Molecular Characterization and Expression of CmobHLH Genes in Pumpkin
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of CmobHLH Genes in Pumpkin
2.2. Phylogenetic Relationship, Gene Structure and Protein–Protein Interaction Networks of CmobHLH Proteins
2.3. Plant Materials, Growth Conditions and Treatments
2.4. Gene Expression Analysis
3. Results
3.1. Identification and Characterization of CmobHLH Family Genes
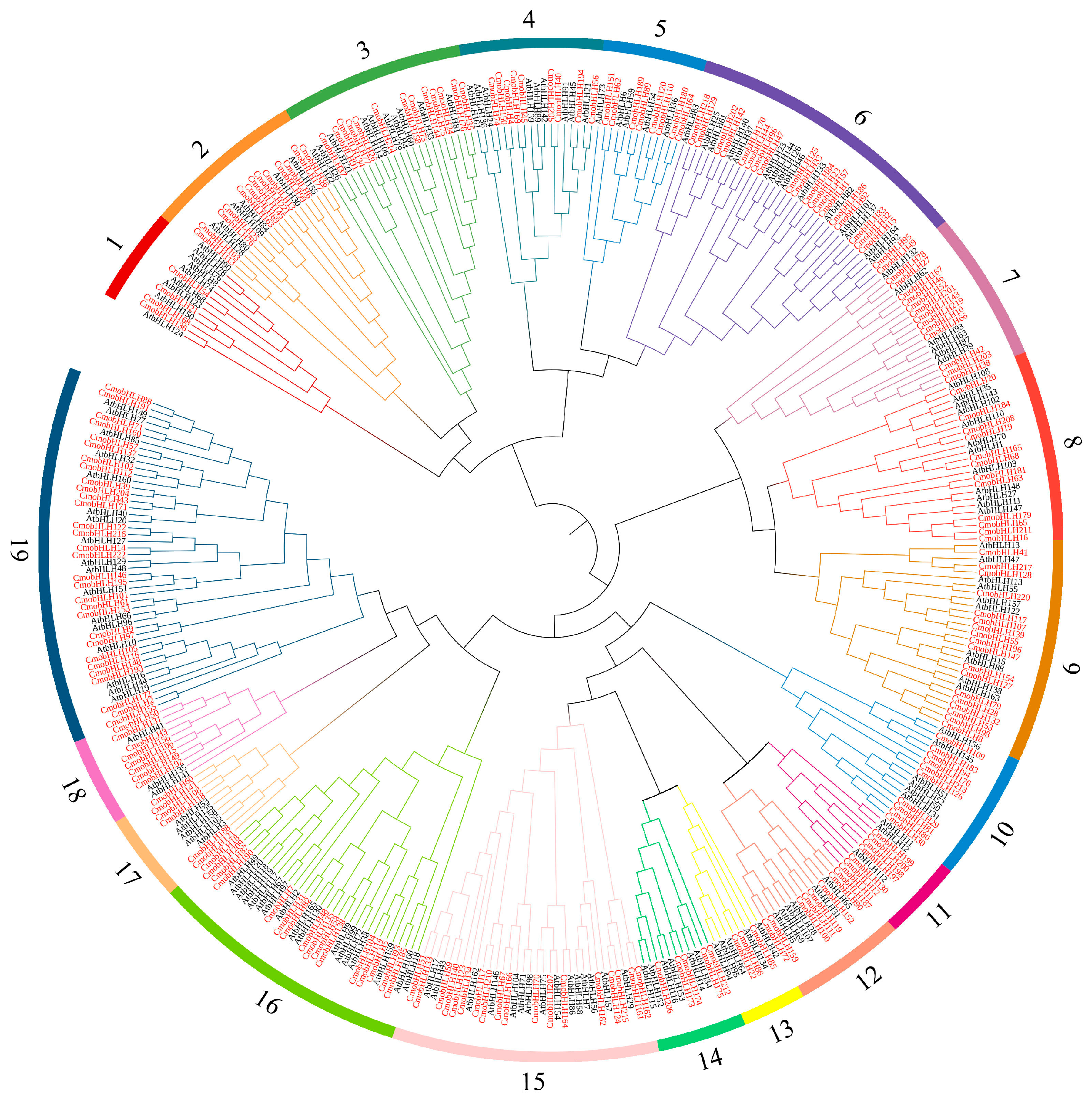
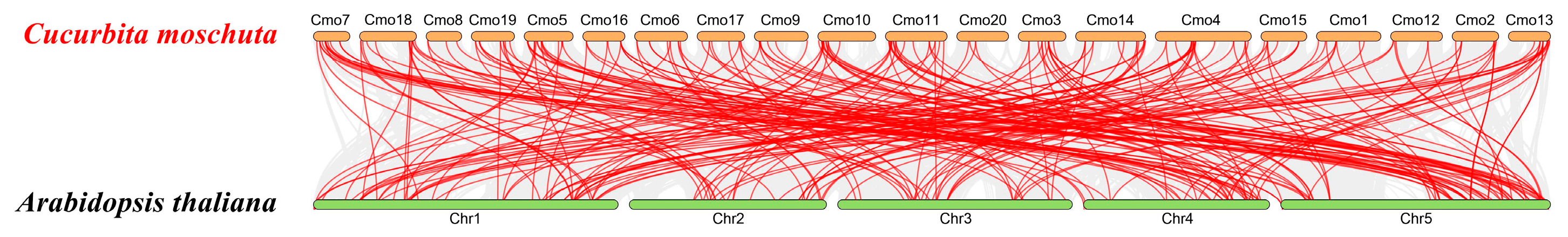
3.2. Phylogenetic Tree and Synteny Analysis of the CmobHLH Proteins
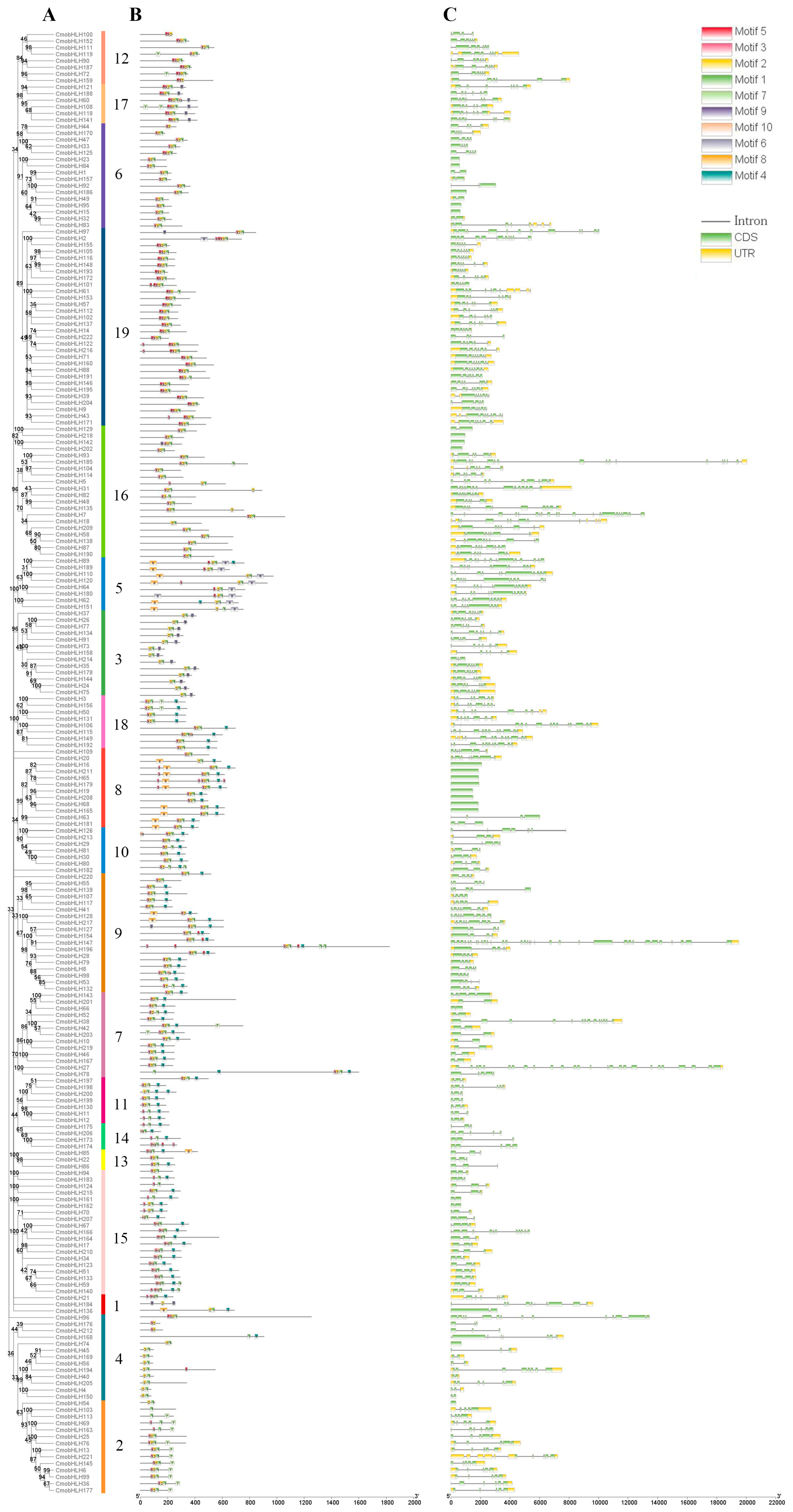
3.3. Conserved Motifs and Gene Structures of CmobHLH Genes
3.4. GO Enrichment and CREs Analysis
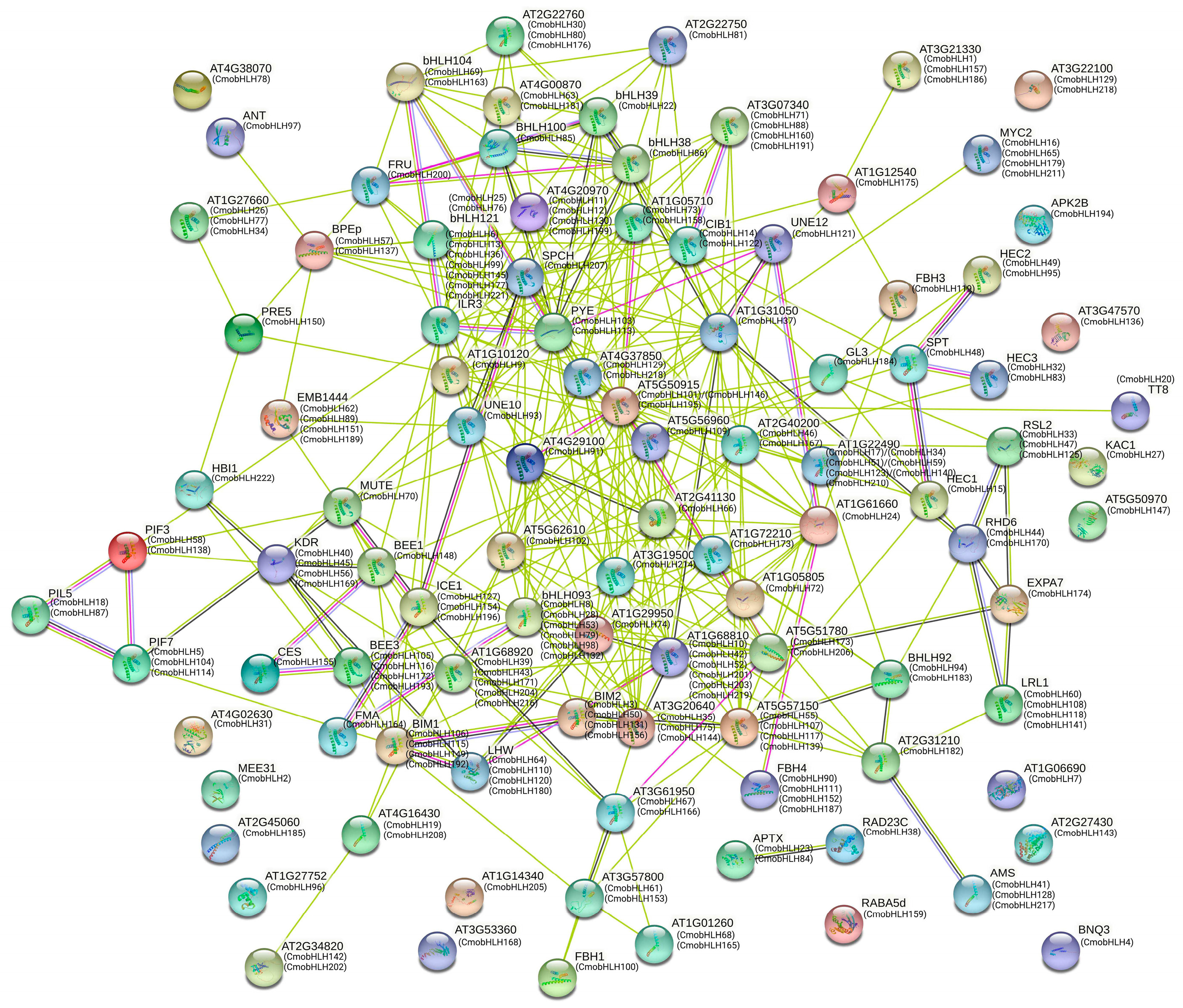
3.5. Protein–Protein Interaction Networks of CmobHLH Proteins
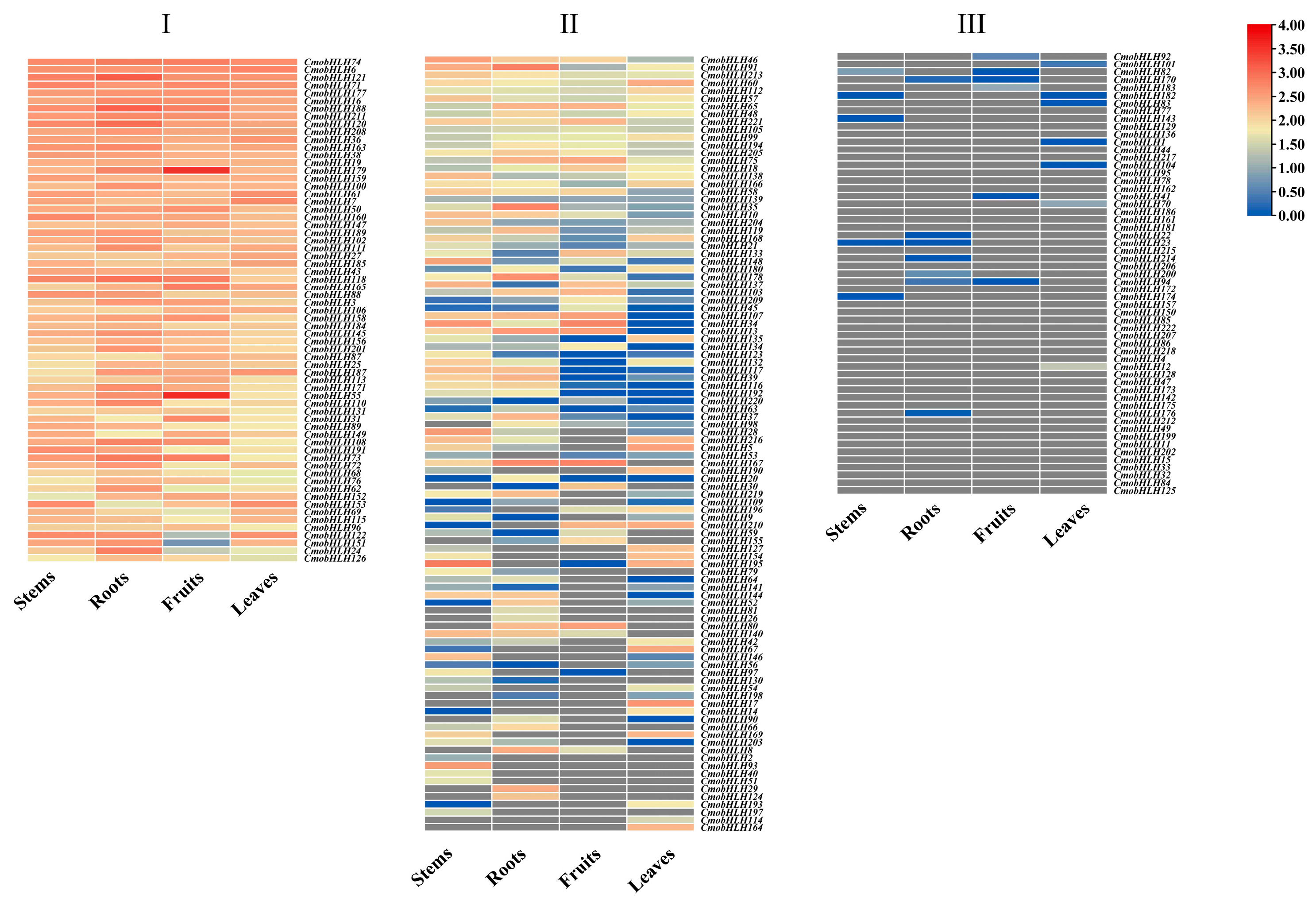
3.6. Tissue Expression of CmobHLH Genes
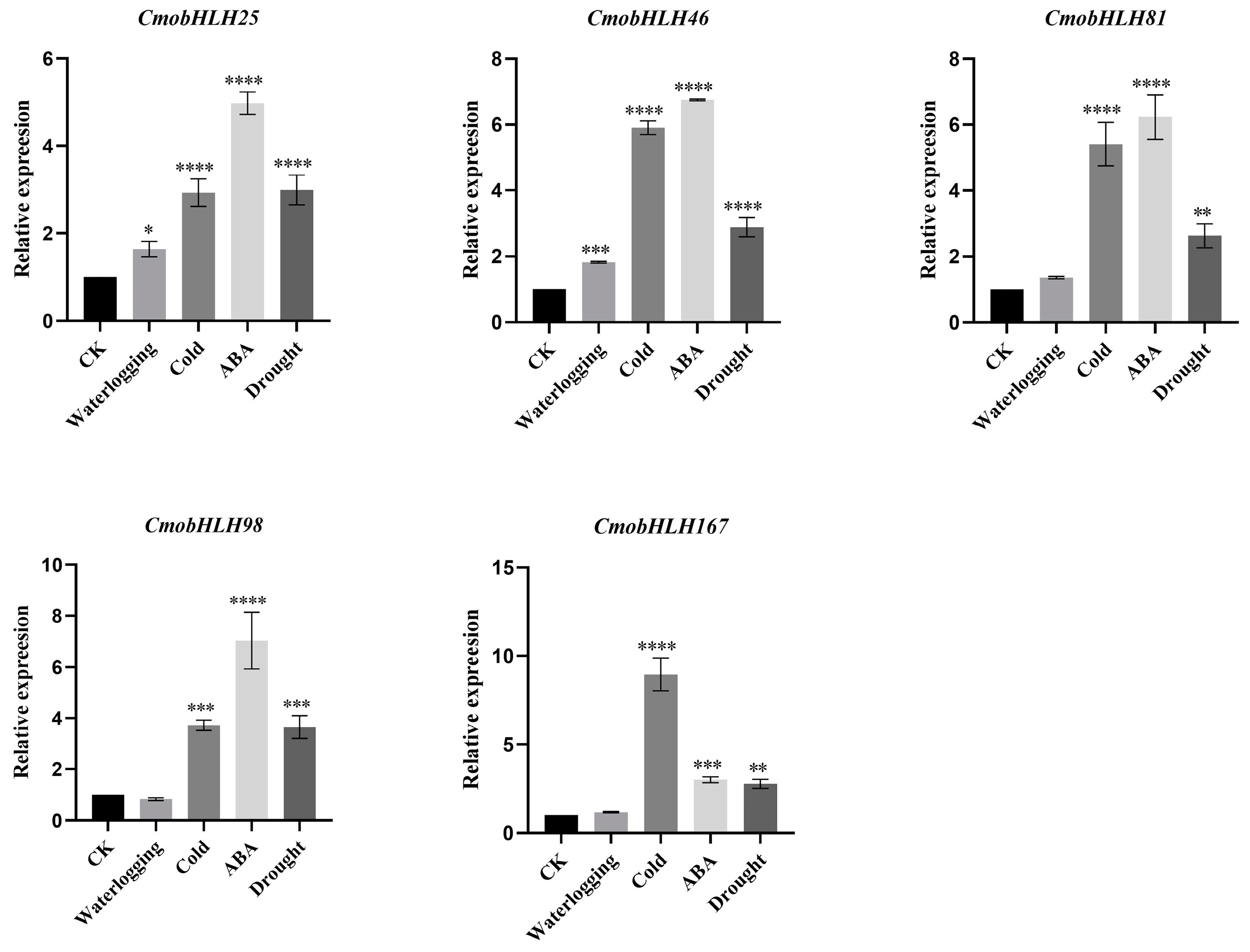
3.7. Expression Analysis of CmobHLH Genes under Abiotic Stresses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Carretero-Paulet, L.; Galstyan, A.; Villanova, I.R.; Martinez-Garcia, J.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef]
- Rehman, S.; Mahmood, T. Functional role of DREB and ERF transcription factors: Regulating stress-responsive network in plants. Acta Physiol. Plant. 2015, 37, 178. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, Y.L.; Kim, S.-U.; Chen, Z.X.; Nie, G.P.; Cheng, S.Y.; Ye, J.B.; Xu, F. Genome-wide identification and characterization of bHLH family genes from Ginkgo biloba. Sci. Rep. 2020, 10, 13723. [Google Scholar] [CrossRef]
- Qian, Y.C.; Zhang, T.Y.; Yu, Y.; Gou, L.P.; Yang, J.T.; Xu, J.; Pi, E. Regulatory mechanisms of bHLH transcription factors in plant adaptive responses to various abiotic stresse. Front. Plant Sci. 2021, 12, 677611. [Google Scholar] [CrossRef] [PubMed]
- Nuno, P.; Liam, D. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 2010, 27, 862–874. [Google Scholar] [CrossRef]
