Overexpression of Cytosolic Glyceraldehyde-3-Phosphate Dehydrogenase 1 Gene Improves Nitrogen Absorption and Utilization in Potato
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation and Detection of Transgenic Potato
2.2. Potato Seedling Culture
2.3. Measurement of Physiological Parameters and Enzyme Activity
2.4. Determination Method for Ion Flux Rate
2.5. Real-Time Reverse Transcription–PCR Analysis of Gene Expression
2.6. Data Processing
3. Results
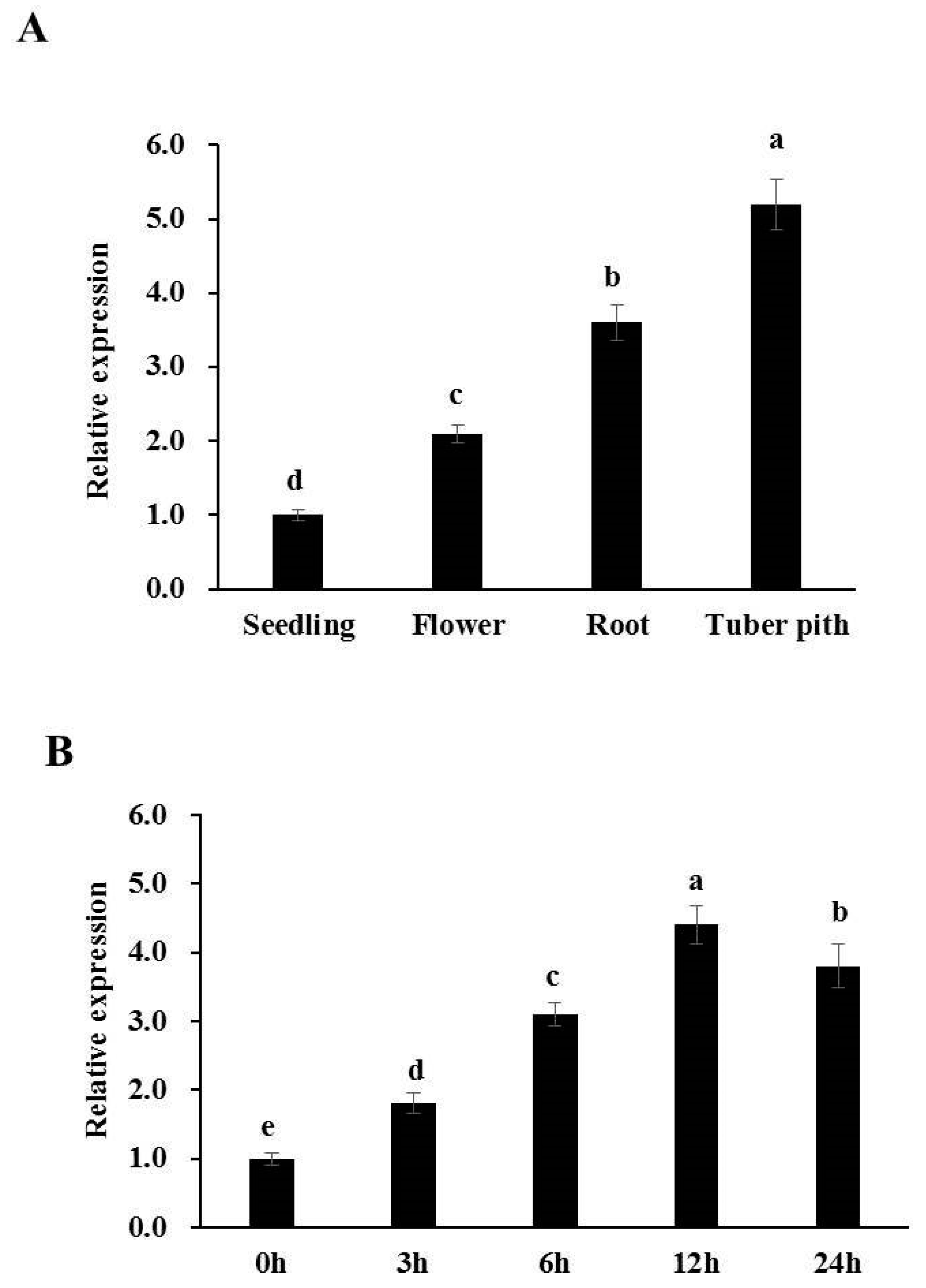
3.1. StGAPC1 Is a Nitrate-Inducible Gene
3.2. Overexpression of StGAPC1 Promotes Seedling Growth under LN Stress
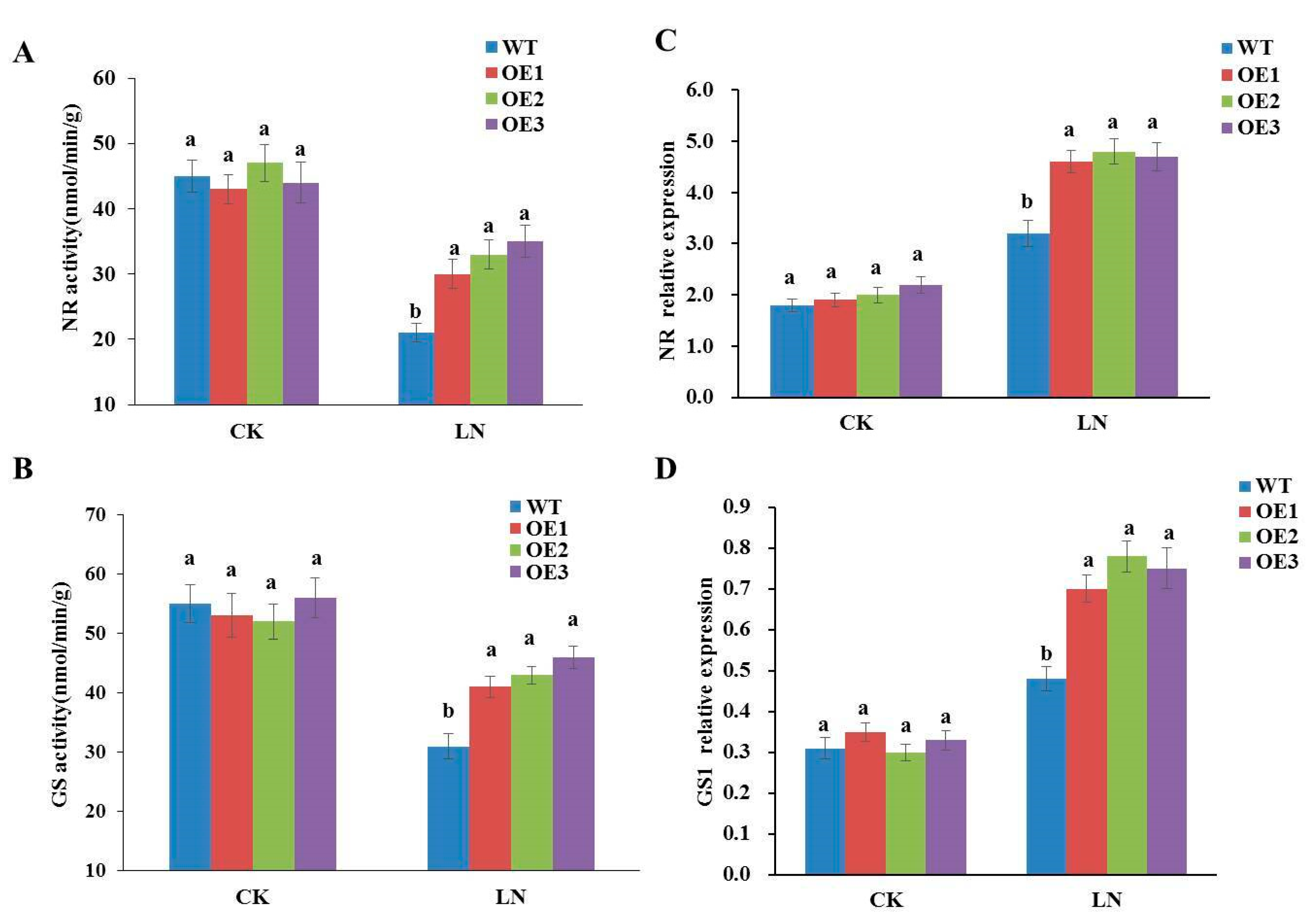
3.3. Overexpression of StGAPC1 Increases Enzyme Activity and Expression Level
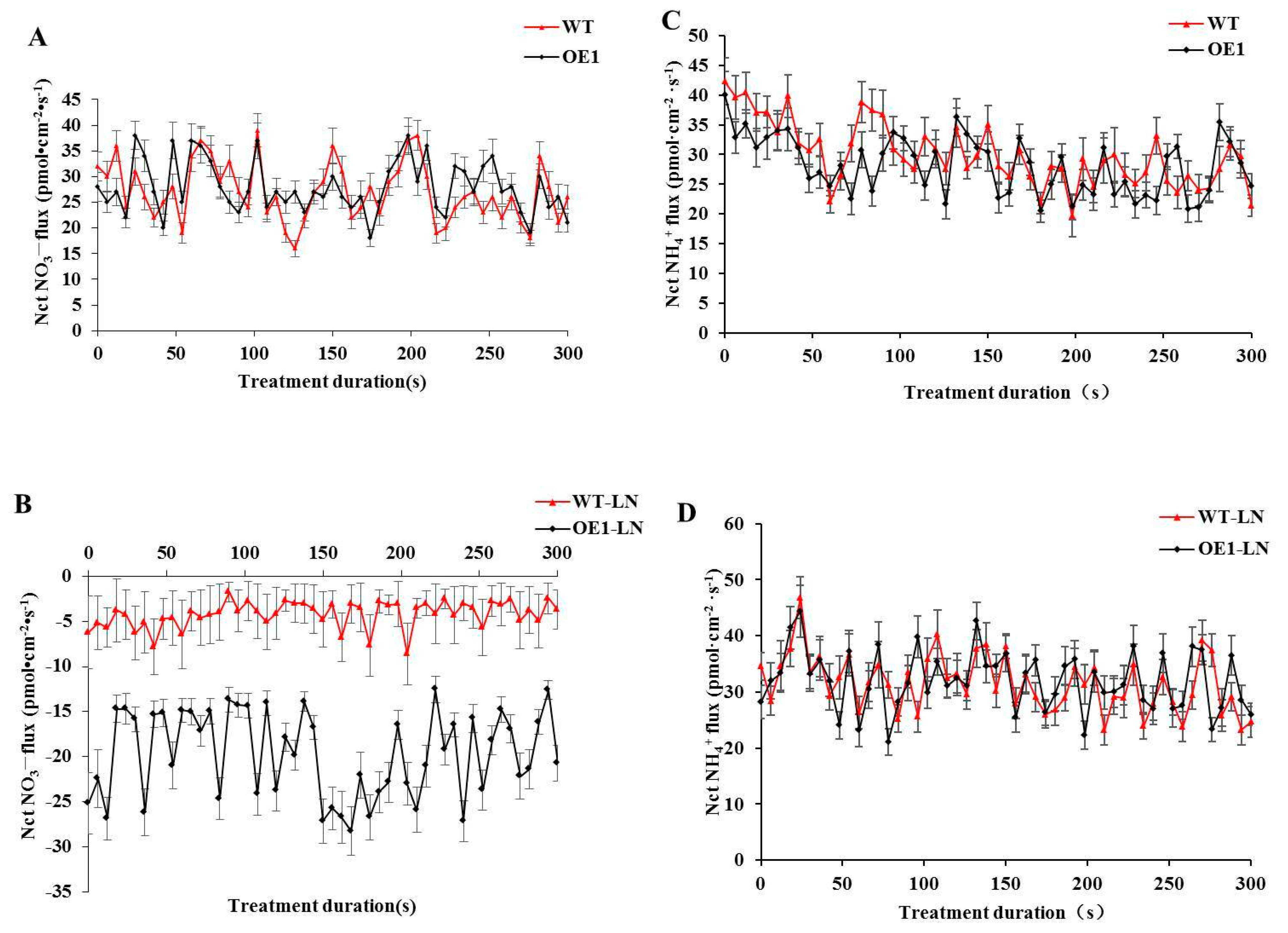
3.4. Overexpression of StGAPC1 Increases the Root NO3− Influx Rates
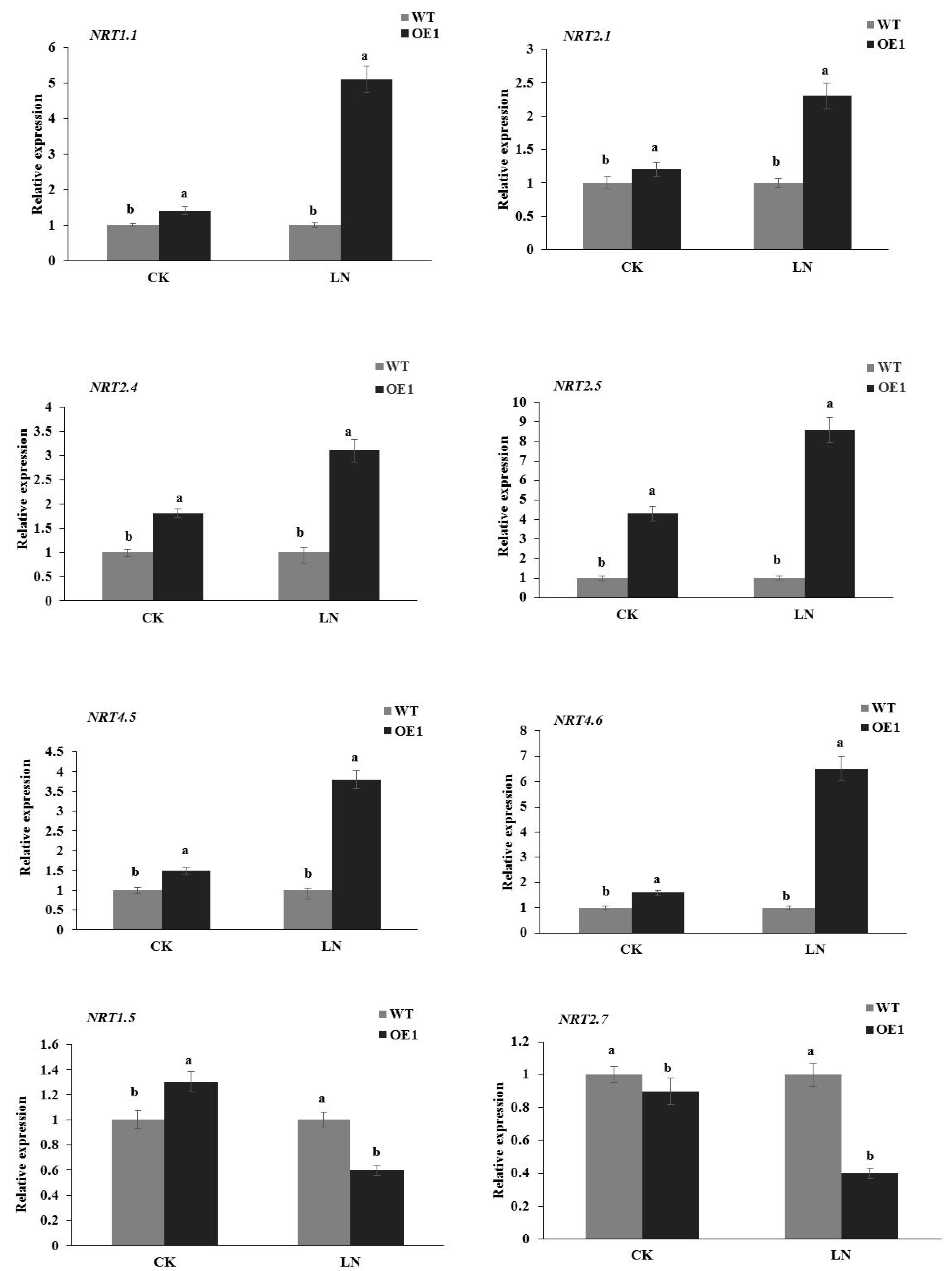
3.5. Overexpression of StGAPC1 Upregulates the Expression of Nitrate Transporters in Roots
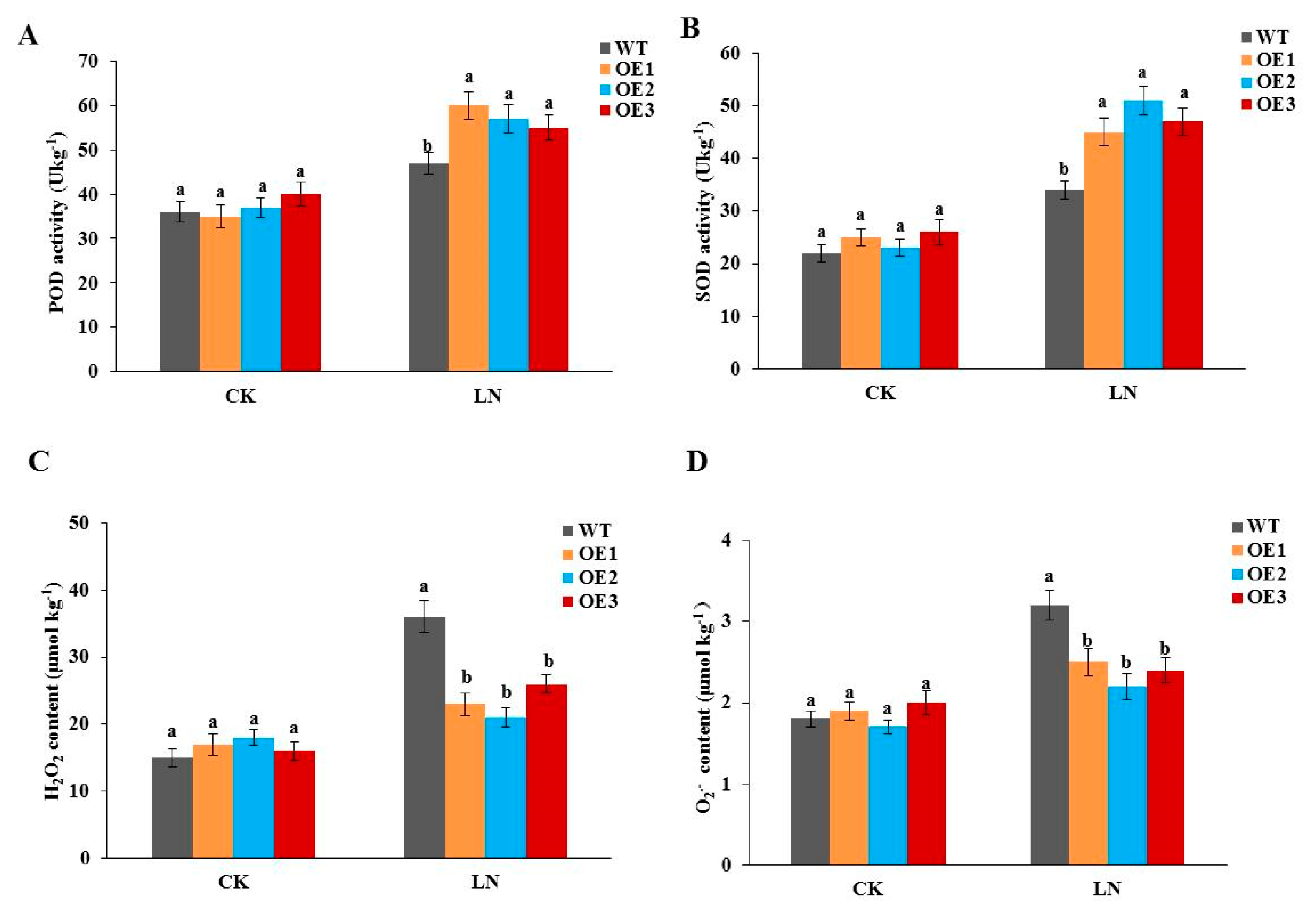
3.6. Overexpressing StGAPC1 Increases ROS System Elimination
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ladha, J.K.; Tirolpadre, A.; Reddy, C.K.; Cassman, K.G.; Verma, S.; Powlson, D.S.; van Kessel, C.; Richter, D.D.B.; Chakraborty, D.; Pathak, H. Global nitrogen budgets in cereals: A 50-year assessment for maize, rice, and wheat production systems. Sci. Rep. 2016, 6, 19355. [Google Scholar] [CrossRef] [PubMed]
- Bobbink, R.; Hicks, K.; Galloway, J.; Spranger, T.; Alkemade, R.; Ashmore, M.; Cinderby, S.; Davidson, E.; Dentener, F.; Emmett, B.; et al. Global assessment of nitrogen deposition effects on terrestrial plant diversity: A synthesis. Ecol. Appl. 2010, 20, 30–59. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, Y.; Chen, K.; Tsay, Y. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.A.A.; Vega, A.; Bouguyon, E.; Krouk, G.; Gojon, A.; Coruzzi, G.; Gutiérrez, R.A.A. Nitrate transport, sensing, and responses in plants. Mol. Plant. 