Genetic Mapping of a Candidate Gene ClIS Controlling Intermittent Stripe Rind in Watermelon
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Phenotypic Measurement
2.2. Marker Development and Fine Mapping
2.3. Prediction and Verification of Candidate Gene
2.4. RNA Extraction, cDNA Synthesis, and qRT-PCR Analysis
3. Results
3.1. Inheritance Features of Intermittent Stripe in Watermelon Rind
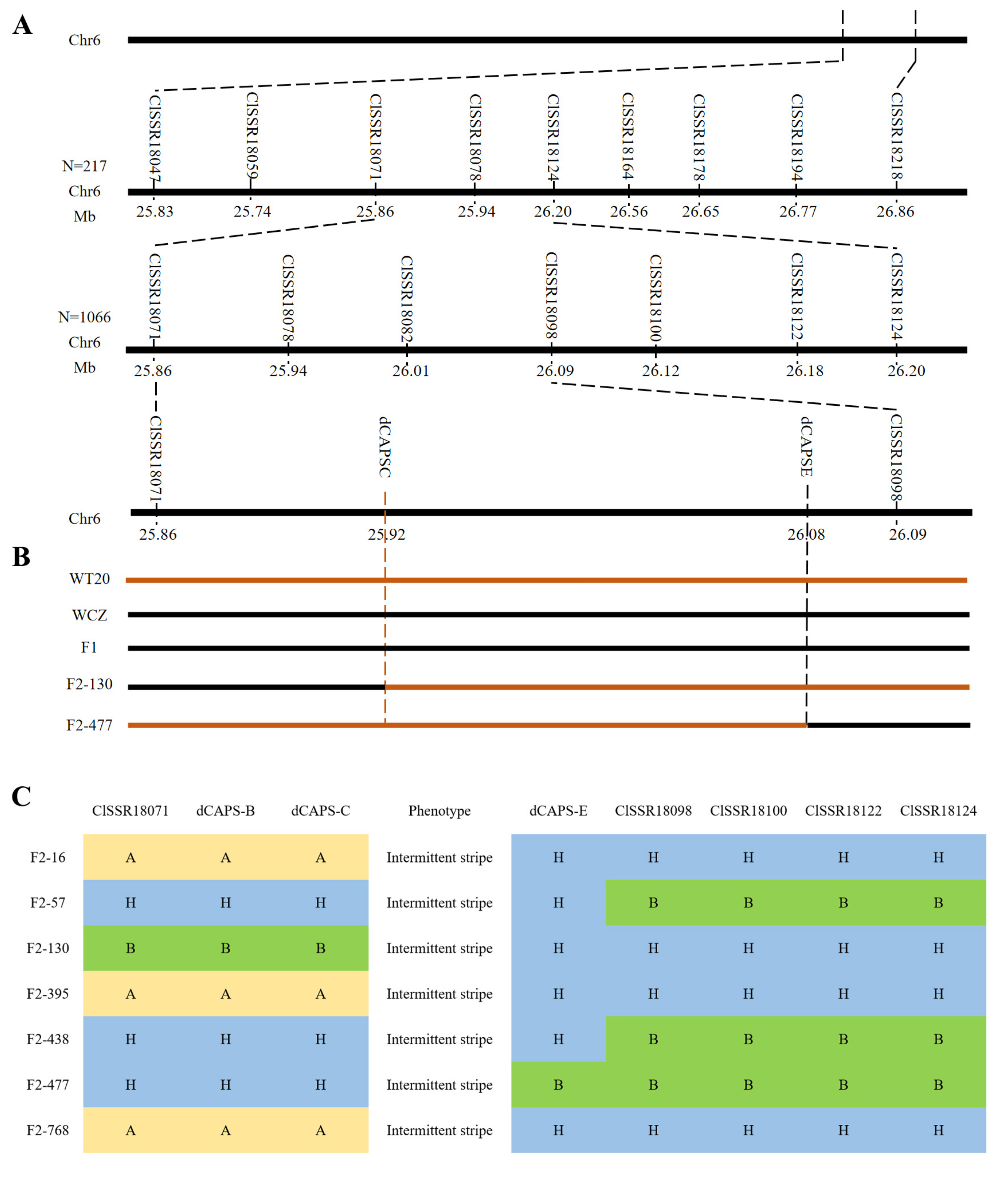
3.2. Fine Mapping of the ClIS Gene
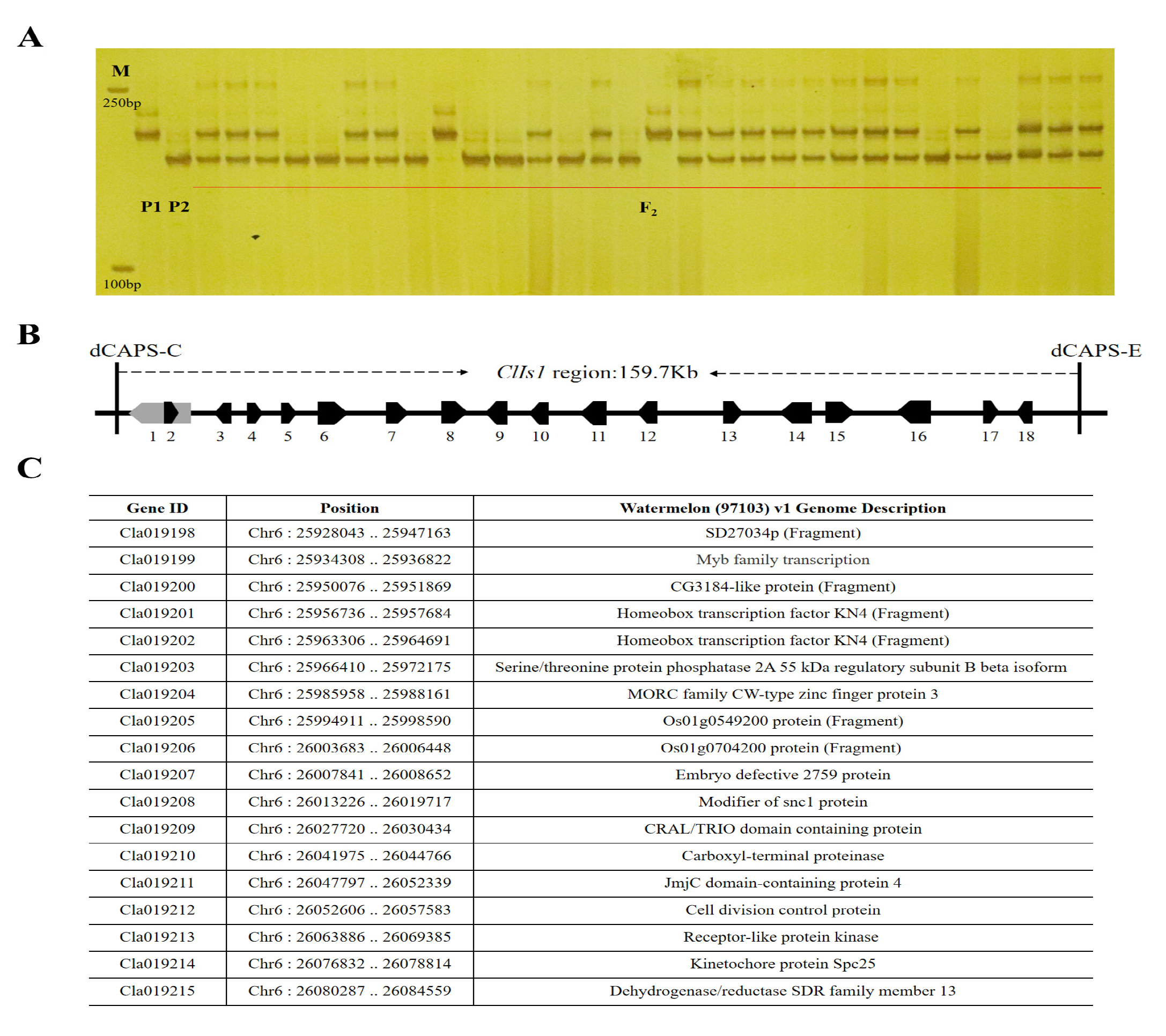
3.3. Sequence Alignment and Gene Identification
3.4. Expression Analysis of ClIS and ClGS Genes
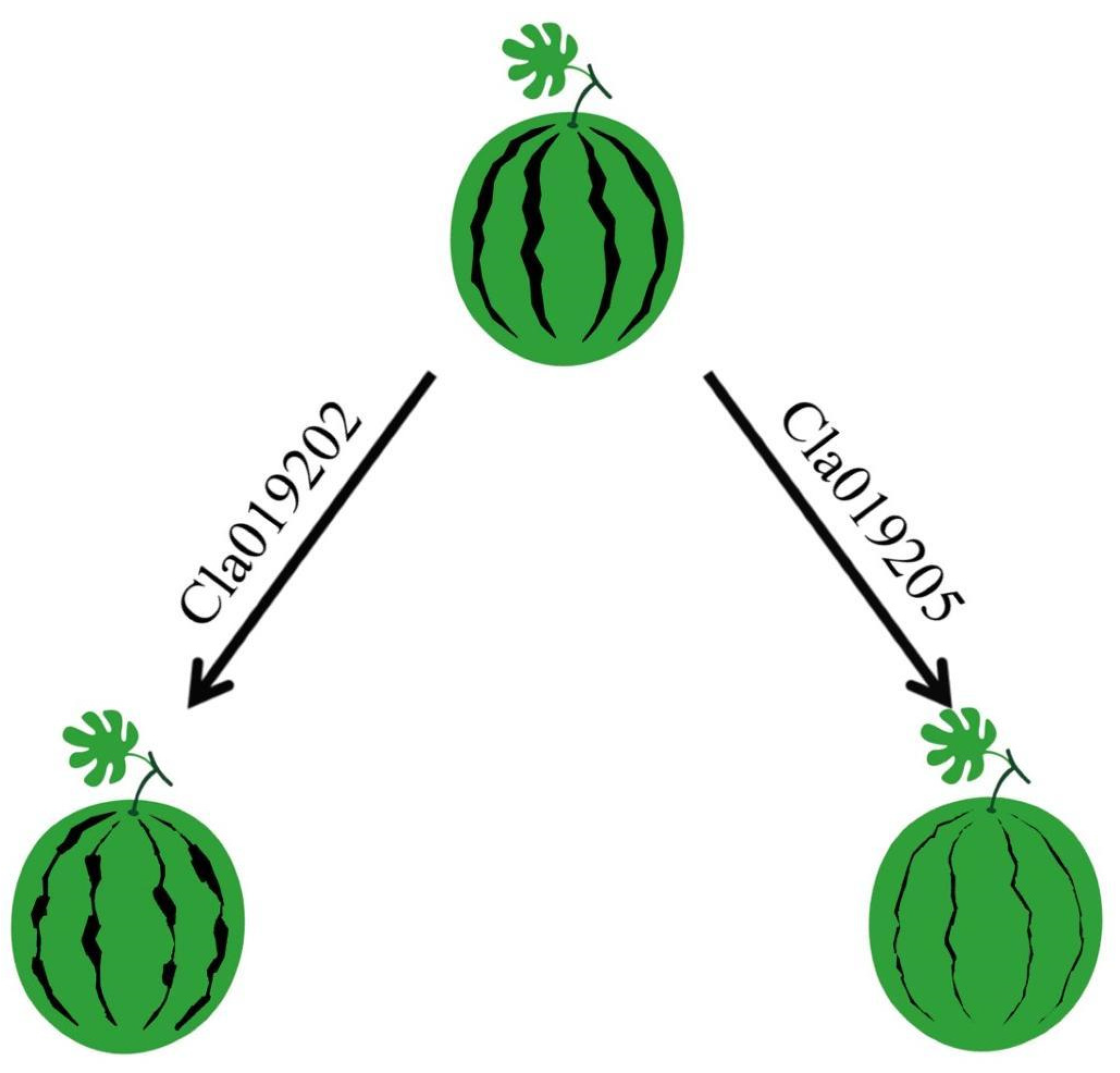
3.5. Formation of Watermelon Stripe Patterns
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Encinas-Viso, F.; Revilla, T.A.; van Velzen, E.; Etienne, R.S. Frugivores and cheap fruits make fruiting fruitful. J. Evol. Biol. 2014, 27, 313–324. [Google Scholar] [CrossRef]
- Pan, Y.; Bradley, G.; Pyke, K.; Ball, G.; Lu, C.; Fray, R. Network inference analysis identifies an APRR2-like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. 2013, 161, 1476–1485. [Google Scholar] [CrossRef] [PubMed]
- Nadakuduti, S.S.; Holdsworth, W.L.; Klein, C.L.; Barry, C.S. KNOX genes influence a gradient of fruit chloroplast development through regulation of GOLDEN2-LIKE expression in tomato. Plant J. 2014, 78, 1022–1033. [Google Scholar] [CrossRef] [PubMed]
- Powell, A.; Nguyen, C.V.; Hill, T.; Cheng, K.L.; Figueroa-Balderas, R.; Aktas, H.; Bennett, A.B. Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 2012, 336, 1711–1715. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Li, C.; Yu, H.; Tao, P.; Yuan, L.; Ye, J.; Zhang, Y. GREEN STRIPE, encoding methylated TOMATO AGAMOUS-LIKE 1, regulates chloroplast development and Chl synthesis in fruit. New Phytol. 2020, 228, 302–317. [Google Scholar] [CrossRef]
- Hao, N.; Du, Y.; Li, H.; Wang, C.; Wang, C.; Gong, S.; Zhou, S.; Wu, T. CsMYB36 is involved in the formation of yellow green peel in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2018, 131, 1659–1669. [Google Scholar] [CrossRef]
