Abstract
Consumer preferences for cooking-related traits are a deciding factor in the success of new cowpea [Vigna unguiculata (L.) Walp] cultivars. Pigment leaching is an undesirable trait for both consumers and producers alike that occurs during the cooking or canning process and has been a goal for improvement efforts through breeding. This study leverages the power of bulked segregant analysis to identify a locus segregating for the pigment-leaching trait in an F2 population of blackeye seed-type cowpea. A single major locus was identified on Vu06 spanning 1.27 Mb, and SNP haplotypes were identified for low and high pigment-leaching bulks. However, further evaluation of accessions that are unrelated to the F2 population or its progenitors suggests that the trait is polygenic, such that low or high leakage is not determined solely by this locus. Parallels were drawn between cowpea and a close relative, black bean (Phaseolus vulgaris L.), to suggest that additional seed coat or cooking-related traits may also be involved in the pigment-leaching trait.
1. Introduction
Cowpea [Vigna unguiculata (L.) Walp], most commonly known as black-eyed pea in the United States, is a warm-season legume and a major source of dietary protein in sub-Saharan Africa while also being produced and consumed worldwide. Cowpea is produced for consumption in a variety of ways; mature seeds can be dried and consumed as whole beans or canned in brine or tomato sauce, and leaves, as well as immature pods and seeds, can be consumed whole or used as a food ingredient [1,2,3]. As a food crop, cowpea is known to have a high nutritional value, but the method through which it is produced for consumption can impact nutrition, especially canning [1,4,5,6,7,8,9]. Canning methods for cowpeas, although economically more expensive than traditional drying methods, have been researched to preserve their natural nutritional value while increasing their shelf lives to reduce potential losses from pests or bacteria [2,7,8,9].
As with many food crops, the visual appeal of cowpea can often influence consumer decisions. Cowpea seed coat color, for example, has been well studied and is an important consumer-related trait, with seed coat color preferences dependent on consumers’ end usage and perceived quality and taste [10,11,12,13]. Cowpea and black bean (Phaseolus vulgaris L.), as well as other grain legumes, have seed coats containing three distinct layers: the palisade cell layer (outermost), the hourglass cell layer (middle), and the parenchymatous cell layer (innermost). Both the palisade and parenchymatous cell layers contain substances that contribute to seed coat pigmentation, but only the palisade layer is known to contain anthocyanins. These anthocyanins can leach from the seed and into the surrounding water or broth during the cooking or canning process.
There have been no previous studies in cowpea regarding the genetic basis for seed coat pigment leaching; however, there have been studies in black bean, a close relative of cowpea that shares a similar seed coat structure [14]. Pigment leaching from seed coats in black bean has been attributed to water imbibition by seeds and subsequent leaching of anthocyanins into the surrounding water [15,16]. Pigment leaching from the seed coat is a trait that has been observed in cowpea grains. Anthocyanins may leach from dark pigmentation on the seed coat or surrounding the hilum of cowpea seeds, also known as the “eye”, causing the surrounding water or broth to become discolored or murky [17]. Consumers may sometimes associate this discoloration and murkiness with the water or broth being dirty.
Pigment leaching is an undesirable trait in grain legumes and warrants further research to better understand its genetic basis. The objective of this study was to use genetic analysis to explore the pigment-leaching trait in blackeye seed-type cowpea. Using an F2 population for bulked segregant analysis, we hoped to determine loci associated with the trait that would enable future work for identifying candidate genes involved in pigment leaching.
2. Materials and Methods
2.1. Plant Materials
A cross between two inbred cowpea lines, ‘Prima’ and ‘California Blackeye 46′ (CB46), was used in the development of a segregating F2 population to study pigment leaching from mature seeds. ‘Prima’ (Nigeria) is a blackeye cultivar that exhibits a high degree of pigment leaching when soaked or cooked in water, while ‘CB46’ (University of California, Davis, USA; Helms et al. 1991) is a California blackeye cultivar observed to exhibit very little pigment leaching. Although from Nigeria, ‘Prima’ is phenotypically similar to California-type cowpeas (early maturing, erect growth, dark foliage, and seed shape and color). It also has closer genetic similarity to California-types and other European cowpeas than to those from West Africa. An F1 plant of this initial cross was self-pollinated to produce an F2 population of 152 progeny that were used in this work.
One hundred fifty-four individuals (152 F2 individuals and two parental lines, ‘Prima’ and ‘CB46’) were planted in 7.5 L pots with UCR Soil Mix II in October 2019 and maintained under greenhouse conditions with irrigation and fertilizer at the University of California, Riverside, CA, USA. Mature pods were collected, dried, and threshed for each plant through May 2020. Young leaf tissues were harvested in November 2019, then stored in plastic Ziplock bags with a silica gel packet for desiccation prior to DNA extraction for genotyping in 2022.