2016, 9, 837–856. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Von, W.N. Ammonium as a signal for physiological and morphological responses in plants. J. Exp. Bot. 2017, 68, 2581–2592. [Google Scholar] [CrossRef] [PubMed]
- Lam, H.M.; Coschigano, K.T.; Oliveira, I.C.; Melo-Oliveira, R.; Coruzzi, G.M. The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 569–593. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.Y.; Duan, Y.K.; Wu, Y.Q.; Zhang, C.H.; Wu, W.L.; Lyu, L.F.; Li, W.L. Physiological and transcriptional responses of carbohydrate and nitrogen metabolism and ion balance in blueberry plants under nitrogen deficiency. Plant Growth Regul. 2023, 101, 519–535. [Google Scholar] [CrossRef]
- Iwamoto, M.; Tagiri, K. MicroRNA-targeted transcription factor gene RDD1 promotes nutrient ion uptake and accumulation in rice. Plant J. 2016, 85, 466–477. [Google Scholar] [CrossRef]
- Jia, Z.; Giehl, R.F.H. The root foraging response under low nitrogen depends on DWARF1-mediated brassinosteroid biosynthesis. Plant Physiol. 2020, 183, 998–1010. [Google Scholar] [CrossRef]
- Tiwari, J.K.; Buckseth, T.; Devi, S.; Varshney, S.; Sahu, S.; Patil, V.U.; Zinta, R.; Ali, N.; Moudgil, V.; Singh, R.K.; et al. Physiological and genome-wide RNA-sequencing analyses identify candidate genes in a nitrogen-use efficient potato cv. Kufri Gaurav. Plant Physiol. Biochem. 2020, 154, 171–183. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Wang, Y.P.; Zhao, Y.F.; Zhang, Y.; Zhang, J.Y.; Ma, H.R.; Han, Y.Z. Transcriptome analysis reveals Nitrogen deficiency induced alterations in leaf and root of three cultivars of potato (Solanum tuberosum L.). PLoS ONE 2020, 15, e0240662. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Hang, J.N.; Wu, B.L.; Wei, X.L.; Zhao, Q.Z.; Fang, Z.M. Co-overexpression of genes for nitrogen transport, assimilation, and utilization boosts rice grain yield and nitrogen use efficiency. Crop J. 2023, 3, 785–799. [Google Scholar] [CrossRef]