- Lun, Y.; Wang, X.; Zhang, C.; Yang, L.; Gao, D.; Chen, H.; Huang, S. A CsYcf54 variant conferring light green coloration in cucumber. Euphytica 2016, 208, 509–517. [Google Scholar] [CrossRef]
- Zhou, Q.; Wang, S.; Hu, B.; Chen, H.; Zhang, Z.; Huang, S. An accumulation and replication of chloroplasts 5 gene mutation confers light green peel in cucumber. J. Integr. Plant Biol. 2015, 57, 936–942. [Google Scholar] [CrossRef]
- Tang, H.; Dong, X.; Wang, J.; Xia, J.; Xie, F.; Zhang, Y.; Wan, Z.J. Fine Mapping and candidate gene prediction for white immature fruit skin in cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2018, 19, 1493. [Google Scholar] [CrossRef]
- Song, M.; Zhang, M.; Cheng, F.; Wei, Q.; Wang, J.; Davoudi, M.; Lou, Q. An irregularly striped rind mutant reveals new insight into the function of PG1β in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2020, 133, 371–382. [Google Scholar] [CrossRef]
- Telias, A.; Lin-Wang, K.; Stevenson, D.E.; Cooney, J.M.; Hellens, R.P.; Allan, A.C.; Hoover, E.; Bradeen, J.M. Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol. 2011, 11, 93. [Google Scholar] [CrossRef] [PubMed]
- Meng, R.; Zhang, J.; An, L.; Zhang, B.; Jiang, X.; Yang, Y.; Zhao, Z. Expression profiling of several gene families involved in anthocyanin biosynthesis in apple (Malus domestica Borkh.) skin during fruit development. J. Plant Growth Regul. 2016, 35, 449–464. [Google Scholar] [CrossRef]
- Huh, J.; Kang, B.; Nahm, S.; Kim, S.; Ha, K.; Lee, M.; Kim, B. A candidate gene approach identified phytoene synthase as the locus for mature fruit color in red pepper (Capsicum spp.). Theor. Appl. Genet. 2001, 102, 524–530. [Google Scholar] [CrossRef]
- Qian, M.; Sun, Y.; Allan, A.C.; Teng, Y.; Zhang, D. The red sport of ‘Zaosu’ pear and its red-striped pigmentation pattern are associated with demethylation of the PyMYB10 promoter. Phytochemistry 2014, 107, 16–23. [Google Scholar] [CrossRef]