- Ke, Y.Z.; Wu, Y.W.; Zhou, H.J.; Chen, P.; Wang, M.M.; Liu, M.M.; Li, P.F.; Yang, J.; Li, J.N.; Du, H. Genome-wide survey of the bHLH super gene family in Brassica napus. BMC Plant Biol. 2020, 20, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Buck, M.J.; Atchley, W.R. Phylogenetic analysis of plant basic helix-loop-helix proteins. J. Mol. Evol. 2003, 56, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Massari, M.E.; Murre, C. Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 2000, 20, 429–440. [Google Scholar] [CrossRef]
- Hiroshi, A.; Takeshi, U.; Takuya, I.; Motoaki, S.; Kazuo, S.; Kazuko, Y.-S. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Wang, R.H.; Li, Y.Y.; Gao, M.G.; Han, M.; Liu, H.L. Genome-wide identification and characterization of the bHLH gene family and analysis of their potential relevance to chlorophyll metabolism in Raphanus sativus L. BMC Genom. 2022, 23, 548. [Google Scholar] [CrossRef]
- Ludwig, S.R.; Habera, L.F.; Dellaporta, S.L.; Wessler, S.R. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc. Natl. Acad. Sci. USA 1989, 86, 7092–7096. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef] [PubMed]
- Li, X.X.; Duan, X.P.; Jiang, H.X.; Sun, Y.J.; Tang, Y.P.; Yuan, Z.; Guo, J.K.; Liang, W.Q.; Chen, L.; Yin, J.Y.; et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006, 141, 1167–1184. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Hu, Z.Z.; Zhao, T.M.; Yang, Y.W.; Chen, T.Z.; Yang, M.L.; Yu, W.G.; Zhang, B.L. Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum). BMC Genom. 2015, 16, 39. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Gao, M.; Huang, L.; Wang, Y.; van Nocker, S.; Wan, R.; Guo, C.; Wang, X.; Gao, H. Identification and expression analysis of the apple (Malus × domestica) basic helix-loop-helix transcription factor family. Sci. Rep. 2017, 7, 28. [Google Scholar] [CrossRef]
- Xi, H.C.; He, Y.J.; Chen, H.Y. Functional characterization of SmbHLH13 in anthocyanin biosynthesis and flowering in eggplant. Hortic. Plant J. 2020, 7, 73–80. [Google Scholar] [CrossRef]
