 61.49 | Chloroplast |
| PgLAC6 | LOC116192848 | XP_031377387.1 | 1763 | Chr1 | 559 | 9.47 | 61.53 | Chloroplast |
| PgLAC7 | LOC116195123 | XP_031379957.1 | 1885 | Chr2 | 556 | 7.06 | 61.22 | Vacuolar membrane |
| PgLAC8 | LOC116197663 | XP_031383717.1 | 1805 | Chr2 | 571 | 8.59 | 62.92 | Chloroplast |
| PgLAC9 | LOC116199437 | XP_031385642.1 | 1890 | Chr3 | 595 | 5.17 | 66.76 | Endoplasmic reticulum |
| PgLAC10 | LOC116199438 | XP_031385643.1 | 1706 | Chr3 | 397 | 5.44 | 44.53 | Cytoskeleton |
| PgLAC11 | LOC116199436 | XP_031385641.1 | 1896 | Chr3 | 601 | 5.05 | 67.30 | Cytosol |
| PgLAC12 | LOC116199364 | XP_031385547.1 | 1806 | Chr3 | 601 | 5.04 | 67.60 | Chloroplast |
| PgLAC13 | LOC116199435 | XP_031385640.1 | 1909 | Chr3 | 602 | 5.67 | 67.59 | Vacuolar membrane |
| PgLAC14 | LOC116202020 | XP_031389389.1 | 1877 | Chr3 | 590 | 5.08 | 66.49 | Chloroplast |
| PgLAC15 | LOC116199704 | XP_031386025.1 | 1784 | Chr3 | 564 | 7.70 | 62.49 | Vacuolar membrane |
| PgLAC16 | LOC116198560 | XP_031384589.1 | 1870 | Chr3 | 591 | 9.38 | 65.15 | Endoplasmic reticulum |
| PgLAC17 | LOC116198765 | XP_031384857.1 | 1876 | Chr3 | 591 | 9.38 | 65.12 | Endoplasmic reticulum |
| PgLAC18 | LOC116199682 | XP_031386005.1 | 1817 | Chr3 | 581 | 9.48 | 64.04 | Chloroplast |
| PgLAC19 | LOC116201632 | XP_031388782.1 | 1837 | Chr3 | 584 | 9.10 | 64.85 | Mitochondrion |
| PgLAC20 | LOC116205486 | XP_031393972.1 | 1668 | Chr4 | 550 | 4.94 | 61.26 | Cytosol |
| PgLAC21 | LOC116205486 | XP_031393974.1 | 1838 | Chr4 | 547 | 4.98 | 60.95 | Cytosol |
| PgLAC22 | LOC116205486 | XP_031393971.1 | 1846 | Chr4 | 595 | 5.10 | 66.46 | Cytosol |
| PgLAC23 | LOC116205995 | XP_031394575.1 | 1887 | Chr4 | 595 | 5.26 | 67.21 | Cytosol |
| PgLAC24 | LOC116202230 | XP_031389549.1 | 1837 | Chr4 | 575 | 5.01 | 63.50 | Vacuolar membrane |
| PgLAC25 | LOC116202230 | XP_031389550.1 | 1747 | Chr4 | 541 | 4.83 | 59.61 | Cytosol |
| PgLAC26 | LOC116202996 | XP_031390491.1 | 1707 | Chr4 | 568 | 6.16 | 62.63 | Vacuolar membrane |
| PgLAC27 | LOC116203824 | XP_031391629.1 | 1790 | Chr4 | 569 | 6.72 | 63.34 | Extracellular |
| PgLAC28 | LOC116204024 | XP_031391913.1 | 1807 | Chr4 | 569 | 5.92 | 62.36 | Cytosol |
| PgLAC29 | LOC116206410 | XP_031395142.1 | 1620 | Chr4 | 519 | 4.51 | 56.86 | Cytosol |
| PgLAC30 | LOC116206410 | XP_031395141.1 | 1765 | Chr4 | 570 | 4.66 | 62.78 | Vacuolar membrane |