- Jiang, B.; Liu, W.; Xie, D.; Peng, Q.; He, X.; Ye, L.; Liang, Z. High-density genetic map construction and gene mapping of pericarp color in wax gourd using specific-locus amplified fragment (SLAF) sequencing. BMC Genom. 2015, 16, 1035. [Google Scholar] [CrossRef]
- Jin, W.; Wang, H.; Li, M.; Wang, J.; Yang, Y.; Zhang, X.; Zhang, K. The R2R3-MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol. J. 2016, 14, 2120–2133. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, J.; Sun, H.; Salse, J.; Lucas, W.J.; Zhang, H.; Xu, Y. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 2013, 45, 51–58. [Google Scholar] [CrossRef]
- Wu, S.; Wang, X.; Reddy, U.; Sun, H.H.; Bao, K.; Gao, L.; Fei, Z. Genome of ‘Charleston Gray’, the principal American watermelon cultivar, and genetic characterization of 1365 accessions in the US National Plant Germplasm System watermelon collection. Plant Biotechnol. J. 2019, 17, 2246–2258. [Google Scholar] [CrossRef]
- Gusmini, G.; Wehner, T.C. Genes determining rind pattern inheritance in watermelon: A review. Hortscience 2005, 40, 1928–1930. [Google Scholar] [CrossRef]
- Gusmini, G.; Wehner, T.C. Qualitative inheritance of rind pattern and flesh color in watermelon. J. Hered. 2006, 97, 177–185. [Google Scholar] [CrossRef]
- Guner, N.; Wehner, T.C. The genes of watermelon. Hortscience 2004, 39, 1175–1182. [Google Scholar] [CrossRef]
- Poole, C. Genetics of cultivated cucurbits. J. Hered. 1944, 35, 122–128. [Google Scholar] [CrossRef]
- Liu, D.; Sun, D.; Liang, J.; Dou, J.; Yang, S.; Zhu, H.; Yang, L. Characterization and bulk segregant analysis of ‘moon and star’ appearance in watermelon. Sci. Hortic. 2021, 285, 110140. [Google Scholar] [CrossRef]
- Weetman, L.M. Inheritance and correlation of shape size, and color in the watermelon. Iowa Agric. Exp. Stn. Res. Bull. 1937, 228, 222–256. [Google Scholar]