An independent group consisting of 14 accessions from the UCR minicore was examined for their pigment-leaching phenotypes. The UCR minicore is a collection of 368 cultivated cowpea accessions that are representative of the genotypic and phenotypic diversity found in cowpea around the world [18]. Accessions were selected based on their genetic similarity to either ‘Prima’ or ‘CB46’. Seed coat phenotypes for these accessions varied, with several exhibiting the same creamy-colored grain and blackeye as ‘Prima’ and ‘CB46’. Mature pods were collected, dried, and threshed from field-grown minicore accessions in October 2019.
2.2. Anthocyanin Leaching Assay
To quantify pigment (anthocyanin) leaching, approximately four grams of dry seed samples from each accession were weighed, counted, and placed in 30 mL Corex tubes (Corning, New York) with 15 mL of water at room temperature. Tubes were subsequently placed into trays of water at 100 °C and set on an electric stove. Seeds were kept in the tray of hot water for 20 min before transferring 0.9 mL of the broth to a 2 mL tube. A total of 0.1 mL of 1.2 M HCl was added to each tube to reach pH ~1.0, where the peak absorbance (A) for anthocyanin is approximately 520 nm. Three aliquots of 0.2 mL from each sample were transferred to a 96-well plate to be measured by an EMax Microplate Reader (Molecular Devices LLC., San Jose, CA, 95134, USA) with wavelength filters. Absorbance readings were recorded at wavelengths of 490 and 650 nm using the SoftMax Pro 6 software (Molecular Devices LLC.), then normalized to account for the relative surface area (SA) of the seeds used. Values were normalized using the following formulae:
where r = radius, estimated on assumptions that the seed shape is spherical and seed density is constant, such that seed mass is proportional to seed volume. Normalized absorbance values were analyzed and plotted, and samples were placed in high- or low-leaching groups based on their value (Figure 1, Table 1).
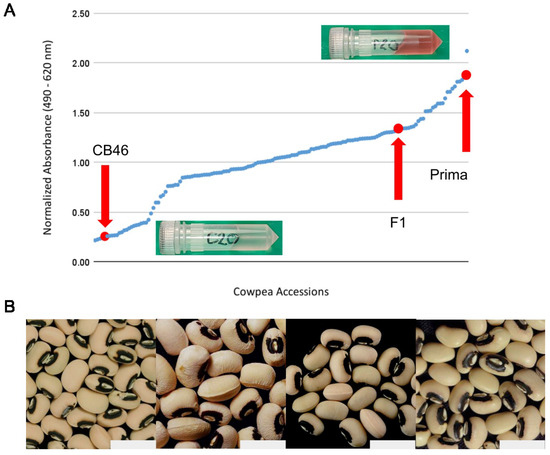
Figure 1.
Mature seed phenotypes of parental, F1, and 152 F2 individuals. (A) One-dimensional scatterplot showing the range of normalized absorbance values indicating the extent of pigment leaching for maternal, paternal, and F1 individuals (red), as well as F2 individuals (blue). Images are included to show the extract color of ‘CB46’ (C20) and ‘Prima’ (P20) after adjustment to pH 1.0. (B) Seed coat phenotypes for (left to right) paternal line, ‘Prima’; maternal line, ‘CB46’; F1 individuals; and F2 individuals. The gray rectangle in each image depicts a scale of 1 cm.

Table 1.
Average mature seed characteristics and absorbance values (pigment leaching) for parental, F1, and bulked F2 individuals. All individuals exhibited an ‘Eye 2′ pattern (creamy-colored grain with a black hilum) as described in Herniter et al. (2019) [10].
2.3. DNA Extraction and Bulked Segregant Analysis
Genomic DNA was extracted from dried leaf samples using the NucleoMag 384 Plant Kit (MACHERY-NAGEL Inc., Pennsylvania, USA). DNA samples were pooled in bulks based on their respective normalized absorbance values for bulked segregant analysis. Low pigment leaching and high pigment leaching bulks were constructed from among the F2 population by pooling equivalent amounts of total genomic DNA from the 23 lowest leakage and 22 highest leakage F2 individuals. Two low leakage pools were constructed from the 23 (12 and 11 individuals) lowest leakage F2 individuals, and two high leakage pools were constructed from the 22 (both sets of 11 individuals) highest leakage F2 individuals. A total of six samples (two low, two high, and the two parental lines) were sent for genotyping.