- Zhu, Y.N.; Zhong, L.H.; Huang, X.M.; Su, W.; Liu, H.C.; Sun, G.W.; Song, S.W.; Chen, R.Y. BcAMT1;5 mediates nitrogen uptake and assimilation in flowering Chinese cabbage and improves plant growth when overexpressed in Arabidopsis. Horticulturae 2023, 9, 43. [Google Scholar] [CrossRef]
- Rius, S.P.; Casati, P.; Iglesias, A.A.; Gomez, C.D.F. Characterization of an Arabidopsis thaliana mutant lacking a cytosolic non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase. Plant Mol. Biol. 2006, 61, 945–957. [Google Scholar] [CrossRef] [PubMed]
- Rius, S.P.; Casati, P.; Iglesias, A.A.; Gomez, C.D.F. Characterization of Arabidopsis lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. Plant Physiol. 2008, 148, 1655–1667. [Google Scholar] [CrossRef] [PubMed]
- Munoz., B.J.; Cascales, M.B.; Mulet, J.M.; Baroja, F.E.; Pozueta, R.J.; Kuhn, J.M.; Segura, J.; Ros, R. Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol. 2009, 151, 541–558. [Google Scholar] [CrossRef] [PubMed]
- Anoman, A.D.; Muñoz, B.J.; Rosa, T.S.; Tornero, M.F.; Serrano, R.; Bueso, E.; Fernie, A.R.; Segura, J.; Ros, R. Plastidial glycolytic glyceraldehyde-3-phosphate dehydrogenase is an important determinant in the carbon and nitrogen metabolism of heterotrophic cells in Arabidopsis. Plant Physiol. 2015, 169, 1619–1637. [Google Scholar] [CrossRef] [PubMed]
- Hajirezaei, M.M.; Peisker, S.; Peisker, M.; Lytovchenko, A.; Fernie, A.R.; Sonnewald, U. The influence of cytosolic phosphorylating glyceraldehyde3-phosphate dehydrogenase (GAPC) on potato tuber metabolism. J. Exp. Bot. 2006, 57, 2363–2377. [Google Scholar] [CrossRef][Green Version]
- Liu, T.F.; Fang, H.; Liu, J.; Reid, S.; Hou, J.; Zhou, T.T.; Tian, Z.D.; Song, B.T.; Xie, C.H. Cytosolic glyceraldehyde-3-phosphate dehydrogenases play crucial roles in controlling cold-induced sweetening and apical dominance of potato (Solanum tuberosum L.) tubers. Plant Cell Environ. 2017, 40, 3043–3054. [Google Scholar] [CrossRef]