- Kumar, R.; Wehner, T.C. Discovery of second gene for solid dark green versus light green rind pattern in watermelon. J. Hered. 2011, 102, 489–493. [Google Scholar] [CrossRef]
- Dou, J.; Lu, X.; Ali, A.; Zhao, S.; Zhang, L.; He, N.; Liu, W. Genetic mapping reveals a marker for yellow skin in watermelon (Citrullus lanatusL.). PLoS ONE 2018, 13, e0200647. [Google Scholar] [CrossRef]
- Wehner, T.C.; Lou, L. Qualitative inheritance of external fruit traits in watermelon. Hortscience 2016, 51, 487–496. [Google Scholar]
- Zhu, H.; Song, P.; Koo, D.H.; Guo, L.; Li, Y.; Sun, S.; Weng, Y.; Yang, L. Genome wide characterization of simple sequence repeats in watermelon genome and their application in comparative mapping and genetic diversity analysis. BMC Genom. 2016, 17, 557. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Y.; Sun, L.; Qiu, G.; Sun, Y.; Zhu, Z.; Wang, X. Construction of a genetic map for Citrullus lanatus based on CAPS markers and mapping of three qualitative traits. Sci. Hortic. 2017, 233, 532–538. [Google Scholar] [CrossRef]
- Li, B.; Zhao, S.; Dou, J.; Ali, A.; Gebremeskel, H.; Gao, L.; Liu, W. Genetic mapping and development of molecular markers for a candidate gene locus controlling rind color in watermelon. Theor. Appl. Genet. 2019, 132, 2741–2753. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, M.; Xu, N.; Yang, S.; Dou, J.; Liu, D.; Sun, S. Fine mapping a ClGS gene controlling dark-green stripe rind in watermelon. Sci. Hortic. 2022, 291, 110583. [Google Scholar] [CrossRef]
- Gama, R.N.; Santos, C.A.; Dias, R.C.; Alves, J.C.; Nogueira, T.O. Microsatellite markers linked to the locus of the watermelon fruit stripe pattern. Genet. Mol. Res. 2015, 14, 269–276. [Google Scholar] [CrossRef]
- Kim, H.; Han, D.; Kang, J.; Choi, Y.; Levi, A.; Lee, G.P.; Park, Y. Sequence characterized amplified polymorphism markers for selecting rind stripe pattern in watermelon (Citrullus lanatusL.). Hortic. Environ. Biotechnol. 2015, 56, 341–349. [Google Scholar] [CrossRef]