- Alsamman, A.M.; Abdelsattar, M.; El Allali, A.; Radwan, K.H.; Nassar, A.E.; Mousa, K.H.; Hussein, A.; Mokhtar, M.M.; Abd El-Maksoud, M.M.; Istanbuli, T.; et al. Genome-wide identification, characterization, and validation of the bHLH transcription factors in grass pea. Front. Genet. 2023, 14, 1128992. [Google Scholar] [CrossRef]
- Han, W.J.; Zhang, Q.; Suo, Y.J.; Li, H.W.; Diao, S.F.; Sun, P.; Huang, L.; Fu, J.M. Identification and expression analysis of the bHLH gene family members in Diospyros kaki. Horticulturae 2023, 9, 380. [Google Scholar] [CrossRef]
- Chang, S.W.; Li, Q.; Huang, B.K.; Chen, W.S.; Tan, H.X. Genome-wide identification and characterisation of bHLH transcription factors in Artemisia annua. BMC Plant Biol. 2023, 23, 63. [Google Scholar] [CrossRef]
- Liu, Z.; Fu, X.A.; Xu, H.; Zhang, Y.X.; Shi, Z.D.; Zhou, G.Z.; Bao, W.L. Comprehensive analysis of bHLH transcription factors in Ipomoea aquatica and its response to anthocyanin biosynthesis. Int. J. Mol. Sci. 2023, 24, 5652. [Google Scholar] [CrossRef]
- Liu, W.W.; Tai, H.H.; Li, S.S.; Gao, W.; Zhao, M.; Xie, C.X.; Li, W.X. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 2014, 201, 1192–1204. [Google Scholar] [CrossRef]
- Zuo, Z.F.; Lee, H.Y.; Kang, H.G. Basic Helix-Loop-Helix transcription factors: Regulators for plant growth development and abiotic stress responses. Int. J. Mol. Sci. 2023, 24, 1419. [Google Scholar] [CrossRef]
- Makkena, S.; Lamb, R.S. The bHLH transcription factor SPATULA is a key regulator of organ size in Arabidopsis thaliana. Plant Signal. Behav. 2013, 8, e24140. [Google Scholar] [CrossRef]
- Niu, N.; Liang, W.; Yang, X.; Jin, W.; Wilson, Z.A.; Hu, J.; Zhang, D. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 2013, 4, 1445. [Google Scholar] [CrossRef]
- Le Hir, R.; Castelain, M.; Chakraborti, D.; Moritz, T.; Dinant, S.; Bellini, C. AtbHLH68 transcription factor contributes to the regulation of ABA homeostasis and drought stress tolerance in Arabidopsis thaliana. Physiol. Plant. 2017, 160, 312–327. [Google Scholar] [CrossRef]