- Li, L.Q.; Lyu, C.C.; Chen, J.; Lu, Y.F.; Yng, S.M.; Ni, S.; Zheng, S.L.; Yu, L.P.; Wang, X.Y.; Wang, Q.; et al. Snakin-2 interacts with cytosolic glyceraldehyde-3-phosphate dehydrogenase 1 to inhibit sprout growth in potato tubers. Hortic. Res. 2022, 9, uhab060. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, X.; Nie, B.; Song, B.; Du, P.; Liu, S.; Li, L.; Zhao, Z. Nitrogen management can inhibit or induce the sprouting of potato tubers: Consequences of regulation tuberization. Postharvest Biol. Technol. 2022, 183, 111722. [Google Scholar] [CrossRef]
- Wang, X.Y.; Li, J.; Zou, X.; Lu, L.M.; Li, L.Q.; Ni, S.; Liu, F. Ectopic expression of AtCIPK23 enhances tolerance against low-K+ stress in transgenic potato. Am. J. Potato Res. 2010, 88, 153–159. [Google Scholar] [CrossRef]
- Ren, Y.Z.; He, X.; Liu, D.C.; Li, J.J.; Zhao, X.Q.; Li, B.; Tong, Y.P.; Zhang, A.M.; Li, Z.S. Major quantitative trait loci for seminal root morphology of wheat seedlings. Mol Breed. 2012, 30, 139–148. [Google Scholar] [CrossRef]
- He, X.; Qu, B.Y.; Li, W.J.; Zhao, X.Q.; Teng, W.; Ma, W.Y.; Ren, Y.Z.; Li, B.; Li, Z.S.; Tong, Y.P. The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol. 2015, 169, 1991–2005. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.Z.; Zhang, F.Z. Activities of nitrate reductase and glutamine synthetase in rice seedlings during cyanide metabolism. Hazard. Mater. 2012, 225, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.M.; Shi, S.L. Physiological and proteomic responses of contrasting alfalfa (Medicago sativa L.) varieties to PEG-induced osmotic stress. Front. Plant Sci. 2018, 9, 242. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Shi, J.; Jiang, C.Z.; Feng, Y.; Cao, Y.; Wang, Q. A short-term carbon dioxide treatment inhibits the browning of fresh-cut burdock. Postharvest Biol. Technol. 2015, 110, 96–102. [Google Scholar] [CrossRef]