- Park, S.W.; Kim, K.T.; Kang, S.C.; Yang, H.B. Rapid and practical molecular marker development for rind traits in watermelon. Hortic. Environ. Biotechnol. 2016, 57, 385–391. [Google Scholar] [CrossRef]
- Yue, Z.; Ma, R.; Cheng, D.; Yan, X.; He, Y.; Wang, C.; Wei, C. Candidate gene analysis of watermelon stripe pattern locus ClSP ongoing recombination suppression. Theor. Appl. Genet. 2021, 134, 3263–3277. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Zhao, S.; Sun, H.; Wang, X.; Wu, S.; Lin, T.; Xu, Y. Resequencing of 414 cultivated and wild watermelon accessions identifies selection for fruit quality traits. Nat. Genet. 2019, 51, 1616–1623. [Google Scholar] [CrossRef]
- Neff, M.M.; Neff, J.D.; Chory, J.; Pepper, A.E. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 1998, 14, 387–392. [Google Scholar] [CrossRef]
- Dou, J.; Yang, H.; Sun, D.; Yang, S.; Sun, S.; Zhao, S.; Yang, L. The branchless gene Clbl in watermelon encoding a TERMINAL FLOWER1 protein regulates the number of lateral branches. Theor. Appl. Genet. 2022, 135, 65–79. [Google Scholar] [CrossRef]
- Kong, Q.; Yuan, J.; Gao, L.; Zhao, L.; Cheng, F.; Huang, Y.; Bie, Z. Evaluation of appropriate reference genes for gene expression normalization during watermelon fruit development. PLoS ONE 2015, 10, e0130865. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Takagi, H.; Abe, A.; Yoshida, K.; Kosugi, S.; Natsume, S.; Mitsuoka, C.; Terauchi, R. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013, 74, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Dou, J.; Zhao, S.; Lu, X.; He, N.; Zhang, L.; Ali, A.; Kuang, H.; Liu, W. Genetic mapping reveals a candidate gene (ClFS1) for fruit shape in watermelon (Citrullus lanatusL.). Theor. Appl. Genet. 2018, 131, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Zhu, C.; Yang, L.; Zhao, W.; Ma, R.; Li, H.; Zhang, X. A point mutation resulting in a 13bp deletion in the coding sequence of Cldf leads to a GA-deficient dwarf phenotype in watermelon. Hortic. Res. 2019, 6, 132. [Google Scholar] [CrossRef]
