Genome-Wide Identification and Expression Pattern Analysis of the TCP Gene Family in Radish (Raphanus sativus L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Treatments
2.2. Identification of RsTCP Genes in Radish Genome
2.3. Protein Properties and Phylogeny Analysis
2.4. Gene Structure, Conserved Motifs, and Cis-Element Analyses
2.5. Synteny Analysis, Chromosomal Localization, and Prediction of rsa-miR319 Target Genes
2.6. Transcription Analysis of RsTCP Genes
2.7. RNA Extraction and RT-qPCR Analysis
3. Results
3.1. Identification and Classification of RsTCP Members in Radish
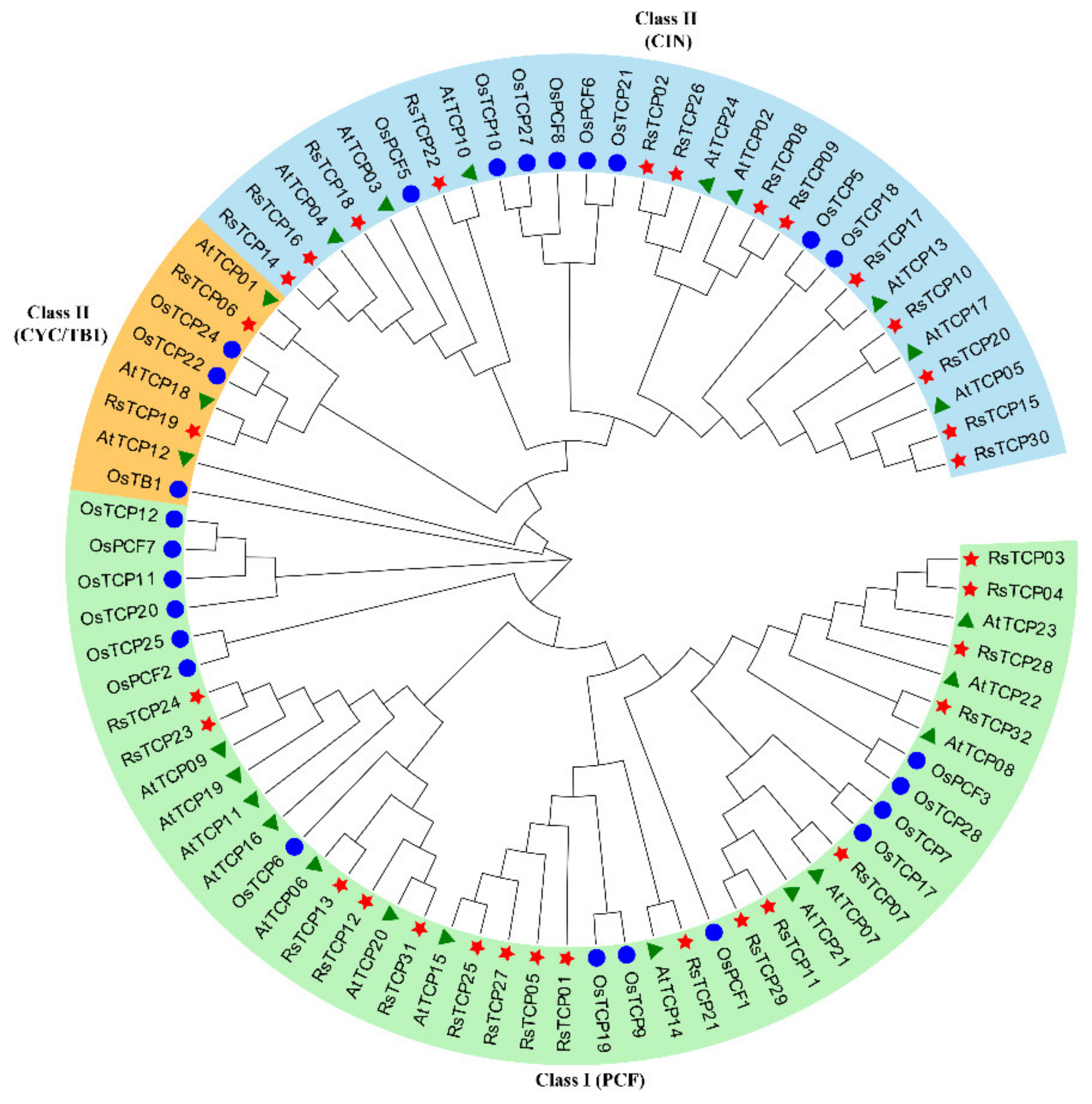
3.2. Phylogenetic Analysis of RsTCP Proteins
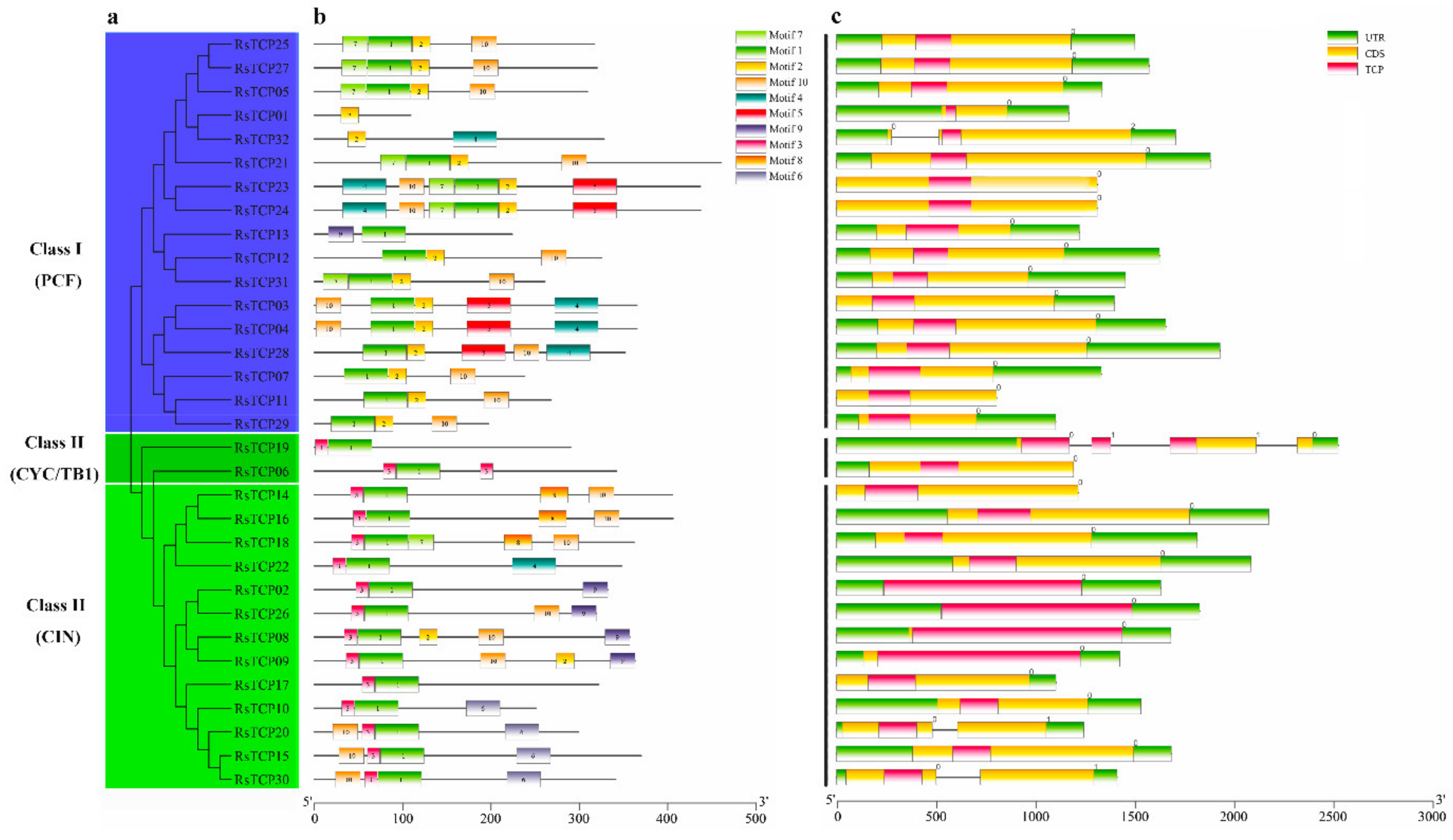
3.3. Motif and Gene Structure Analyses
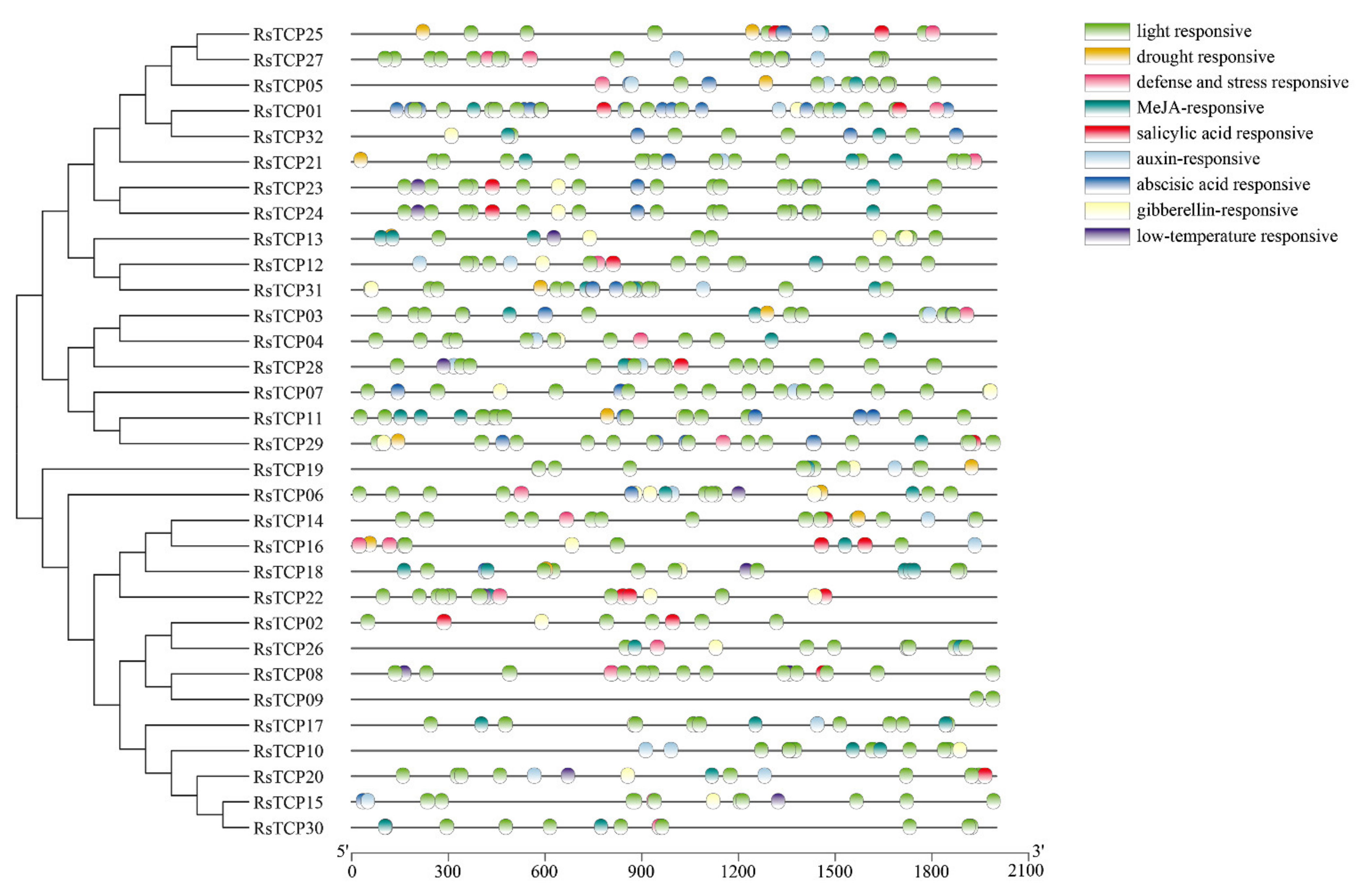
3.4. Cis-Element Analysis of RsTCP Genes
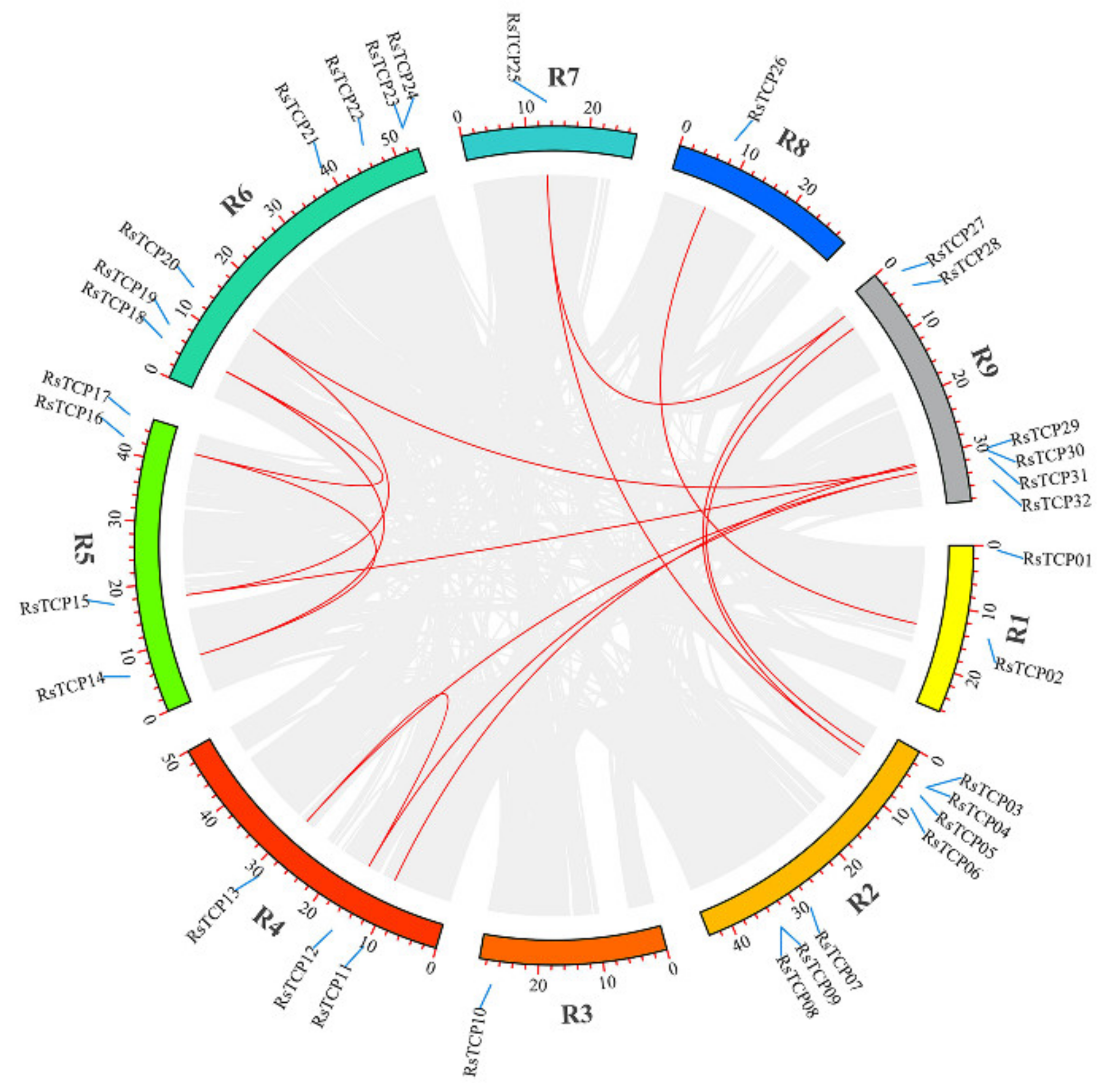
3.5. Chromosomal Localization and Gene Distribution Analysis
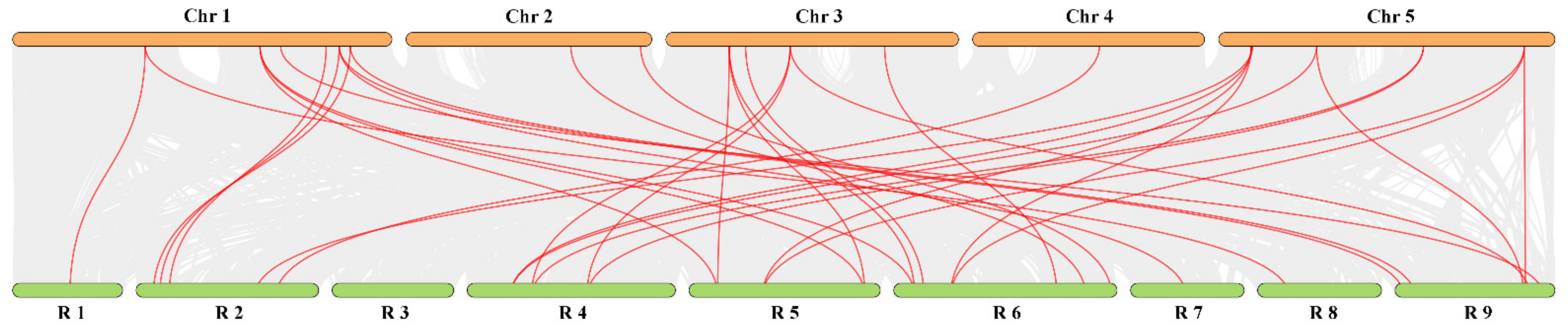