- Yang, D.; Ma, N.; Liu, Z.; Ma, X.; Zhao, S.; Meng, Q. Suppression of tomato SlGGP aggravates methyl viologen-mediated oxidative stress. Biol. Plant. 2016, 60, 677–685. [Google Scholar] [CrossRef]
- Khan, M.U.; Li, P.H.; Amjad, H.; Khan, A.Q.; Arafat, Y.; Waqas, M.; Li, Z.; Noman, A.; Islam, W.; Wu, L.K.; et al. Exploring the potential of overexpressed OsCIPK2 rice as a nitrogen utilization efficient crop and analysis of its associated rhizo-compartmental microbial communities. Int. J. Mol. Sci. 2019, 20, 3636. [Google Scholar] [CrossRef]
- Kumar, P.; Pandey, S.K.; Singh, B.P.; Singh, S.V.; Kumar, D. Effect of N rate on growth, yield, economics and crisps quality of Indian potato processing cultivars. Potato Res. 2007, 50, 143–155. [Google Scholar] [CrossRef]
- Soualiou, S.; Duan, F.Y.; Li, X.; Zhou, W.B. Nitrogen supply alleviates cold stress by increasing photosynthesis and nitrogen assimilation in maize seedlings. J. Exp. Bot. 2023, 74, 3142–3162. [Google Scholar] [CrossRef]
- Hu, B.; Wang, W.; Chen, J.J.; Liu, Y.Q.; Chu, C.C. Genetic improvement toward nitrogen-use efficiency in rice: Lessons and perspectives. Mol. Plant 2023, 1, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, Y.; Mochizuki, S.; Koiwai, H.; Kondo, K.; Kishimoto, K.; Katoh, E.; Minami, E. Rice ubiquitin ligase EL5 prevents root meristematic cell death under high nitrogen conditions and interacts with a cytosolic GAPDH. Plant Signal. Behav. 2015, 10, e990801. [Google Scholar] [CrossRef]
- Lu, S.P.; Yao, S.B.; Wang, G.L.; Guo, L.; Zhou, Y.M.; Hong, Y.Y.; Wang, X.M. Phospholipase De enhances Braasca napus growth and seed production in response to nitrogen availability. Plant Biotechnol. J. 2016, 14, 926–937. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.N.; Chai, X.F.; Gao, B.B.; Deng, C.L.; Günther, C.S.; Wu, T.; Zhang, X.Z.; Xu, X.F.; Han, Z.H.; Wang, Y. Multiple-omics reveal the role of transcription factor bHLH130 during low nitrogen in apple. Plant Physiol. 2023, 191, 1305–1323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Han, Z.J.; Lu, Y.; Zhao, Y.F.; Wang, Y.P.; Zhang, J.Y.; Ma, H.R.; Han, Y.Z. Genome-wide identification, structural and gene expression analysis of the nitrate transporters (NRTs) family in potato (Solanum tuberosum L.). PLoS ONE 2021, 21, 0257383. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.C.; Xuan, H.M.; Yang, Y.Y.; Wang, L.N.; Wei, L.T.; Wang, Y.H.; Kang, G.Z. Transcription analysis of genes encoding the wheat root transporter NRT1 and NRT2 families during nitrogen starvation. J. Plant Growth Regul. 2014, 33, 837–848. [Google Scholar] [CrossRef]
- Xing, J.P.; Wang, Y.B.; Yao, Q.Q.; Zhang, Y.S.; Zhang, M.C.; Li, Z.H. Brassinosteroids modulate nitrogen physiological response and promote nitrogen uptake in maize (Zea mays L.). Crop J. 2022, 1, 166–176. [Google Scholar] [CrossRef]
- Liu, Q.; Wu, K.; Song, W.Z.; Zhong, N.; Wu, Y.Z.; Fu, X.D. Improving crop nitrogen use efficiency toward sustainable green revolution. Annu. Rev. Plant Biol. 2022, 73, 523–550. [Google Scholar] [CrossRef]
- Ma, X.H.; Nian, J.Q.; Yu, H.; Zhang, F.X.; Feng, T.P.; Kou, L.Q.; Zhang, J.; Wang, D.F.; Li, H.W.; Cheng, L.C.; et al. Link glucose signaling to nitrogen utilization by the OsHXK7-ARE4 complex in rice. Dev. Cell. 2023, 58, 1489–1501. [Google Scholar] [CrossRef]
- Krouk, G.; Lacombe, B.; Bielach, A.; Walker, F.P.; Malinska, K.; Mounier, E.; Hoyerova, K.; Tillard., P.; Leon, S.; Ljung, K.; et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell. 2010, 18, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Kanno, Y.; Hanada, A.; Chiba, Y.; Ichikawa, T.; Nakazawa, M.; Matsui, M.; Koshiba, T.; Kamiya, Y.; Seo, M. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. USA 2012, 109, 9653–9658. [Google Scholar] [CrossRef] [PubMed]