- Gebremeskel, H.; Dou, J.; Li, B.; Zhao, S.; Muhammad, U.; Lu, X.; He, N.; Liu, W. Molecular mapping and candidate gene analysis for GA3 responsive short internode in watermelon (Citrullus lanatus). Int. J. Mol. Sci. 2020, 21, 290. [Google Scholar] [CrossRef]
- Liao, N.; Hu, Z.; Li, Y.; Hao, J.; Chen, S.; Xue, Q.; Zhang, M. Ethylene-responsive factor 4 is associated with the desirable rind hardness trait conferring cracking resistance in fresh fruits of watermelon. Plant Biotechnol. J. 2020, 18, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zhu, K.; Sun, Q.; Zhang, W.; Wang, X.; Cao, H.; Deng, X. Natural variation in CCD4 promoter underpins species-specific evolution of red coloration in Citrus peel. Mol. Plant 2019, 12, 1294–1307. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Duan, S.; Kang, Q.; Liu, D.; Yang, S.; Niu, H.; Zhu, H.; Sun, S.; Hu, J.; Dou, J.; et al. Genetic Mapping of a Candidate Gene ClIS Controlling Intermittent Stripe Rind in Watermelon. Horticulturae 2023, 9, 263. https://doi.org/10.3390/horticulturae9020263
Wang Y, Duan S, Kang Q, Liu D, Yang S, Niu H, Zhu H, Sun S, Hu J, Dou J, et al. Genetic Mapping of a Candidate Gene ClIS Controlling Intermittent Stripe Rind in Watermelon. Horticulturae. 2023; 9(2):263. https://doi.org/10.3390/horticulturae9020263
Chicago/Turabian StyleWang, Yinping, Shixiang Duan, Qishuai Kang, Dongming Liu, Sen Yang, Huanhuan Niu, Huayu Zhu, Shouru Sun, Jianbin Hu, Junling Dou, and et al. 2023. "Genetic Mapping of a Candidate Gene ClIS Controlling Intermittent Stripe Rind in Watermelon" Horticulturae 9, no. 2: 263. https://doi.org/10.3390/horticulturae9020263
APA StyleWang, Y., Duan, S., Kang, Q., Liu, D., Yang, S., Niu, H., Zhu, H., Sun, S., Hu, J., Dou, J., & Yang, L. (2023). Genetic Mapping of a Candidate Gene ClIS Controlling Intermittent Stripe Rind in Watermelon. Horticulturae, 9(2), 263. https://doi.org/10.3390/horticulturae9020263





