BrDHC1, a Novel Putative DEAD-Box Helicase Gene, Confers Drought Tolerance in Transgenic Brassica rapa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
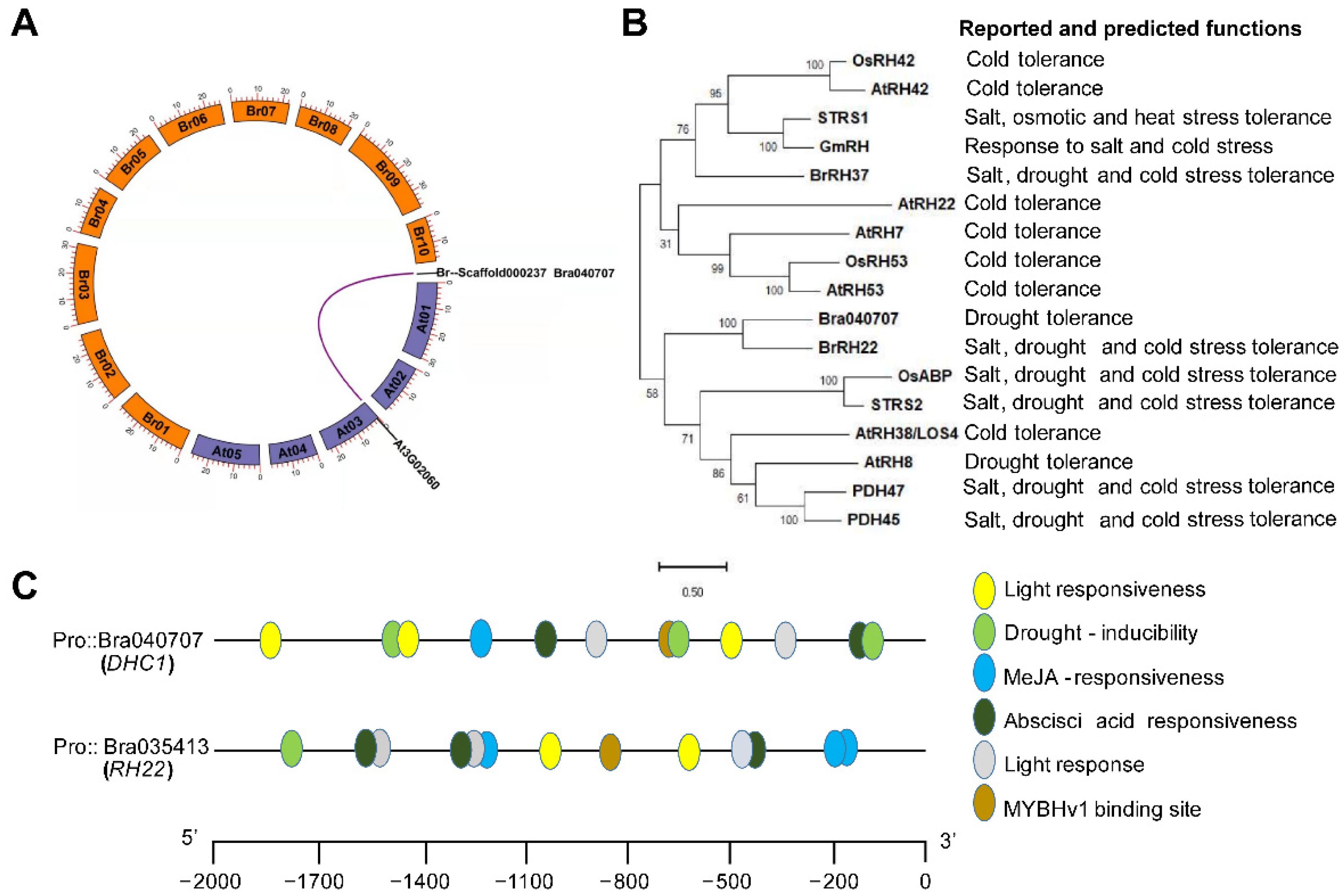
2.2. Collinearity and Evolution Analysis
2.3. Cis-Acting Regulatory Elements Analysis
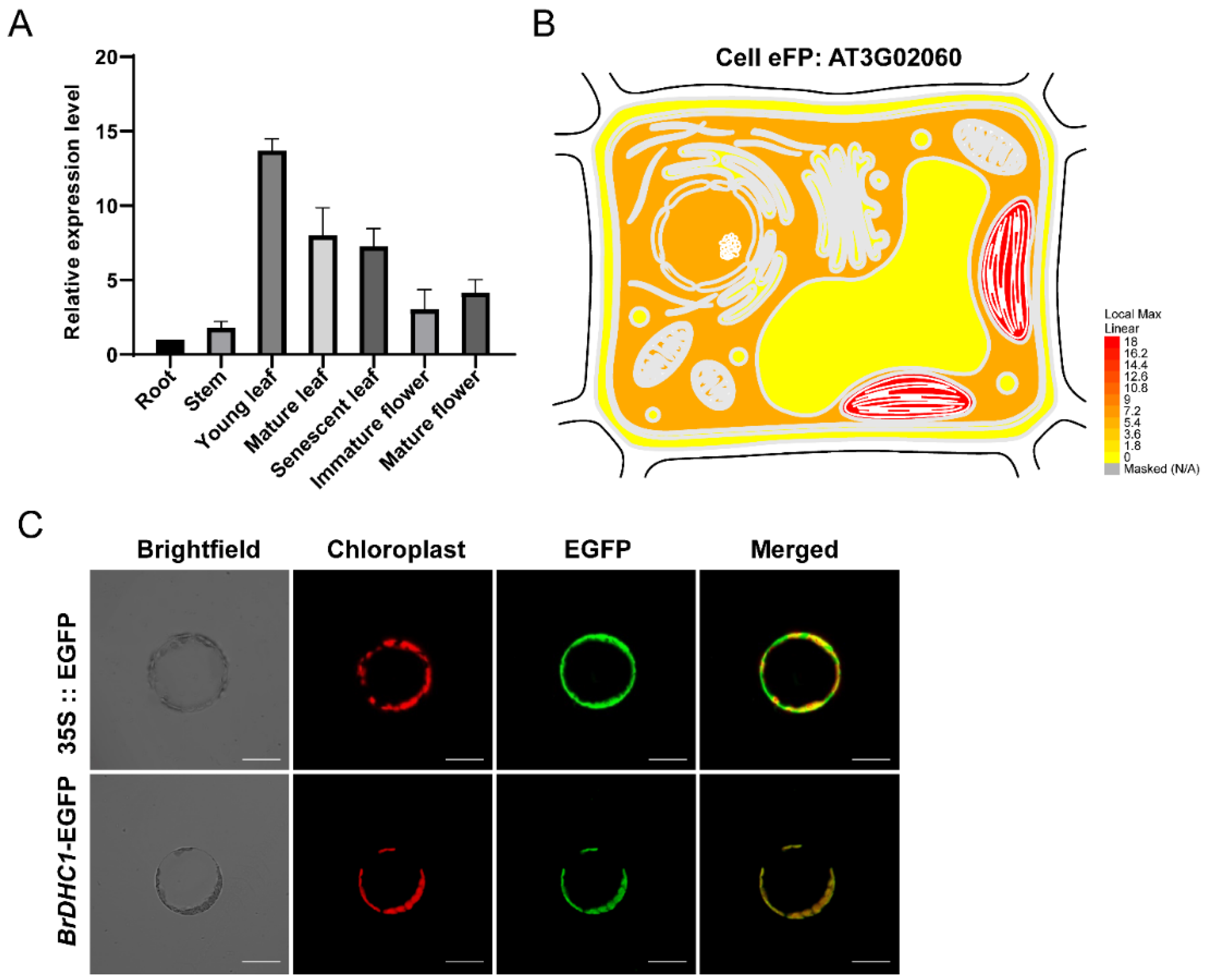
2.4. Subcellular Location and Production of Transgenic Plants
2.5. Drought Stress Treatment and Phenotype Observation
2.6. Measurement of Stomatal Opening Rate
2.7. Relative Water Content (RWC) Determination
2.8. Determination of Chlorophyll Content
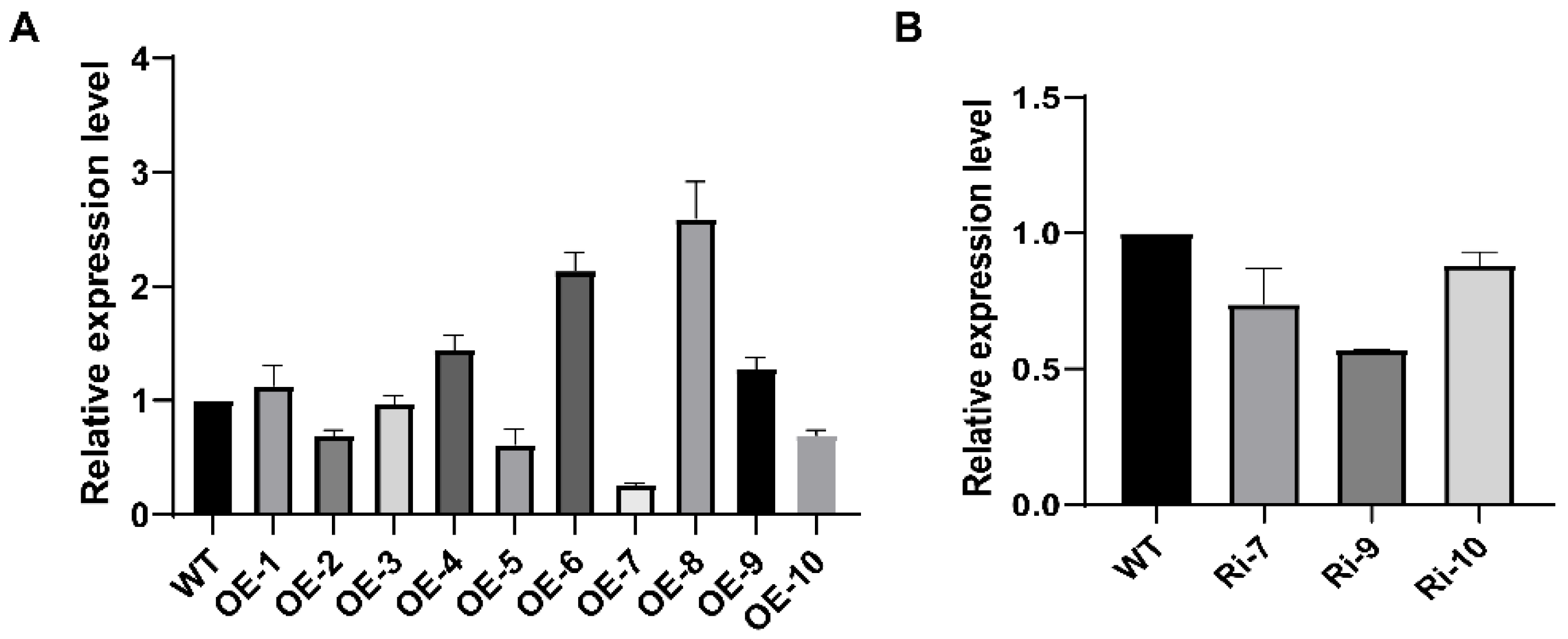
2.9. Quantitative Real-Time PCR Analysis
2.10. Statistical Analysis
3. Results
3.1. Evolutionary Analysis of Gene Collinear and DEAD-Box Helicase Members
3.2. Expression Pattern Analysis and Subcellular Localization of BrDHC1 in B. rapa
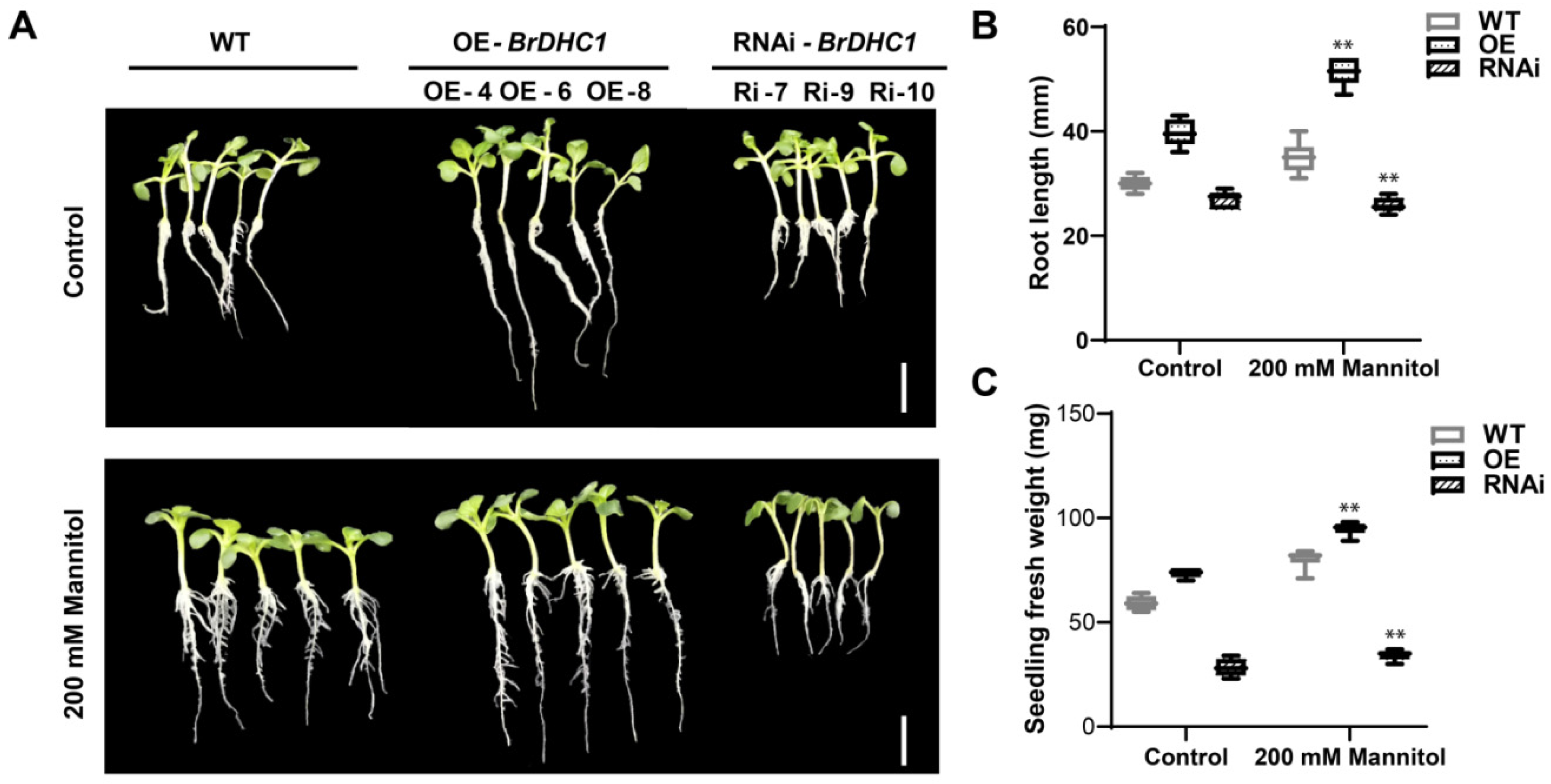
3.3. BrDHC1 Regulates Root Development of Seedlings under Drought Stress
3.4. BrDHC1 Regulates Stomatal Aperture under Osmotic Stress
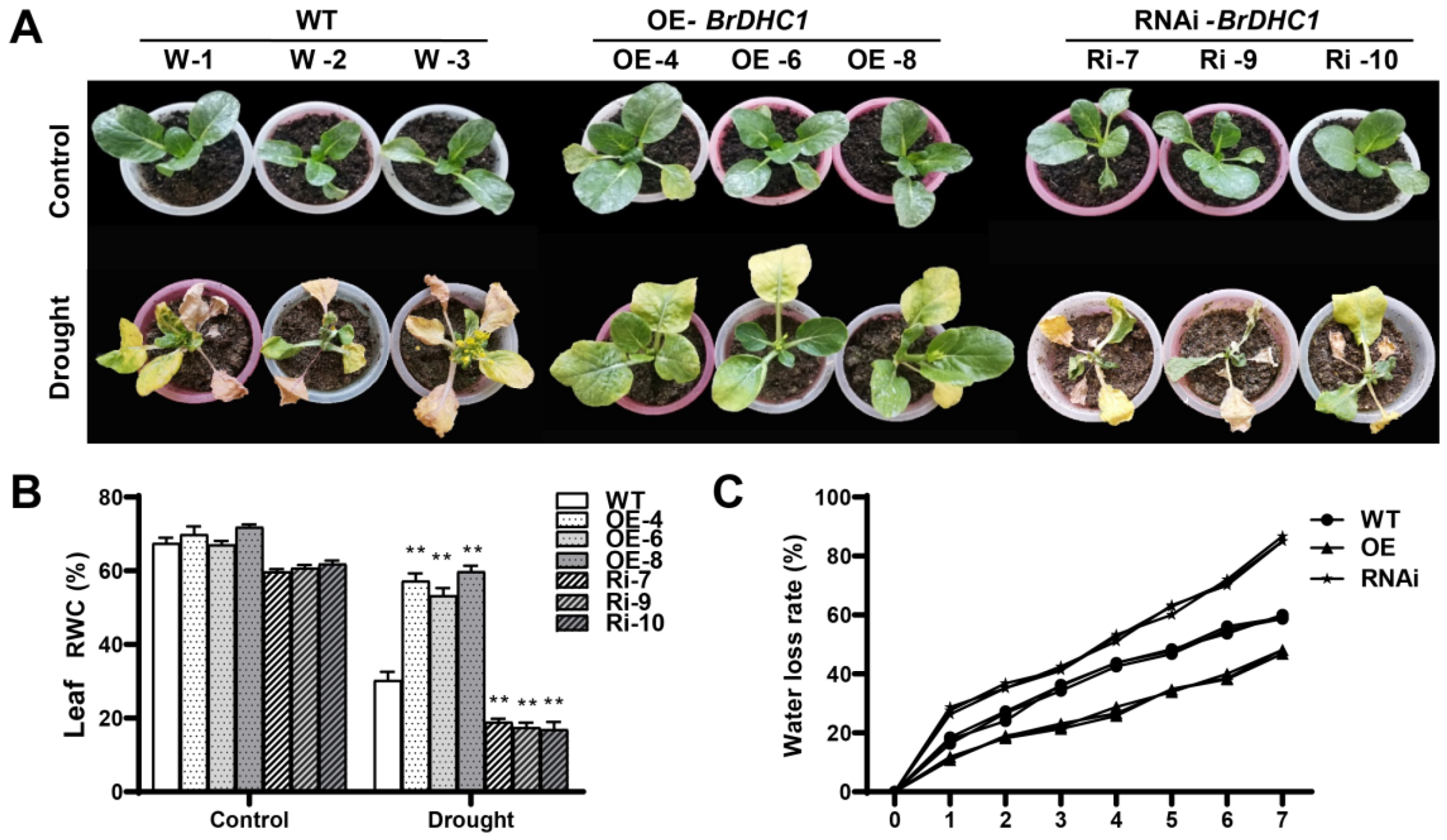
3.5. BrDHC1 Improves the Drought Resistance of B. rapa
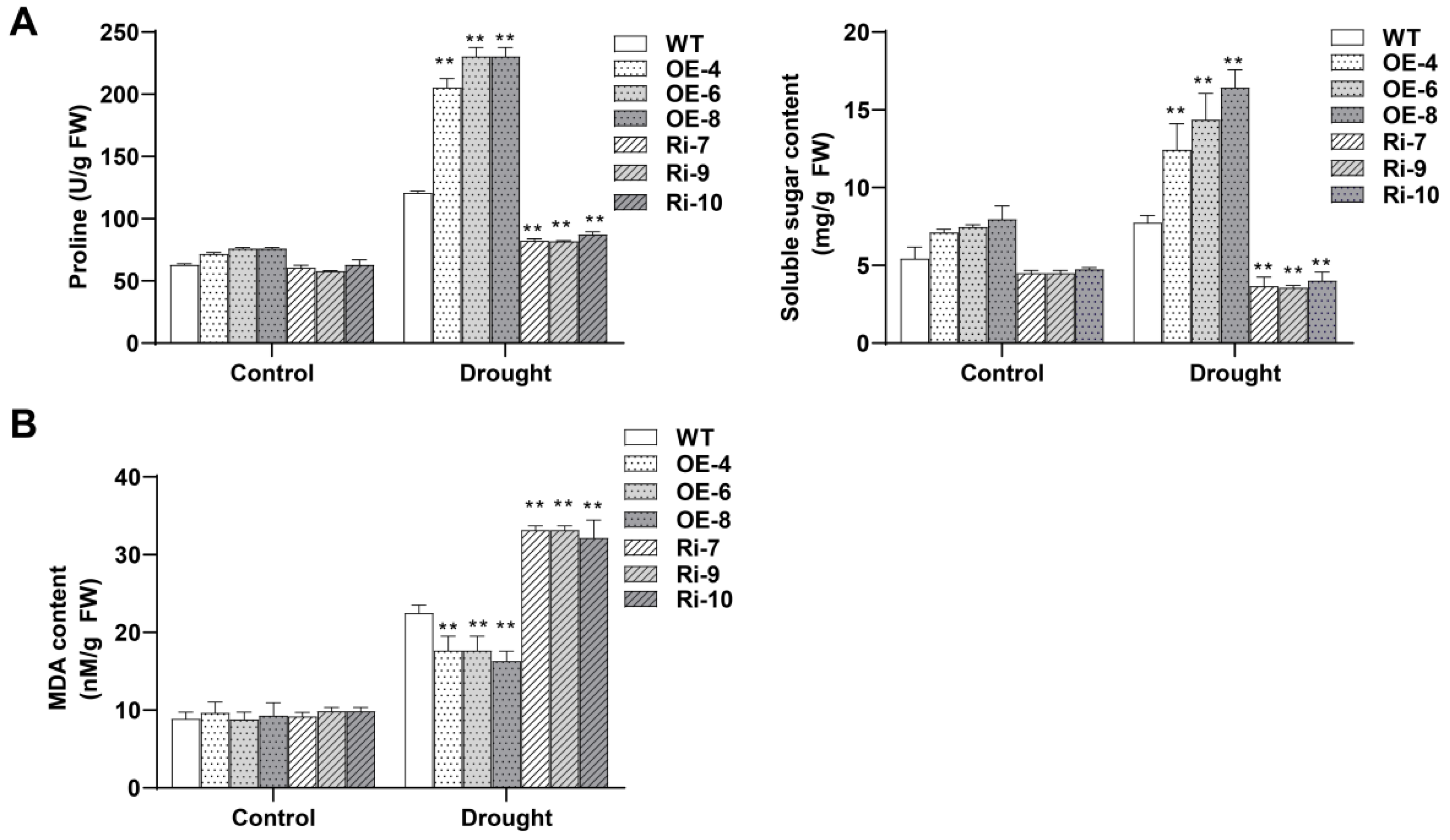
3.6. BrDHC1 Regulates Photosynthesis and Antioxidant Enzyme Activity of B. rapa under Drought Stress
3.7. BrDHC1 Regulates Osmotic Regulators and Malondialdehyde Contents of B. rapa under Drought Stress
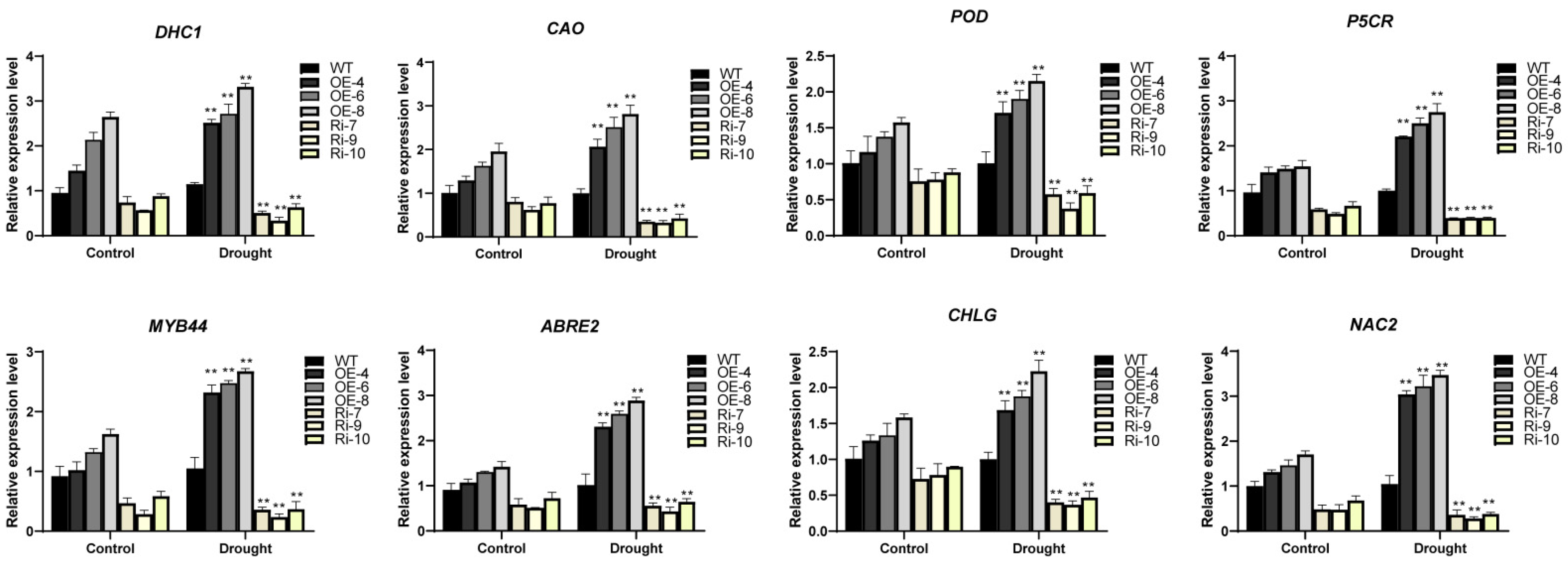
3.8. BrDHC1 Confers Drought Stress with the Synergy of Stress-Related Genes
4. Discussion
4.1. Expression Localization of BrDHC1 Aids Understanding Gene Function
4.2. BrDHC1 Positively Regulates Drought Stress
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Primer Name | Primer Sequence (5′ to 3′) |
|---|---|
| BrDHC1-2469-F | AACTGCAGATGACATCCTTGCTCCTCCCAAATCC |
| BrDHC1-2469-R | GGGGTACCGTATTTGATAAGAGCAGGGAGTGATG |
| RNAi-BrDHC1-F | TTCCGGTGATTATGTGGTGC |