| PgLAC31 | LOC116206444 | XP_031395180.1 | 1789 | Chr4 | 564 | 8.89 | 62.93 | Peroxisome |
| PgLAC32 | LOC116205510 | XP_031393999.1 | 1881 | Chr4 | 591 | 9.78 | 65.91 | Chloroplast |
| PgLAC33 | LOC116206011 | XP_031394594.1 | 1809 | Chr4 | 565 | 5.21 | 63.18 | Chloroplast |
| PgLAC34 | LOC116206011 | XP_031394592.1 | 2074 | Chr4 | 599 | 5.21 | 66.79 | Endoplasmic reticulum |
| PgLAC35 | LOC116206011 | XP_031394593.1 | 2074 | Chr4 | 599 | 5.21 | 66.79 | Endoplasmic reticulum |
| PgLAC36 | LOC116206216 | XP_031394887.1 | 1885 | Chr4 | 614 | 5.04 | 68.98 | Chloroplast |
| PgLAC37 | LOC116206933 | XP_031395630.1 | 1852 | Chr5 | 587 | 9.90 | 65.32 | Chloroplast |
| PgLAC38 | LOC116209331 | XP_031398793.1 | 1873 | Chr5 | 563 | 9.31 | 61.69 | Vacuolar membrane |
| PgLAC39 | LOC116207137 | XP_031395863.1 | 1816 | Chr5 | 571 | 8.76 | 63.02 | Chloroplast |
| PgLAC40 | LOC116209617 | XP_031399176.1 | 1813 | Chr5 | 568 | 9.12 | 62.73 | Chloroplast |
| PgLAC41 | LOC116209616 | XP_031399174.1 | 1853 | Chr5 | 568 | 9.4 | 62.76 | Chloroplast |
| PgLAC42 | LOC116209508 | XP_031399023.1 | 1874 | Chr5 | 595 | 5.08 | 67.17 | Chloroplast |
| PgLAC43 | LOC116209509 | XP_031399024.1 | 1876 | Chr5 | 595 | 4.94 | 67.03 | Vacuolar membrane |
| PgLAC44 | LOC116209332 | XP_031398797.1 | 1784 | Chr5 | 512 | 4.77 | 57.58 | Cytosol |
| PgLAC45 | LOC116209332 | XP_031398799.1 | 1812 | Chr5 | 506 | 5.24 | 57.14 | Chloroplast |
| PgLAC46 | LOC116209332 | XP_031398796.1 | 1739 | Chr5 | 531 | 4.77 | 60.24 | Cytoskeleton |
| PgLAC47 | LOC116209332 | XP_031398795.1 | 1910 | Chr5 | 588 | 5.1 | 66.57 | Cytosol |
| PgLAC48 | LOC116209332 | XP_031398794.1 | 1925 | Chr5 | 593 | 4.86 | 67.00 | Peroxisome |
| PgLAC49 | LOC116209566 | XP_031399095.1 | 1812 | Chr5 | 585 | 4.87 | 65.04 | Vacuolar membrane |
| PgLAC50 | LOC116210060 | XP_031399722.1 | 1857 | Chr6 | 578 | 8.6 | 63.75 | Chloroplast |
| PgLAC51 | LOC116213035 | XP_031403701.1 | 1851 | Chr7 | 593 | 7.29 | 65.69 | Chloroplast |
| PgLAC52 | LOC116189420 | XP_031374936.1 | 1708 | Chr8 | 481 | 6.41 | 52.59 | Nucleus |
| PgLAC53 | LOC116189420 | XP_031374934.1 | 1808 | Chr8 | 576 | 6.78 | 63.14 | Chloroplast |
| PgLAC54 | LOC116187280 | XP_031371794.1 | 1855 | Chr8 | 607 | 5.5 | 68.71 | Chloroplast |
| PgLAC55 | LOC116189029 | XP_031374373.1 | 1596 | Chr8 | 603 | 5.05 | 67.58 | Extracellular |
| PgLAC56 | LOC116189029 | XP_031374374.1 | 1910 | Chr8 | 543 | 4.91 | 61.46 | Cytosol |