3.6. Evolution Analysis of the RsTCP Genes and Target Site Analysis of rsa-miR319
3.7. Characterization of Deduced RsTCP Proteins
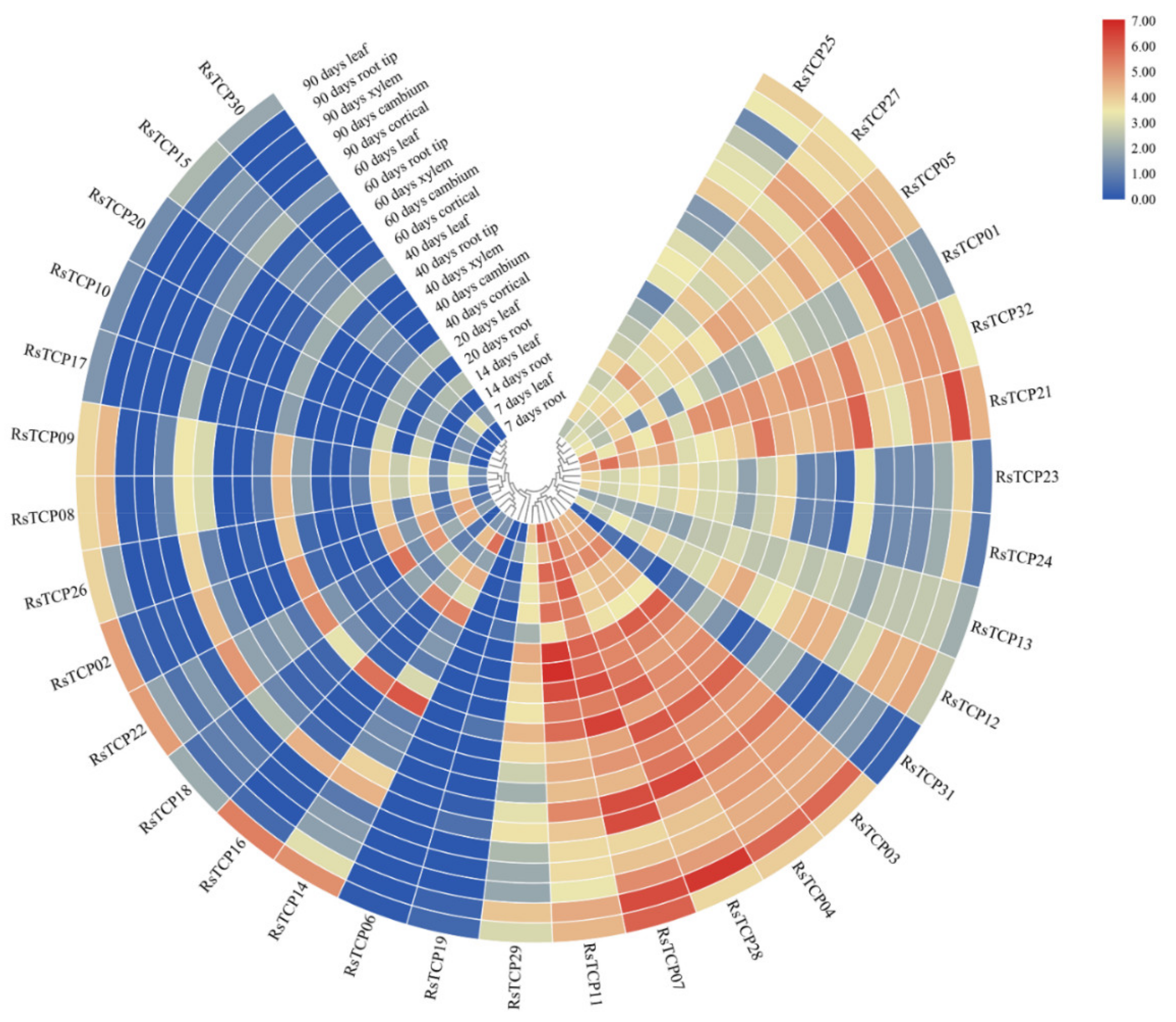
3.8. Transcription Profiling of RsTCP Genes in Radish
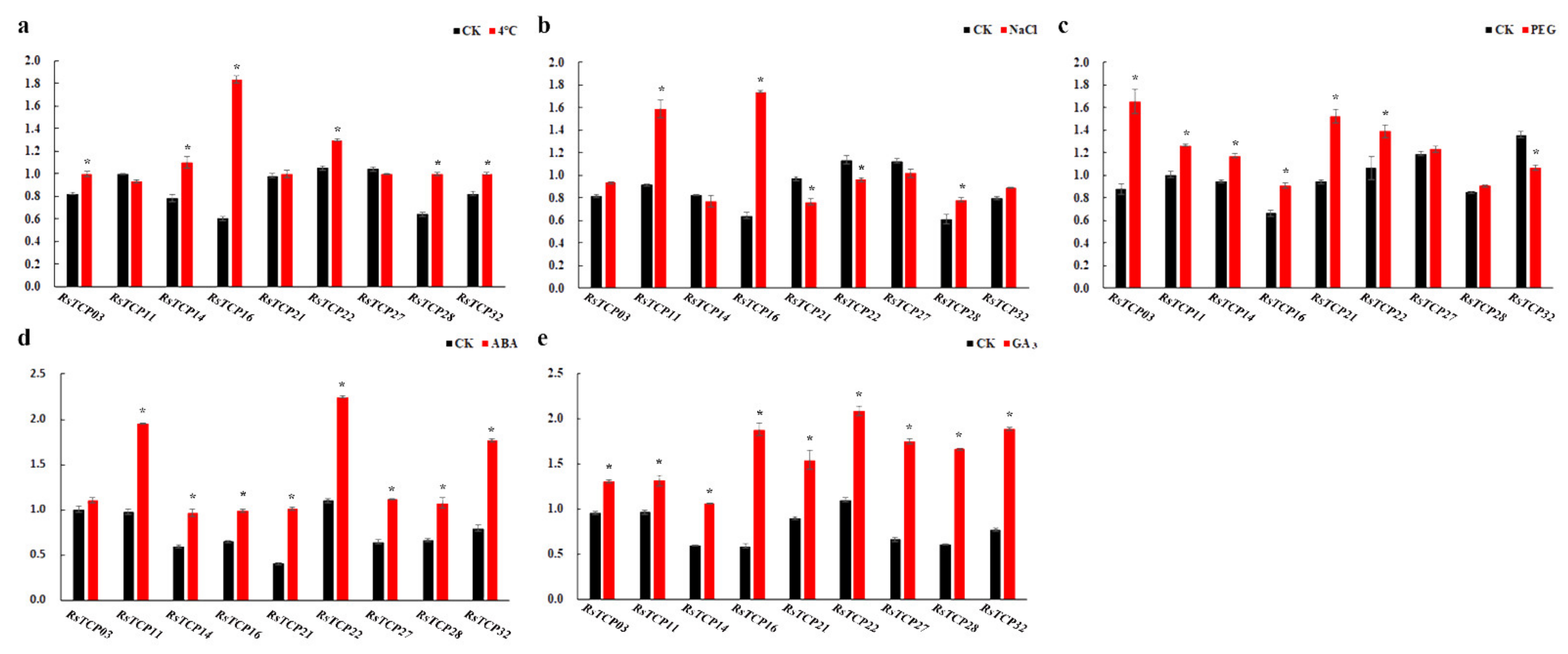
3.9. Transcription of RsTCP Genes in Response to Environmental Shocks and Phytohormones
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paul, M.V.; Iyer, S.; Amerhauser, C.; Lehmann, M.; Van Dongen, J.T.; Geigenberger, P. Oxygen Sensing via the Ethylene Response Transcription Factor RAP2.12 Affects Plant. Metabolism and Performance under Both Normoxia and Hypoxia. Plant Physiol. 2016, 172, 141–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehti-Shiu, M.D.; Panchy, N.; Wang, P.; Uygun, S.; Shiu, S.-H. Diversity, expansion, and evolutionary novelty of plant DNA-binding transcription factor families. Biochim. Biophys. Acta Gene Reg. Mech. 2017, 1860, 3–20. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Cochet, F.; Ponnaiah, M.; Lebreton, S.; Matheron, L.; Pionneau, C.; Boudsocq, M.; Resentini, F.; Huguet, S.; Blázquez, M.; et al. The MPK8-TCP14 pathway promotes seed germination in Arabidopsis. Plant J. 2019, 100, 677–692. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, H.; Mou, M.; Chen, Y.; Xiang, S.; Chen, L.; Yu, D. Arabidopsis Class II TCP Transcription Factors Integrate with the FT-FD Module to Control Flowering. Plant Physiol. 2019, 181, 97–111. [Google Scholar] [CrossRef]
- Lan, J.; Zhang, J.; Yuan, R.; Yu, H.; An, F.; Sun, L.; Chen, H.; Zhou, Y.; Qian, W.; He, H.; et al. TCP transcription factors suppress cotyledon trichomes by impeding a cell differentiation-regulating complex. Plant Physiol. 2021, 186, 434–451. [Google Scholar] [CrossRef]
- Gastaldi, V.; Lucero, L.E.; Ferrero, L.V.; Ariel, F.D.; Gonzalez, D.H. Class-I TCP Transcription Factors Activate the SAUR63 Gene Subfamily in Gibberellin-Dependent Stamen Filament Elongation. Plant Physiol. 