Genotyping was performed at the University of Southern California Molecular Genomics Core facility (Los Angeles, California, USA) using the Cowpea iSelect Consortium Array, which assays nearly 50,000 SNPs [19]. The University of Southern California Molecular Genomics Core uses an Illumina iScan Reader for SNP data production when using the Illumina iSelect Cowpea Consortium Array. SNPs were called in the GenomeStudio software v2.0.5 (Illumina Inc., San Diego, California, USA) using a custom cluster file as previously described in Munoz-Amatriain et al. (2017) [19].
2.4. Trait Mapping
Mapping of the pigment-leaching trait was achieved by generating the genotype calls of both parental lines and each bulked DNA pool using GenomeStudio v.2.0.5. The output from GenomeStudio produced a list of SNP markers, which was then filtered for polymorphic markers using Microsoft Excel. Markers that were found to be polymorphic between the parents were retained, then sorted based on their physical positions. These positions were based on the pseudochromosomes available from Phytozome (phytozome.net) [20]. The genotypes of each population were visually examined using Microsoft Excel and the Flapjack software [21]. The parental and bulked genotypes were examined for polymorphic regions where a bulk was homozygous. The workspace and data used in GenomeStudio can be found in Data S1. Polymorphic SNP data for the parental lines and F2 bulks can be found in Table S1.
Comparisons between the parental lines and UCR minicore genotypes were performed in Flapjack. Lines were sorted by similarity to either ‘CB46’ or ‘Prima’ at the identified region of interest on Vu06. Minicore genotypes that had high levels of similarity to either ‘CB46’ or ‘Prima’ were noted.
3. Results
3.1. Inheritance of Pigment Leaching from ‘Prima’ and ‘CB46’
Pigment leaching from cowpea seed coats was measured in the ‘CB46’ ‘Prima’ cowpea F2 population and parental cultivars. Anthocyanins were quantified via spectrophotometry because of their role as the major component in pigment leaching that results in water discoloration [22]. The visual appearance of seeds of the two cultivars is quite similar, both having a slightly rough, white, or creamy-colored seed coat with black pigmentation surrounding the hilum to form an “eye”. When cooked or canned, ‘Prima’ exhibits a high degree of pigment leaching, causing the surrounding water or brine to become dark and murky. ‘CB46’ has very little pigment leaching. This difference was the primary consideration in choosing these cultivars to develop the F2 population. No visual differences were noted in the F1 or F2 individuals when compared to either of the parental lines.
F1 seed from the initial ‘CB46’ ‘Prima’ cross was tested for pigment leaching as well. Normalized absorbance values showed high levels of pigment leaching intermediate between the two parental cultivars but closer to that of ‘Prima’. Significant differences in pigment leaching were observed among the F2 population. The F2 individuals showed a continuous distribution of phenotypes and were assigned to lowest or highest pigment-leaching groups based on their normalized absorbance values. A cutoff for ‘low’ leaching was set at a normalized absorbance value of 0.49. Transgressive segregation for pigment leaching occurred for both the low and high limits of the parental cultivars. Of the 152 F2 individuals, we observed 23 with the lowest pigment-leaching phenotype, which formed a more discreet group than other F2 individuals (Figure 1). To test if the F2 population segregates in a Mendelian fashion, we performed a Chi-square test , which led us to conclude that the pigment leaching trait is not controlled entirely by a single gene.
3.2. Genetic Markers and Haplotype Analysis
Genetic markers were screened in the ‘CB46’ ‘Prima’ F2 population. Of the 51,128 SNPs assayed using the Cowpea iSelect Consortium Array, a total of 8221 polymorphic SNP markers with known positions on chromosomes Vu01 through Vu11 were identified between the two parental lines. From these polymorphic markers, a single homozygous region was identified on chromosome Vu06 in both of the low-leaching bulks. This region on Vu06 spans the distance between 22,042,272 bp and 23,317,263 bp (1,274,991 bp), contains 127 polymorphic SNP markers, and has the same haplotype as ‘CB46’ in the low-leaching bulks. The high-leaching bulks were heterozygous at SNP loci across this region (Table S1). Additionally, 159 gene models were found in this region based on annotation data from Lonardi et al. 2019 [20].
In the hopes of leveraging the past literature regarding genetic associations for pigment leaching in black bean, we attempted to identify regions of synteny between the two legumes. Given the genetic relatedness between cowpea and black bean, we compared the identified region from cowpea to the corresponding region of synteny in black bean using the ZZBrowse tool from the Legume Information System [23]. We found that the syntenic region for the identified low-leakage locus in cowpea on chromosome Vu06 did not correspond with any known locus associated with seed traits in black bean.
Following trait mapping using the F2 population, we examined additional cowpea accessions from the University of California, Riverside (UCR) minicore, whose genotyping data were already available [18]. The genotype data for the minicore accessions were examined at the pigment-leaching locus on chromosome Vu06. Minicore accessions were identified with haplotypes matching ‘CB46’ or ‘Prima’. We found 15 accessions matching the low pigment leaching haplotype of ‘CB46’, and 12 accessions matching the high pigment leaching haplotype of ‘Prima’ (Figures S1 and S2). Multiple accessions were also identified that shared haplotypes with a high degree of similarity (> 90%) to either ‘CB46’ or ‘Prima’.