- Wang, W.S.; Zhu, J.; Lu, Y.T. Overexpression of AtbHLH112 suppresses lateral root emergence in Arabidopsis. Funct. Plant Biol. FPB 2014, 41, 342–352. [Google Scholar] [CrossRef]
- Sun, H.; Fan, H.J.; Ling, H.Q. Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genom. 2015, 16, 9. [Google Scholar] [CrossRef]
- Tan, C.; Qiao, H.L.; Ma, M.; Wang, X.; Tian, Y.Y.; Bai, S.; Hasi, A. Genome-wide identification and characterization of melon bHLH transcription factors in regulation of fruit development. Plants 2021, 10, 2271. [Google Scholar] [CrossRef]
- Zhao, M.R.; Li, J.; Zhu, L.; Chang, P.; Li, L.L.; Zhang, L.Y. Identification and characterization of MYB-bHLH-WD40 regulatory complex members controlling anthocyanidin biosynthesis in blueberry fruits development. Genes 2019, 10, 496. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, B.; Deyholos, M.K. Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Mol. Genet. Genom. MGG 2009, 282, 503–516. [Google Scholar] [CrossRef]
- Gu, X.Y.; Gao, S.X.; Li, J.; Song, P.Y.; Zhang, Q.; Guo, J.f.; Wang, X.Y.; Han, X.Y.; Wang, X.J.; Zhu, Y.; et al. The bHLH transcription factor regulated gene OsWIH2 is a positive regulator of drought tolerance in rice. Plant Physiol. Biochem. 2021, 169, 269–279. [Google Scholar] [CrossRef]
- Zhao, Q.; Fan, Z.H.; Qiu, L.N.; Che, Q.Q.; Wang, T.; Li, Y.Y.; Wang, Y.Z. MdbHLH130, an apple bHLH transcription factor, confers water stress resistance by regulating stomatal closure and ROS homeostasis in transgenic tobacco. Front. Plant Sci. 2020, 11, 543696. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Xiang, X.H.; Liu, D.; Yang, A.; Wang, Y.Y. Tobacco transcription factor NtbHLH123 confers tolerance to cold stress by regulating the NtCBF pathway and reactive oxygen species homeostasis. Front. Plant Sci. 2018, 9, 381. [Google Scholar] [CrossRef] [PubMed]
- Li, J.T.; Wang, L.N.; Zhu, W.; Wang, N.A.; Xin, H.P.; Li, S.H. Characterization of two VvICE1 genes isolated from ‘Muscat Hamburg’ grapevine and their effect on the tolerance to abiotic stresses. Sci. Hortic. 2014, 165, 266–273. [Google Scholar] [CrossRef]