2020, 182, 2096–2110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cubas, P.; Lauter, N.; Doebley, J.; Coen, E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999, 18, 215–222. [Google Scholar] [CrossRef] [Green Version]
- Navaud, O.; Dabos, P.; Carnus, E.; Tremousaygue, D.; Hervé, C. TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 2007, 65, 23–33. [Google Scholar] [CrossRef]
- Parapunova, V.; Busscher, M.; Busscher-Lange, J.; Lammers, M.; Karlova, R.; Bovy, A.G.; Angenent, G.C.; de Maagd, R.A. Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 2014, 14, 157. [Google Scholar] [CrossRef] [Green Version]
- Shi, P.; Guy, K.M.; Wu, W.; Fang, B.; Yang, J.; Zhang, M.; Hu, Z. Genome-wide identification and expression analysis of the ClTCP transcription factors in Citrullus lanatus. BMC Plant Biol. 2016, 16, 85. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.F.; Chen, Y.Y.; Hsiao, Y.Y.; Shen, C.Y.; Hsu, J.L.; Yeh, C.M.; Mitsuda, N.; Ohme-Takagi, M.; Liu, Z.J.; Tsai, W.C. Genome-wide identification and characterization of TCP genes involved in ovule development of Phalaenopsis equestris. J. Exp. Bot. 2016, 67, 5051–5066. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Hu, S.; Yu, Q.; Wang, C.; Yang, Y.; Sun, H.; Yang, Y.; Sun, X. Genome-Wide Identification and Characterization of BrrTCP Transcription Factors in Brassica rapa ssp. rapa. Front Plant Sci. 2017, 8, 1588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, K.; Ni, Z.; Qu, Y.; Cai, Y.; Yang, Z.; Sun, G.; Chen, Q. Genome-wide identification and expression analyses of TCP transcription factor genes in Gossypium barbadense. Sci. Rep. 2018, 8, 14526. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.M.; Wang, M.M.; Yang, J.; Wen, J.; Guo, P.C.; Wu, Y.W.; Ke, Y.Z.; Li, P.F.; Li, J.N.; Du, H. Evolutionary and Comparative Expression Analyses of TCP Transcription Factor Gene Family in Land Plants. Int. J. Mol. Sci. 2019, 20, 