To test the minicore accessions with matching or highly similar haplotypes, we selected 7 minicore accessions that carry the low pigment-leaching haplotype and 7 minicore accessions that carry the high pigment-leaching haplotype (Figure 2). Among the selected accessions for the low haplotype, 6 had the blackeye seed type, and 1 had a full black seed coat, similar to that of black bean. Among the selected accessions for the high haplotype, 3 had the blackeye seed type, and 4 had a full black seed coat. The selected accessions were then assessed against ‘CB46’ and ‘Prima’ using the leaching assay method described in this paper. Results from this assay did not support a conclusion that leakage is controlled entirely by the locus identified from the ‘CB46’ ‘Prima’ F2 population (Figure 3, Table 2). Two minicore accessions that carry a high pigment-leaching haplotype (TVu-3657 and TVu-13979) had absorbance values in a similar range to the F2 individuals used in the low pigment-leaching bulks and ‘CB46’ parental line. Conversely, one minicore accession that carried predominantly the low pigment-leaching haplotype (UCR5353) had an absorbance value higher than those of the low pigment-leaching bulks and higher than that of ‘Prima’. These findings suggest that the region identified on Vu06 is only one part of the apparently polygenic control of seed pigment leakage in cowpea. For example, other visible variables in the seed coats of the minicore accessions (Figure 3) include color, pattern, and texture, any of which could potentially have epistatic effects on anthocyanin leakage. Additional minicore accessions were tested whose seed coat patterns differed from the blackeye or full black seed types and were consistent with these findings (Figure S3, Table S2).
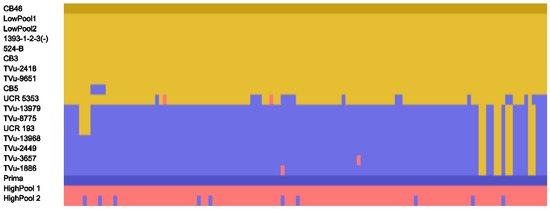
Figure 2.
Evaluating UCR minicore accessions for haplotypes matching the ‘Prima’ or ‘CB46’ parental lines at the 1.27 Mb locus on Vu06. Loci in yellow matched the haplotype of ‘CB46’, while those in blue matched ‘Prima’. Loci in red/pink were heterozygous or had missing data at that locus. Each row represents the genotype call for one accession. There are 127 columns, where each column represents a SNP call from the 1.27 Mb region in order of physical position based on genotype data from Munoz et al. [18]. Haplotypes of ‘Prima’, ‘CB46’, bulks, and a subset of 14 minicore accessions are shown. ‘Prima’ and ‘CB46’ are darker shades of blue and yellow, respectively, to emphasize the reference haplotypes when determining similarity in Flapjack [21].
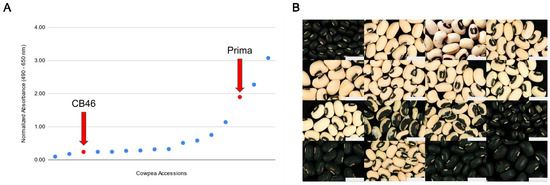
Figure 3.
Mature seed phenotypes of UCR minicore individuals. (A) One-dimensional scatterplot showing the range of normalized absorbance values indicating the extent of pigment-leaching values for ‘CB46’ and ‘Prima’ (red), as well as UCR minicore accessions (blue). Each dot represents one accession, ordered from lowest to highest normalized absorbance. (B) Images demonstrating differences in seed coat patterns. The gray rectangle in each image depicts a scale of 1 cm. Row 1: (left to right) TVu-3657, TVu-9651, CB46, CB3; Row 2: TVu-2418, TVu-13979, CB5, 1393-1-2-3(-); Row 3: 524-B, UCR193, TVu-8775, TVu-1886; Row 4: TVu-2449, Prima, TVu-13968, UCR5353.

Table 2.
Average seed characteristics and pigment-leaching values for 14 UCR minicore individuals with haplotypes matching ‘Prima’ or ‘CB46’. Accessions in the table were ordered to match that of Figure 3. The haplotype for each accession was determined by comparing minicore accessions to ‘CB46’ and ‘Prima’ for similarity. Individuals with “Eye” seed coat patterns had white coat colors and pigmentation surrounding the hilum. Individuals with “Full” seed coat patterns were primarily one color as listed in “Seed coat Color”. No distinct hilum pigmentation was identifiable for “Full” types. *Indicates predominantly the indicated haplotype for UCR5353.