- Sun, K.L.; Wang, H.Y.; Xia, Z.L. The maize bHLH transcription factor bHLH105 confers manganese tolerance in transgenic tobacco. Plant Sci. 2018, 280, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ying, Y.H.; Narsai, R.; Ye, L.X.; Zheng, L.Q.; Tian, J.L.; Whelan, J.; Shou, H.X. Identification of OsbHLH133 as a regulator of iron distribution between roots and shoots in Oryza sativa. Plant Cell Environ. 2013, 36, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Caili, F.; Huan, S.; Quanhong, L. A review on pharmacological activities and utilization technologies of pumpkin. Plant Foods Hum. Nutr. 2006, 61, 73–80. [Google Scholar] [CrossRef]
- Guo, W.L.; Chen, B.H.; Guo, Y.Y.; Chen, X.J.; Li, Q.F.; Yang, H.L.; Li, X.Z.; Zhou, J.G.; Wang, G.Y. Expression of Pumpkin CmbHLH87 Gene Improves Powdery Mildew Resistance in Tobacco. Front. Plant Sci. 2020, 11, 163. [Google Scholar] [CrossRef]
- Poole, R.L. The TAIR database. Methods Mol. Biol. 2007, 406, 179–212. [Google Scholar] [CrossRef]
- Zheng, Y.; Wu, S.; Bai, Y.; Sun, H.H.; Jiao, C.; Guo, S.G.; Zhao, K.; Blanca, J.; Zhang, Z.H.; Huang, S.W.; et al. Cucurbit Genomics Database (CuGenDB): A central portal for comparative and functional genomics of cucurbit crops. Nucleic Acids Res. 2019, 47, D1128–D1136. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.N.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2017, 5, e11335. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 9. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Peer, Y.V.d.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2020, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Torii, K.U. Hormonal and environmental signals guiding stomatal development. BMC Biol. 2018, 16, 21. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Song, Y.H.; Josephson-Day, A.R.; Miller, R.J.; Breton, G.; Olmstead, R.G.; Imaizumi, T. Flowering bHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 3582–3587. [Google Scholar] [CrossRef]