| RNAi-BrDHC1-R | CGAGACCCGTTTGGACTCAT |
| BrDHC1(RNAi)-BP-F | GGGGACAAGTTTGTACAAAAAAGCAGGCT TTCCGGTGATTATGTGGTGC |
| BrDHC1(RNAi)-BP-R | GGGGACAAGTTTGTACAAAAAAGCAGGCT CGAGACCCGTTTGGACTCAT |
| PDK(RNAi)-F | GACGAAGAAGATAAAAGTTGAGAGT |
| PDK(RNAi)-R | ACCTTGTTTATTCATGTTCGACTAA |
| EGFP-F | ATGGTGAGCAAGGGCGAG |
| EGFP-R | GCTCTTACTTGTACAGCTCGTC |
| β-actin-F | ATCAACTACCAGCCTCCAAC |
| β-actin-R | CTGCTGTGTTGTTGCTGATC |
| CAO-F | AATGCCCTTACCACGGATGG |
| CAO-R | AGGTCCATTACAAGCTCGGC |
| CHLG-F | GCTTTGGGAGGGTCCTTGTT |
| CHLG-R | ATCGCTATTCCCAACCCAGC |
| POD-F | GGTACGTGCTACACCTGGAC |
| POD-R | GCATCCACCCTGAAAGTCG |
| P5CR-F | AAGGCCATCACGGAAGTGAG |
| P5CR-R | CAACAAGTGCTCCGTCTT |
| NAC2-F | ACTGCATATCCCTTGGCGAG |
| NAC2-R | AGATTCCCACCAGGTTGCAC |
| MYB44-F | TCGGGAAAATCGTGTCGGTT |
| MYB44-R | CTTCAGCGTCGAGTTCCAGT |
| ABRE2-F | CTCGACCAGAAACCTTCCCC |
| ABRE2-R | GAAGATTGCGCTGCGTGTAG |
Appendix B
References
- Xiong, L.; Schumaker, K.S.; Zhu, J.K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14 (Suppl. 1), S165–S183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seki, M.; Umezawa, T.; Urano, K.; Shinozaki, K. Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 2007, 10, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [Green Version]
- Umezawa, T.; Fujita, M.; Fujita, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Engineering drought tolerance in plants: Discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 2006, 17, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Gupta, A.; Azooz, M.M.; Sharma, S.; Ahmad Parvaiz Dames, J. Chapter 4—Genetic approaches to improve salinity tolerance in plants. In Salt Stress in Plants: Signalling, Omics and Adaptations; Springer: New York, NY, USA, 2013; pp. 63–78. [Google Scholar] [CrossRef]