4. Discussion
Reducing or eliminating pigment leaching from seed coats is an important goal for breeders developing new blackeye cowpea cultivars. ‘Prima’ and ‘CB46’ lack pigmentation in both the outer and innermost cell layers of the seed coat, aside from the black “eye”, resulting in a uniform white or creamy-colored grain that is often considered desirable to consumers. Anthocyanin absorbance values of the F2 population varied, with the lowest leaching group being distinctly separate from the majority. This low-leaching group constituted only about 1/6 of the total population, not 1/4, as one might expect from a single major locus where low leakage results only from a homozygous recessive state. The frequency of low pigment-leaching individuals in the F2 population being significantly less than 25% may have several explanations. A simple possibility is segregation distortion. Alternatively, an epistatic factor may be segregating in the F2 population, such that the effect of the low pigment-leaching haplotype on Vu06 can be masked by an unlinked epistatic allele that is present in ‘Prima’. An epistatic interaction was found in cowpea seed coat pattern development by Herniter et al. [10]. Based on the F1 phenotype, we can conclude that high pigment leaching is partially dominant over low pigment leaching at the locus on Vu06. It should be noted, however, that examination of pigment leaching in the additional minicore accessions demonstrated that this trait is not controlled solely by the Vu06 locus identified in the F2 population [Table 2, Figure 3]. Differences are readily apparent in the color, pattern, and texture of the seed coats of the minicore accessions, which potentially also factor into pigment leaching as a polygenic trait (Table 2, Figures S1 and S3). Future work is necessary to apply these findings in cowpea with different seed coat traits and backgrounds.
We examined the locus on Vu06 for gene models that may assist in describing our findings. Within the 1.27 Mb region (22,042,272–23,317,263 bp), we identified 159 gene models, of which 84 had functional annotations, using the Vigna unguiculata v1.2 reference genome available from Phytozome [20] (Table S3). A threshold of transcripts per million (TPM) ≥ 2 was used to filter for ‘expressed’ genes. It was noted that 67 of the 84 genes with functional annotations were found to be expressed in the seed, using data from the Legume Information System (LIS) gene expression atlas [23]. The LIS gene expression atlas contains RNA-seq data for seeds at 8, 10, 14, and 18 days after pollination. Expression levels varied for each of these genes at the different seed developmental stages. Gene expression at these different stages may coincide with the development of certain seed traits involved with pigment leaching, aligned with observations by Herniter et al. regarding seed coat pattern development [10]. Finer mapping of the trait determinant was attempted by comparing the GenomeStudio theta values of SNP calls in the two low bulks versus ‘CB46’. The result was a 59 kb region between 22,745,594 bp and 22,804,595 bp defined by a run of 11 consecutive SNPs with theta values very close to those of ‘CB46’ (Data S1). This region contains seven genes (Vigun06g096100, Vigun06g096200, Vigun06g096300, Vigun06g096400, Vigun06g096500, Vigun06g096600, and Vigun06g096700), four of which are both annotated and expressed in the seed (Vigun06g096100, Vigun06g096200, Vigun06g096300, and Vigun06g096400) and could conceivably be related to pigment leaching. For example, this region includes a gene (Vigun06g096300) that encodes an ankyrin repeat family protein, which is expressed in leaf and flower tissue and very early in seed development. Ankyrin repeat proteins are involved in protein stability, folding, and binding specificity and, as such, could conceivably influence processes related to cell wall integrity [24]. Ankyrin repeat family proteins have also been shown to be involved in anthocyanin synthesis [25,26]. This and other genes within the region on Vu06 identified in this work provide numerous testable hypotheses for further studies to explore the mechanistic basis of pigment leaching in cowpea seeds.
In this work, we sought to understand the genetic basis of pigment leaching in blackeye seed-type cowpea. Previous work involving anthocyanin leaching in legumes was conducted in black bean to identify QTL associated with traits for better canning quality and color retention [22]. Several seed coat and cooking-related traits have been suggested to have an influence on pigment leaching. The general hypothesis for increased levels of pigment leaching revolves around compromised seed coat integrity resulting from genetics or cooking processes. For example, higher pigment leaching in black bean has been associated with higher levels of water imbibition as well as the asp gene [14,15,27]. Additionally, seeds that require longer cooking times are associated with higher amounts of pectin in the seed coat, which increases the structural integrity of the seed coat. Salts, longer cooking times, and higher cooking temperatures may also be factors that affect pigment leaching and have been addressed in the literature for cowpea as well as other legumes [7,8,28,29,30]. The similarities in seed coat structure of black bean may be translatable to cowpea, especially given the extensive synteny between common bean and cowpea genomes [19,20].