- Schuster, C.; Gaillochet, C.; Lohmann, J.U. Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Development 2015, 142, 3343–3350. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.H.; Zhao, Z.Z.; He, J.X. Brassinosteroid signaling in plant–microbe interactions. Int. J. Mol. Sci. 2018, 19, 4091. [Google Scholar] [CrossRef]
- Wu, Y.Y.; Wu, S.H.; Wang, X.Q.; Mao, T.Y.; Bao, M.Z.; Zhang, J.W.; Zhang, J. Genome-wide identification and characterization of the bHLH gene family in an ornamental woody plant Prunus mume. Hortic. Plant J. 2022, 8, 531–544. [Google Scholar] [CrossRef]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2004, 39, 366–380. [Google Scholar] [CrossRef]
- Naval, M.; Gil Muñoz, F.; Lloret, A.; Badenes, M.; Ríos, G. Molecular characterization of a TTG1-like gene expressed in persimmon fruit. Acta Hortic. 2017, 1172, 3359–3362. [Google Scholar] [CrossRef]
- Gao, F.; Robe, K.; Gaymard, F.; Izquierdo, E.; Dubos, C. The transcriptional control of iron homeostasis in plants: A tale of bHLH transcription factors? Front. Plant Sci. 2019, 10, 6. [Google Scholar] [CrossRef]
- Shupei, R.; Yuru, T.; Xinli, X.; Yue, L.; Jinhuan, C. Chromosome doubling mediates superior drought tolerance in Lycium ruthenicum via abscisic acid signaling. Hortic. Res. 2020, 7, 007. [Google Scholar]
- Hao, Y.Q.; Zong, X.M.; Ren, P.; Qian, Y.Q.; Fu, A.G. Basic Helix-Loop-Helix (bHLH) Transcription Factors Regulate a Wide Range of Functions in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 7152. [Google Scholar] [CrossRef] [PubMed]
- Long, T.A.; Tsukagoshi, H.; Busch, W.; Lahner, B.; Salt, D.E.; Benfey, P.N. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 2010, 22, 2219–2236. [Google Scholar] [CrossRef]
- Feng, Y.; Zeng, S.L.; Yan, J.P.; Li, K.Z.; Xu, H.N. Genome-wide analysis and expression of MYC family genes in tomato and the functional identification of slmyc1 in response to salt and drought stress. Agronomy 2023, 13, 757. [Google Scholar] [CrossRef]