- Fang, Y.; Xiong, L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell. Mol. Life Sci. 2015, 72, 673–689. [Google Scholar] [CrossRef]
- Daszkowska-Golec, A.; Szarejko, I. Open or close the gate-stomata action under the control of phytohormones in drought stress conditions. Front. Plant Sci. 2013, 4, 138. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Wang, Z.; Liao, D.; Raza, M.A.; Wang, B.; Zhang, J.; Chen, J.; Feng, L.; Wu, X.; Liu, C.; et al. Uptake and utilization of nitrogen, phosphorus and potassium as related to yield advantage in maize-soybean intercropping under different row configurations. Sci. Rep. 2020, 10, 9504. [Google Scholar] [CrossRef]
- Zhuang, J.; Wang, Y.; Chi, Y.; Zhou, L.; Chen, J.; Zhou, W.; Song, J.; Zhao, N.; Ding, J. Drought stress strengthens the link between chlorophyll fluorescence parameters and photosynthetic traits. PeerJ 2020, 8, e10046. [Google Scholar] [CrossRef]
- Kalaji, H.M.; Bąba, W.; Gediga, K.; Goltsev, V.; Samborska, I.A.; Cetner, M.D.; Dimitrova, S.; Piszcz, U.; Bielecki, K.; Karmowska, K.; et al. Chlorophyll fluorescence as a tool for nutrient status identification in rapeseed plants. Photosynth. Res. 2018, 136, 329–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gebre, M.G.; Earl, H.J. Soil water deficit and fertilizer placement effects on root biomass distribution, soil water extraction, water use, yield, and yield components of Soybean [Glycine max (L.) Merrill.] grown in 1-m rooting Columns. Front. Plant Sci. 2021, 12, 581127. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S.; Panda, P.; Sahoo, L.; Panda, S.K. Reactive oxygen species signaling in plants under abiotic stress. Plant Signal. Behav. 2013, 8, e23681. [Google Scholar] [CrossRef] [Green Version]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Hanin, M.; Brini, F.; Ebel, C.; Toda, Y.; Takeda, S.; Masmoudi, K. Plant dehydrins and stress tolerance: Versatile proteins for complex mechanisms. Plant Signal. Behav. 