| PgLAC57 | LOC116189768 | XP_031375356.1 | 1762 | Unplaced Scaffold | 564 | 8.54 | 63.07 | Peroxisome |
| Species | I | II | III | IV | V | VI | VII | VIII | Total | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Arabidopsis thaliana | 2 | 4 | 4 | 3 | 3 | 1 | 0 | 0 | 17 | [11] |
| Camellia sinensis | 7 | 6 | 4 | 12 | 12 | 2 | 0 | 0 | 43 | [16] |
| Citrus sinensis | 9 | 7 | 3 | 1 | 6 | 0 | 1 | 0 | 27 | [14] |
| Glycine max | 15 | 13 | 8 | 8 | 49 | 0 | 0 | 0 | 93 | [28] |
| Oryza sativa | 7 | 2 | 5 | 5 | 11 | 0 | 0 | 0 | 30 | [10] |
| Panicum virgatum | 9 | 4 | 8 | 9 | 19 | 0 | 0 | 0 | 49 | [29] |
| Populus trichocarpa | 12 | 12 | 6 | 12 | 6 | 5 | 0 | 0 | 53 | [12] |
| Prunus persica | 14 | 4 | 1 | 12 | 17 | 0 | 0 | 0 | 48 | [13] |
| Punica granatum | 7 | 6 | 1 | 4 | 5 | 34 | 0 | 0 | 57 | This study |
| Pyrus bretschneideri | 10 | 10 | 2 | 11 | 1 | 7 | 0 | 0 | 41 | [15] |
| Salvia miltiorrhiza | 9 | 3 | 2 | 5 | 10 | 32 | 1 | 3 | 65 | [30] |
| Solanum melongena | 4 | 7 | 8 | 4 | 1 | 1 | 3 | 20 | 48 | [17] |
| Sorghum bicolor | 4 | 3 | 4 | 8 | 8 | 0 | 0 | 0 | 27 | [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, J.; Yao, J.; Tong, R.; Wang, S.; Li, M.; Song, C.; Wan, R.; Jiao, J.; Zheng, X. Genome-Wide Identification of Laccase Gene Family from Punica granatum and Functional Analysis towards Potential Involvement in Lignin Biosynthesis. Horticulturae 2023, 9, 918. https://doi.org/10.3390/horticulturae9080918
Shi J, Yao J, Tong R, Wang S, Li M, Song C, Wan R, Jiao J, Zheng X. Genome-Wide Identification of Laccase Gene Family from Punica granatum and Functional Analysis towards Potential Involvement in Lignin Biosynthesis. Horticulturae. 2023; 9(8):918. https://doi.org/10.3390/horticulturae9080918
Chicago/Turabian StyleShi, Jiangli, Jianan Yao, Ruiran Tong, Sen Wang, Ming Li, Chunhui Song, Ran Wan, Jian Jiao, and Xianbo Zheng. 2023. "Genome-Wide Identification of Laccase Gene Family from Punica granatum and Functional Analysis towards Potential Involvement in Lignin Biosynthesis" Horticulturae 9, no. 8: 918. https://doi.org/10.3390/horticulturae9080918
APA StyleShi, J., Yao, J., Tong, R., Wang, S., Li, M., Song, C., Wan, R., Jiao, J., & Zheng, X. (2023). Genome-Wide Identification of Laccase Gene Family from Punica granatum and Functional Analysis towards Potential Involvement in Lignin Biosynthesis. Horticulturae, 9(8), 918. https://doi.org/10.3390/horticulturae9080918