- Kiribuchi, K.; Jikumaru, Y.; Kaku, H.; Minami, E.; Hasegawa, M.; Kodama, O.; Seto, H.; Okada, K.; Nojiri, H.; Yamane, H. Involvement of the Basic Helix-Loop-Helix transcription factor RERJ1 in wounding and drought stress responses in rice plants. J. Agric. Chem. Soc. Jpn. 2005, 69, 1042–1044. [Google Scholar] [CrossRef]
- Fan, M.; Bai, M.Y.; Kim, J.G.; Tina, W.; Eunkyoo, O.; Lawrence, C.; Ho, P.C.; Seung-Hyun, S.; Seong-Ki, K.; Beth, M.M.; et al. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell 2014, 26, 828–841. [Google Scholar] [CrossRef] [PubMed]
- Song, S.S.; Qi, T.C.; Fan, M.; Zhang, X.; Gao, H.; Huang, H.; Wu, D.W.; Guo, H.W.; Xie, D.X. The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genetics 2017, 9, e1003653. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.H.; Xu, H.L.; Zhao, H.G.; Liu, H.; Xu, L.; Liang, Z.S. Genome-wide investigation of bHLH genes and expression analysis under salt and hormonal treatments in Andrographis paniculata. Ind. Crops Prod. 2022, 183, 114928. [Google Scholar] [CrossRef]
- Goossens, J.; Mertens, J.; Goossens, A. Role and functioning of bHLH transcription factors in jasmonate signalling. J. Exp. Bot. 2017, 68, 1333–1347. [Google Scholar] [CrossRef]
- Song, M.Y.; Wang, H.M.; Wang, Z.; Huang, H.T.; Chen, S.W.; Ma, H.Q. Genome-wide characterization and analysis of bHLH transcription factors related to anthocyanin biosynthesis in Fig (Ficus carica L.). Front. Plant Sci. 2021, 12, 730692. [Google Scholar] [CrossRef]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, T.; Han, J.; Ren, Z.H. Genome-wide identification and characterization of cucumber bHLH family genes and the functional characterization of CsbHLH041 in NaCl and ABA tolerance in Arabidopsis and cucumber. BMC Plant Biol. 2020, 20, 272. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.J.; Wang, Z.Q.; Fu, Z.K.; Liu, D.N.; Yin, Y.L.; Li, H.G.; Gu, C.S. Genome-wide analysis of basic Helix-Loop-Helix family genes and expression analysis in response to drought and salt stresses in Hibiscus hamabo Sieb. et Zucc. Int. J. Mol. Sci. 2021, 22, 8748. [Google Scholar] [CrossRef] [PubMed]