2011, 6, 1503–1509. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Mo, Y.; Wang, Y.; Yang, R.; Zheng, J.; Liu, C.; Li, H.; Ma, J.; Zhang, Y.; Wei, C.; Zhang, X. Regulation of plant growth, photosynthesis, antioxidation and osmosis by an Arbuscular Mycorrhizal fungus in watermelon seedlings under well-watered and drought conditions. Front. Plant Sci. 2016, 7, 644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linder, P. DEAD-box proteins: A family affair—Active and passive players in RNP-remodeling. Nucleic Acids Res. 2006, 34, 4168–4180. [Google Scholar] [CrossRef]
- Vashisht, A.A.; Tuteja, N. Stress responsive DEAD-box helicases: A new pathway to engineer plant stress tolerance. J. Photochem. Photobiol. B Biol. 2006, 84, 150–160. [Google Scholar] [CrossRef]
- Mcglynn, P. Helicases that underpin replication of protein-bound DNA in Escherichia coli. Biochem. Soc. Trans. 2011, 39, 606–610. [Google Scholar] [CrossRef] [Green Version]
- Tuteja, N.; Tuteja, R. Unraveling DNA helicases, motif, structure, mechanism and function. Eur. J. Biochem. 2004, 271, 1849–1863. [Google Scholar] [CrossRef] [PubMed]
- Jankowsky, E. RNA helicases at work: Binding and rearranging. Trends Biochem. Sci. 2011, 36, 19–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umate, P.; Tuteja, R.; Tuteja, N. Genome-wide analysis of helicase gene family from rice and Arabidopsis: A comparison with yeast and human. Plant Mol. Biol. 2010, 73, 449–465. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, X.; Shen, X.; Chen, R.; Zhu, C.; Zhang, Z.; Chen, Y.; Lin, W.; Xu, X.; Lin, Y.; et al. Genome-wide identification and characterization of DEAD-box helicase family associated with early somatic embryogenesis in Dimocarpus longan Lour. J. Plant Physiol. 2021, 258–259, 153364. [Google Scholar] [CrossRef]