The results based on the bulked-segregant analysis are promising for addressing pigment leaching in cowpea cultivars with a blackeye background. The locus on Vu06 was the only identified region of homozygosity in the two low pigment-leaching phenotypic bulks. The identification of a low pigment-leaching haplotype of ‘CB46’ and a high pigment-leaching haplotype of ‘Prima’ on Vu06 provides breeders with a packet of genetic markers to assist in selection against high pigment leaching when breeding for additional traits in blackeye cultivars. A recent example of marker-assisted selection utilized ‘CB46’ as the recurrent parent to develop of a new aphid-resistant cultivar, ‘CB77′ [31], which we now know also carries the low pigment-leaching haplotype of CB46. Although candidate genes relating to pigment leaching were not emphasized in this study, the narrowing of the trait determinant on Vu06 to a region containing only 159 genes, approximately half of which have function annotations, provides a launching point for further exploration of the mechanistic basis of pigment leaching [20,23].
5. Conclusions
In conclusion, this work delimited a locus controlling cowpea seed coat pigment leaching to a region of chromosome Vu06 spanning 1,274,991 bp, providing a packet of SNP markers defining low- and high-leaching haplotypes present in low-leaching CB46 and high-leaching Prima ‘blackeye’ seed-type cultivars. Initial steps were taken to explore the trait determinant in this locus that may contribute to the pigment-leaching trait. Additional work, however, would be required before considering these as candidate genes. The identification of this locus and the SNP markers defining two haplotypes should assist cowpea breeders in developing more consumer-accepted cultivars by enabling marker-assisted selection for this genetic component of the seed coat pigment leaching trait.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae9070739/s1. Figure S1: Pigment leaching of ‘Prima’, ‘CB46’, and UCR minicore accessions. Figure S2: Evaluating UCR minicore accessions for haplotypes matching the ‘Prima’ or ‘CB46’ parental lines at the 1.27 Mb locus on Vu06. Figure S3: Mature seed phenotypes of UCR minicore individuals. Table S1: SNP data from ‘CB46’, ‘Prima’, and the F2 bulks. Table S2: Average seed characteristics and pigment leaching values for 25 UCR minicore individuals with haplotypes matching ‘Prima’ or ‘CB46’. Table S3: List of unique genes spanning the identified pigment leaching locus (22,042,272–23,317,263 bp) on chromosome Vu06. Data S1: GenomeStudio workspace for exploring pigment leaching in cowpea.
Author Contributions
Conceptualization, C.S.B., B.L.H., P.R. and T.J.C.; methodology, K.P. and T.J.C.; validation, C.S.B.; formal analysis, C.S.B. and T.J.C.; investigation, C.S.B., K.P. and T.J.C.; resources, B.L.H., P.R., J.R.P.S. and T.J.C.; writing—original draft preparation, C.S.B. and T.J.C.; writing—review and editing, C.S.B., B.L.H., P.R., J.R.P.S., K.P. and T.J.C.; visualization, C.S.B.; supervision, T.J.C.; project administration, T.J.C.; funding acquisition, T.J.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Hatch Project CA-R-BPS-5306-H, and USAID Feed the Future Innovation Labs for Legume Systems Research (Cooperative Agreement 7200AA18LE00003) and Collaborative Research on Grain Legumes (Cooperative Agreement EDH-A-00-07-00005).
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article. Publicly available SNP data for accessions in the UCR minicore are available in the supplementary material (Table S2) of Muñoz-Amatriaín et al. 2021 [doi:10.1002/leg3.95].