- Zhan, H.; Liu, H.Z.; Ai, W.F.; Han, X.Y.; Wang, Y.; Lu, X.J. Genome-Wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in Mongolian Oak (Quercus mongolica). Curr. Issues Mol. Biol. 2023, 45, 1127–1148. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Yang, H.; Lai, D.L.; He, A.L.; Xue, G.X.; Feng, L.; Chen, L.; Cheng, X.B.; Ruan, J.J.; Yan, J.; et al. Genome-wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in sorghum [Sorghum bicolor (L.) Moench]. BMC Genom. 2021, 22, 415. [Google Scholar] [CrossRef]
- Wang, Z.; Jia, C.H.; Wang, J.Y.; Miao, H.X.; Liu, J.H.; Chen, C.; Yang, H.X.; Xu, B.Y.; Jin, Z.Q. Genome-wide analysis of basic Helix-Loop-Helix transcription factors to elucidate candidate genes related to fruit ripening and stress in banana (Musa acuminata L. AAA Group, cv. Cavendish). Front. Plant Sci. 2020, 11, 650. [Google Scholar] [CrossRef]
- Fan, Y.; Lai, D.L.; Yang, H.; Xue, G.X.; He, A.L.; Chen, L.; Feng, L.; Ruan, J.J.; Xiang, D.B.; Yan, J.; et al. Genome-wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in foxtail millet (Setaria italica L.). BMC Genom. 2021, 22, 778. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ao, W.; Luo, W.; Xu, W.; Wang, X.; Liu, J.; Sun, Y. Molecular Characterization and Expression of CmobHLH Genes in Pumpkin. Horticulturae 2023, 9, 648. https://doi.org/10.3390/horticulturae9060648
Ao W, Luo W, Xu W, Wang X, Liu J, Sun Y. Molecular Characterization and Expression of CmobHLH Genes in Pumpkin. Horticulturae. 2023; 9(6):648. https://doi.org/10.3390/horticulturae9060648
Chicago/Turabian StyleAo, Wenhong, Weirong Luo, Wenchen Xu, Xudong Wang, Junjun Liu, and Yongdong Sun. 2023. "Molecular Characterization and Expression of CmobHLH Genes in Pumpkin" Horticulturae 9, no. 6: 648. https://doi.org/10.3390/horticulturae9060648
APA StyleAo, W., Luo, W., Xu, W., Wang, X., Liu, J., & Sun, Y. (2023). Molecular Characterization and Expression of CmobHLH Genes in Pumpkin. Horticulturae, 9(6), 648. https://doi.org/10.3390/horticulturae9060648