- Wan, R.; Liu, J.; Yang, Z.; Zhu, P.; Cao, Q.; Xu, T. Genome-wide identification, characterisation and expression profile analysis of DEAD-box family genes in sweet potato wild ancestor Ipomoea trifida under abiotic stresses. Genes Genom. 2020, 42, 325–335. [Google Scholar] [CrossRef]
- Wang, Y.; Liao, J.; Wu, J.; Huang, H.; Yuan, Z.; Yang, W.; Wu, X.; Li, X. Genome-wide identification and characterization of the soybean DEAD-Box gene family and expression response to Rhizobia. Int. J. Mol. Sci. 2022, 23, 1120. [Google Scholar] [CrossRef]
- Xu, R.; Zhang, S.; Lu, L.; Cao, H.; Zheng, C. A genome-wide analysis of the RNA helicase gene family in Solanum lycopersicum. Gene 2013, 513, 128–140. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhu, Y.; Liu, Z.J.; Ouyang, S. The emerging roles of the DDX41 protein in immunity and diseases. Protein Cell 2017, 8, 83–89. [Google Scholar] [CrossRef] [Green Version]
- Owttrim, G.W. RNA helicases: Diverse roles in prokaryotic response to abiotic stress. RNA Biol. 2013, 10, 96–110. [Google Scholar] [CrossRef] [Green Version]
- Tuteja, N.; Tuteja, R. Prokaryotic and eukaryotic DNA helicases, essential molecular motor proteins for cellular machinery. Eur. J. Biochem. 2004, 271, 1835–1848. [Google Scholar] [CrossRef]
- Wang, D.; Qin, B.; Li, X.; Tang, D.; Zhang, Y.; Cheng, Z.; Xue, Y. Nucleolar DEAD-box RNA hlicase TOGR1 regulates thermotolerant growth as a pre-rRNA chaperone in rice. PLoS Genet. 2016, 12, e1005844. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, G.; Kang, H. Chloroplast or mitochondria-targeted DEAD-box RNA helicases play essential roles in organellar RNA metabolism and abiotic stress responses. Front. Plant Sci. 2017, 8, 871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macovei, A.; Vaid, N.; Tula, S.; Tuteja, N. A new DEAD-box helicase ATP-binding protein (OsABP) from rice is responsive to abiotic stress. Plant Signal. Behav. 