- Tal, I.; Zhang, Y.; Jørgensen, M.E.; Pisanty, O.; Barbosa, I.C.R.; Zourelidou, M.; Regnault, T.; Crocoll, C.; Olsen, C.E.; Weinstain, R.; et al. The Arabidopsis NPF3 protein is a GA transporter. Nat. Commun. 2016, 7, 11486. [Google Scholar] [CrossRef]
- Safi, A.; Medici, A.; Szponarski, W.; Marshall-Colon, A.; Ruffel, S.; Gaymard, F.; Coruzzi, G.; Lacombe, B.; Krouk, G. GARP transcription factors and reactive oxygen species are regulators of Arabidopsis nitrogen starvation response. J. Exp. Bot. 2021, 72, 3881–3901. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, H.; Wang, Y.; Teng, R.M.; Liu, J.Y.; Lin, S.J.; Zhuang, J. Cytosolic ascorbate peroxidase 1 modulates ascorbic acid metabolism through cooperating with nitrogen regulatory protein P-II in tea plant under nitrogen deficiency stress. Genomics 2020, 112, 3497–3503. [Google Scholar] [CrossRef]
- Chu, X.Q.; Wang, J.G.; Li, M.Z.; Zhang, S.J.; Gao, Y.Y.; Fan, M.; Han, C.; Xiang, F.N.; Li, G.Y.; Wang, Y.; et al. HBI transcription factor-mediated ROS homeostasis regulates nitrate signal transduction. Plant Cell 2021, 33, 3004–3021. [Google Scholar] [CrossRef] [PubMed]
- Hancock, J.T.; Henson, D.; Nyirenda, M.; Desikan, R.; Harrison, J.; Lewis, M.; Hughes, J.; Neill, S.J. Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiol. Biochem. 2005, 43, 828–835. [Google Scholar] [CrossRef]
- Zhang, X.H.; Rao, X.L.; Shi, H.T.; Li, R.J.; Lu, Y.T. Overexpression of a cytosolic glyceraldehyde-3-phosphatedehydrogenase gene OsGAPC3 confers salt tolerance in rice. Plant Cell Tiss. Organ Cult. 2011, 107, 1–11. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, Z.Y.; Ji, H.K.; Zhou, Y.; Yang, S.S. TaWRKY40 transcription factor positively regulate the expression of TaGAPC1 to enhance drought tolerance. BMC Genom. 2019, 20, 795. [Google Scholar] [CrossRef]