Acknowledgments
We thank Anthony Hall and Jeffrey Ehlers for their comments and discussions regarding the pigment-leaching phenotype. We also thank Alyssa Abuda, Sheila Close, and Michael Utterback for assistance in maintaining the plants, harvesting pods, threshing, and seed counting to help keep the work moving forward during COVID-19 times, and Ira Herniter for high-quality photographs of cowpea seeds.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Singh, B.B. Cowpea: The Food Legume of the 21st Century; ASA, CSSA, and SSSA Books; American Society of Agronomy and Soil Science Society of America: Madison, WI, USA, 2014; ISBN 978-0-89118-622-9. [Google Scholar]
- Taiwo, K.A.; Akanbi, C.T.; Ajibola, O.O. Establishing Processing Conditions for Canning Cowpea Seeds in Tomato Sauce. Int. J. Food Sci. Technol. 1997, 32, 313–324. [Google Scholar] [CrossRef]
- Kapravelou, G.; Martínez, R.; Andrade, A.M.; López Chaves, C.; López-Jurado, M.; Aranda, P.; Arrebola, F.; Cañizares, F.J.; Galisteo, M.; Porres, J.M. Improvement of the Antioxidant and Hypolipidaemic Effects of Cowpea Flours (Vigna unguiculata) by Fermentation: Results of in Vitro and in Vivo Experiments. J. Sci. Food Agric. 2015, 95, 1207–1216. [Google Scholar] [CrossRef]
- Bessada, S.M.F.; Barreira, J.C.M.; Oliveira, M.B.P.P. Pulses and Food Security: Dietary Protein, Digestibility, Bioactive and Functional Properties. Trends Food Sci. Technol. 2019, 93, 53–68. [Google Scholar] [CrossRef]
- Adebooye, O.C.; Singh, V. Physico-Chemical Properties of the Flours and Starches of Two Cowpea Varieties (Vigna unguiculata (L.) Walp.). Innov. Food Sci. Emerg. Technol. 2008, 9, 92–100. [Google Scholar] [CrossRef]
- Moreno, L.d.A.; de Oliveira, G.R.F.; Batista, T.B.; Bossolani, J.W.; Ducatti, K.R.; Guimarães, C.C.; da Silva, E.A.A. Quality of Cowpea Seeds: A Food Security Strategy in the Tropical Environment. PLoS ONE 2022, 17, e0276136. [Google Scholar] [CrossRef]
- Afoakwa, E.O.; Yenyi, S.E.; Sakyi-Dawson, E. Response Surface Methodology for Optimizing the Pre-Processing Conditions during Canning of a Newly Developed and Promising Cowpea (Vigna unguiculata) Variety. J. Food Eng. 2006, 73, 346–357. [Google Scholar] [CrossRef]
- Carvalho dos Santos, W.P.; Weste Nano, R.M.; de Oliveira, F.S.; Maia, L.C.; de Souza Miranda, K.E.; Campos, I.A.L. Evaluation of the Effects of Canning Variables on the Mineral Composition of Canned Cowpeas (Vigna unguiculata L. Walp.) Using Multi-Response Analysis. Food Sci. Technol. Int. 2023. [Google Scholar] [CrossRef]
- Hama, M.O.; Amadou, I.; Daou, C.; Zhang, M. Optimization of the Preparation Treatment to Obtain the Desired Quality of Canned Cowpea (Vigna unguiculata, TN 5-78) Variety Grown in the Sahel Region. Songklanakarin J. Sci. Technol. 2020, 42, 477–724. [Google Scholar]
- Herniter, I.A.; Lo, R.; Muñoz-Amatriaín, M.; Lo, S.; Guo, Y.-N.; Huynh, B.-L.; Lucas, M.; Jia, Z.; Roberts, P.A.; Lonardi, S.; et al. Seed Coat Pattern QTL and Development in Cowpea (Vigna unguiculata [L.] Walp.). Front. Plant Sci. 2019, 10, 1346. [Google Scholar] [CrossRef]
- Kostyla, A.S.; Clydesdale, F.M.; McDaniel, M.R. The Psychophysical Relationships between Color and Flavor∗. Crit. Rev. Food Sci. Nutr. 1978, 10, 303–321. [Google Scholar] [CrossRef] [PubMed]
- Simonne, A.H.; Weaver, D.B.; Wei, C. Immature Soybean Seeds as a Vegetable or Snack Food: Acceptability by American Consumers. Innov. Food Sci. Emerg. Technol. 2000, 1, 289–296. [Google Scholar] [CrossRef]
- Mishili, F.J.; Fulton, J.; Shehu, M.; Kushwaha, S.; Marfo, K.; Jamal, M.; Kergna, A.; Lowenberg-DeBoer, J. Consumer Preferences for Quality Characteristics along the Cowpea Value Chain in Nigeria, Ghana, and Mali. Agribusiness 2009, 25, 16–35. [Google Scholar] [CrossRef]
- Lush, W.M.; Evans, L.T. The Seed Coats of Cowpeas and Other Grain Legumes: Structure in Relation to Function. Field Crops Res. 1980, 3, 267–286. [Google Scholar] [CrossRef]