2012, 7, 1138–1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, E.; Cho, C.W.; Yun, B.H.; Choi, H.K.; So, H.A.; Lee, S.W.; Lee, J.H. Molecular cloning and characterization of the soybean DEAD-box RNA helicase gene induced by low temperature and high salinity stress. Gene 2009, 443, 91–99. [Google Scholar] [CrossRef]
- Nawaz, G.; Lee, K.; Park, S.J.; Kim, Y.O.; Kang, H. A chloroplast-targeted cabbage DEAD-box RNA helicase BrRH22 confers abiotic stress tolerance to transgenic Arabidopsis plants by affecting translation of chloroplast transcripts. Plant Physiol. Biochem. 2018, 127, 336–342. [Google Scholar] [CrossRef]
- Shivakumara, T.N.; Sreevathsa, R.; Dash, P.K.; Sheshshayee, M.S.; Papolu, P.K.; Rao, U.; Tuteja, N.; UdayaKumar, M. Overexpression of Pea DNA Helicase 45 (PDH45) imparts tolerance to multiple abiotic stresses in chili (Capsicum annuum L.). Sci. Rep. 2017, 7, 2760. [Google Scholar] [CrossRef]
- Vashisht, A.A.; Pradhan, A.; Tuteja, R.; Tuteja, N. Cold- and salinity stress-induced bipolar pea DNA helicase 47 is involved in protein synthesis and stimulated by phosphorylation with protein kinase C. Plant J. 2005, 44, 76–87. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, W.; Stanley, B.A.; Assmann, S.M. Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell 2008, 20, 3210–3226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Browne, M.; Yardimci, N.T.; Scoffoni, C.; Jarrahi, M.; Sack, L. Prediction of leaf water potential and relative water content using terahertz radiation spectroscopy. Plant Direct 2020, 4, e00197. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yin, G.; Sun, H.; Wang, H.; Chen, S.; Senthilnath, J.; Wang, J.; Fu, Y. Scaling effects on chlorophyll content estimations with RGB camera mounted on a UAV platform using machine-learning methods. Sensors 2020, 20, 5130. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wu, L.M.; Fang, Y.; Yang, H.N.; Bai, L.Y. Effects of drought-stress on seed germination and growth physiology of quinclorac-resistant echinochloa crusgalli. PLoS ONE 2019, 14, e0214480. [Google Scholar] [CrossRef]
- Liu, X.; Li, L.; Li, M.; Su, L.; Lian, S.; Zhang, B.; Li, X.; Ge, K.; Li, L. AhGLK1 affects chlorophyll biosynthesis and photosynthesis in peanut leaves during recovery from drought. Sci. Rep. 2018, 8, 2250. [Google Scholar] [CrossRef] [Green Version]
- Hatzig, S.; Zaharia, L.I.; Abrams, S.; Hohmann, M.; Legoahec, L.; Bouchereau, A.; Nesi, N.; Snowdon, R.J. Early osmotic adjustment responses in drought-resistant and drought-sensitive oilseed rape. J. Integr. Plant Biol. 2014, 56, 797–809. [Google Scholar] [CrossRef]
- Herath, V.; Verchot, J. Insight into the bZIP gene family in solanum tuberosum: Genome and transcriptome analysis to understand the roles of gene diversification in spatiotemporal gene expression and function. Int. J. Mol. Sci. 2020, 22, 253. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, A.; Li, X.; Lu, C. The role of chloroplast gene expression in plant responses to environmental stress. Int. J. Mol. Sci. 2020, 21, 6082. [Google Scholar] [CrossRef]
- Nishiyama, R.; Le, D.T.; Watanabe, Y.; Matsui, A.; Tanaka, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. Transcriptome analyses of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS ONE 2012, 7, e32124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muhammad, I.; Shalmani, A.; Ali, M.; Yang, Q.H.; Ahmad, H.; Li, F.B. Mechanisms regulating the dynamics of photosynthesis under abiotic stresses. Front. Plant Sci. 2020, 11, 615942. [Google Scholar] [CrossRef] [PubMed]