- Han, S.J.; Wang, Y.; Zheng, X.Y.; Jia, Q.; Zhao, J.P.; Bai, F.; Hong, Y.G.; Liu, Y.L. Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in nicotiana benthamiana. Plant Cell. 2015, 27, 1318–1331. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, Y.; Zhou, D.X. Rice NAD+-dependent histone deacetylase OsSRT1 represses glycolysis and regulates the moonlighting function of GAPDH as a transcriptional activator of glycolytic genes. Nucleic Acids Res. 2017, 45, 12241–12255. [Google Scholar] [CrossRef]
- Kim, S.C.; Guo, L.; Wang, X. Nuclear moonlighting of cytosolic glyceraldehyde-3-dehydrogenase regulates Arabidopsis response to heat stress. Nat. Commun. 2020, 11, 3439. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Song, J.; Zhuang, X.; Lu, Y.; Wang, Q.; Yang, S.; Lu, L.; Wang, X.; Li, L. Overexpression of Cytosolic Glyceraldehyde-3-Phosphate Dehydrogenase 1 Gene Improves Nitrogen Absorption and Utilization in Potato. Horticulturae 2023, 9, 1105. https://doi.org/10.3390/horticulturae9101105
Liu J, Song J, Zhuang X, Lu Y, Wang Q, Yang S, Lu L, Wang X, Li L. Overexpression of Cytosolic Glyceraldehyde-3-Phosphate Dehydrogenase 1 Gene Improves Nitrogen Absorption and Utilization in Potato. Horticulturae. 2023; 9(10):1105. https://doi.org/10.3390/horticulturae9101105
Chicago/Turabian StyleLiu, Jingrui, Jun Song, Xiaoyu Zhuang, Yifei Lu, Qiang Wang, Shimin Yang, Liming Lu, Xiyao Wang, and Liqin Li. 2023. "Overexpression of Cytosolic Glyceraldehyde-3-Phosphate Dehydrogenase 1 Gene Improves Nitrogen Absorption and Utilization in Potato" Horticulturae 9, no. 10: 1105. https://doi.org/10.3390/horticulturae9101105
APA StyleLiu, J., Song, J., Zhuang, X., Lu, Y., Wang, Q., Yang, S., Lu, L., Wang, X., & Li, L. (2023). Overexpression of Cytosolic Glyceraldehyde-3-Phosphate Dehydrogenase 1 Gene Improves Nitrogen Absorption and Utilization in Potato. Horticulturae, 9(10), 1105. https://doi.org/10.3390/horticulturae9101105