- Bushey, S.M. Water Uptake and Its Relationship to Pigment Leaching in Black Beans (Phaseolus vulgaris L.). In Reports of Bean Improvement Cooperative and National Dry Bean Council Research Conference; USDA: Washington, DC, USA, 2000; pp. 104–105. [Google Scholar]
- Beninger, C.W.; Hosfield, G.L.; Bassett, M.J.; Owens, S. Chemical and Morphological Expression of the B and Asp Seedcoat Genes in Phaseolus vulgaris. J. Am. Soc. Hortic. Sci. 2000, 125, 52–58. [Google Scholar] [CrossRef]
- Mann, A. Coloration of the Seed Coat of Cowpeas. J. Agr. Res. 1914, 2, 33–56. [Google Scholar]
- Muñoz-Amatriaín, M.; Lo, S.; Herniter, I.A.; Boukar, O.; Fatokun, C.; Carvalho, M.; Castro, I.; Guo, Y.-N.; Huynh, B.-L.; Roberts, P.A.; et al. The UCR Minicore: A resource for cowpea research and breeding. Legume Sci. 2021, 3, e95. [Google Scholar] [CrossRef]
- Muñoz-Amatriaín, M.; Mirebrahim, H.; Xu, P.; Wanamaker, S.I.; Luo, M.; Alhakami, H.; Alpert, M.; Atokple, I.; Batieno, B.J.; Boukar, O.; et al. Genome Resources for Climate-Resilient Cowpea, an Essential Crop for Food Security. Plant J. 2017, 89, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Lonardi, S.; Muñoz-Amatriaín, M.; Liang, Q.; Shu, S.; Wanamaker, S.I.; Lo, S.; Tanskanen, J.; Schulman, A.H.; Zhu, T.; Luo, M.-C.; et al. The Genome of Cowpea (Vigna unguiculata [L.] Walp.). Plant J. 2019, 98, 767–782. [Google Scholar] [CrossRef]
- Milne, I.; Shaw, P.; Stephen, G.; Bayer, M.; Cardle, L.; Thomas, W.T.B.; Flavell, A.J.; Marshall, D. Flapjack—Graphical Genotype Visualization. Bioinformatics 2010, 26, 3133–3134. [Google Scholar] [CrossRef]
- Cichy, K.A.; Fernandez, A.; Kilian, A.; Kelly, J.D.; Galeano, C.H.; Shaw, S.; Brick, M.; Hodkinson, D.; Troxtell, E. QTL Analysis of Canning Quality and Color Retention in Black Beans (Phaseolus vulgaris L.). Mol. Breed. 2014, 33, 139–154. [Google Scholar] [CrossRef]
- Berendzen, J.; Brown, A.V.; Cameron, C.T.; Campbell, J.D.; Cleary, A.M.; Dash, S.; Hokin, S.; Huang, W.; Kalberer, S.R.; Nelson, R.T.; et al. The Legume Information System and Associated Online Genomic Resources. Legume Sci. 2021, 3, e74. [Google Scholar] [CrossRef]
- Wolf, D.; Hofbrucker-MacKenzie, S.A.; Izadi, M.; Seemann, E.; Steiniger, F.; Schwintzer, L.; Koch, D.; Kessels, M.M.; Qualmann, B. Ankyrin Repeat-Containing N-Ank Proteins Shape Cellular Membranes. Nat. Cell Biol. 2019, 21, 1191–1205. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.; Shin, D.H.; Cho, M.-H.; Kim, T.-L.; Bhoo, S.H.; Hahn, T.-R. An Ankyrin Repeat Protein Is Involved in Anthocyanin Biosynthesis in Arabidopsis. Physiol. Plant. 2011, 142, 314–325. [Google Scholar] [CrossRef]
- Toppino, L.; Barchi, L.; Mercati, F.; Acciarri, N.; Perrone, D.; Martina, M.; Gattolin, S.; Sala, T.; Fadda, S.; Mauceri, A.; et al. A New Intra-Specific and High-Resolution Genetic Map of Eggplant Based on a RIL Population, and Location of QTLs Related to Plant Anthocyanin Pigmentation and Seed Vigour. Genes 2020, 11, 745. [Google Scholar] [CrossRef]
- Lamprecht, H. Zur Genetik Von Phaseolus Vulgaris Xvii—Xviii. Hereditas 1940, 26, 292–304. [Google Scholar] [CrossRef]
- Otobe, K.; Watanabe, S.; Harada, K. Analysis of QTLs for the Micromorphology on the Seed Coat Surface of Soybean Using Recombinant Inbred Lines. Seed Sci. Res. 2015, 25, 409–415. [Google Scholar] [CrossRef]
- Uzogara, S.G.; Morton, I.D.; Daniel, J.W. Quality Changes and Mineral Content of Cowpea (Vigna unguiculata L. Walp.) Seeds Processed with ‘Kanwa’ Alkaline Salt. Food Chem. 1988, 30, 1–18. [Google Scholar] [CrossRef]
- Deorukhkar, A.; Ananthanarayan, L. Effect of Thermal Processing Methods on Flavonoid and Isoflavone Content of Decorticated and Whole Pulses. J. Food Sci. Technol. 2021, 58, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Huynh, B.-L.; Duong, T.; Clark, N.E.; Long, R.; Light, S.E.; Dahlquist-Willard, R.M.; Ehlers, J.D.; Close, T.J.; Roberts, P.A. Registration of Aphid-Resistant ‘California Blackeye 77’ Cowpea. J. Plant Regist. 2022, 16, 13–20. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).