- Nowicka, B.; Ciura, J.; Szymanska, R.; Kruk, J. Improving photosynthesis, plant productivity and abiotic stress tolerance-current trends and future perspectives. J. Plant Physiol. 2018, 231, 415–433. [Google Scholar] [CrossRef] [PubMed]
- Gururani, M.A.; Venkatesh, J.; Tran, L.S. Regulation of photosynthesis during abiotic stress-induced photoinhibition. Mol. Plant 2015, 8, 1304–1320. [Google Scholar] [CrossRef] [Green Version]
- Meher Shivakrishna, P.; Ashok, R.K.; Manohar, R.D. Effect of PEG-6000 imposed drought stress on RNA content, relative water content (RWC), and chlorophyll content in peanut leaves and roots. Saudi J. Biol. Sci. 2018, 25, 285–289. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, G.; Gu, H.; Jiang, W.; Tian, Z.; Shi, G.; Chen, W.; Tian, B.; Wei, X.; Zhang, L.; Wei, F.; et al. BrDHC1, a Novel Putative DEAD-Box Helicase Gene, Confers Drought Tolerance in Transgenic Brassica rapa. Horticulturae 2022, 8, 707. https://doi.org/10.3390/horticulturae8080707
Cao G, Gu H, Jiang W, Tian Z, Shi G, Chen W, Tian B, Wei X, Zhang L, Wei F, et al. BrDHC1, a Novel Putative DEAD-Box Helicase Gene, Confers Drought Tolerance in Transgenic Brassica rapa. Horticulturae. 2022; 8(8):707. https://doi.org/10.3390/horticulturae8080707
Chicago/Turabian StyleCao, Gangqiang, Huihui Gu, Wenjing Jiang, Zhaoran Tian, Gongyao Shi, Weiwei Chen, Baoming Tian, Xiaochun Wei, Luyue Zhang, Fang Wei, and et al. 2022. "BrDHC1, a Novel Putative DEAD-Box Helicase Gene, Confers Drought Tolerance in Transgenic Brassica rapa" Horticulturae 8, no. 8: 707. https://doi.org/10.3390/horticulturae8080707
APA StyleCao, G., Gu, H., Jiang, W., Tian, Z., Shi, G., Chen, W., Tian, B., Wei, X., Zhang, L., Wei, F., & Xie, Z. (2022). BrDHC1, a Novel Putative DEAD-Box Helicase Gene, Confers Drought Tolerance in Transgenic Brassica rapa. Horticulturae, 8(8), 707. https://doi.org/10.3390/horticulturae8080707