3591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, H.; Chen, Y.; Du, H.; Zhang, L.; Zhang, K.; He, H.; Pan, J.; Cai, R.; Wang, G. Genome-Wide Identification and Characterization of the TCP Gene Family in Cucumber (Cucumis sativus L.) and Their Transcriptional Responses to Different Treatments. Genes 2020, 11, 1379. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, P.; Das, G.M.; Joseph, A.P.; Chatterjee, N.; Srinivasan, N.; Nath, U. Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell. 2010, 22, 1174–1189. [Google Scholar] [CrossRef] [Green Version]
- Danisman, S.; Wal, F.V.D.; Dhondt, S.; Waites, R.; de Folter, S.; Bimbo, A.; van Dijk, A.D.; Muino, J.M.; Cutri, L.; Dornelas, M.C.; et al. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 2012, 159, 1511–1523. [Google Scholar] [CrossRef] [Green Version]
- Howarth, D.G.; Donoghue, M.J. Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. USA 2006, 103, 9101–9106. [Google Scholar] [CrossRef] [Green Version]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef]
- Nicolas, M.; Cubas, P. TCP factors: New kids on the signaling block. Curr. Opin. Plant Biol. 2016, 33, 33–41. [Google Scholar] [CrossRef]
- Ju, Y.; Guo, L.; Cai, Q.; Ma, F.; Zhu, Q.Y.; Zhang, Q.; Sodmergen. Arabidopsis JINGUBANG Is a Negative Regulator of Pollen Germination That Prevents Pollination in Moist Environments. Plant Cell. 2016, 28, 2131–2146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.Y.; Zhao, P.M.; Cheng, H.Q.; Han, L.B.; Wu, X.M.; Gao, P.; Wang, H.Y.; Yang, C.L.; Zhong, N.Q.; Zuo, J.R.; et al. The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiol. 2013, 162, 1669–1680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Zhang, D.; Li, J. TCP1 Modulates DWF4 Expression via Directly Interacting with the GGNCCC Motifs in the Promoter Region of DWF4 in Arabidopsis thaliana. J. Genet. Genom. 2015, 42, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Guan, P.; Ripoll, J.J.; Wang, R.; Vuong, L.; Bailey-Steinitz, L.J.; Ye, D.; Crawford, N.M. Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. USA 2017, 114, 2419–2424. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Yang, Q.; Liao, X.; Tian, Y.; Zhang, F.; Zhang, L.; Liu, Q. A natural antisense RNA improves chrysanthemum cold tolerance by regulating the transcription factor DgTCP1. Plant Physiol. 2022, kiac267. [Google Scholar] [CrossRef]
- Almeida, D.M.; Gregorio, G.B.; Oliveira, M.M.; Saibo, N.J. Five novel transcription factors as potential regulators of OsNHX1 gene expression in a salt tolerant rice genotype. Plant Mol. Biol. 2017, 93, 61–77. [Google Scholar] [CrossRef]
- Xu, L.; Wang, Y.; Zhai, L.; Xu, Y.; Wang, L.; Zhu, X.; Gong, Y.; Yu, R.; Limera, C.; Liu, L. Genome-wide identification and characterization of cadmium-responsive microRNAs and their target genes in radish (Raphanus sativus L.) roots. J. Exp. Bot. 2013, 64, 4271–4287. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.M.; Kim, N.; Ahn, B.O.; Oh, M.; Chung, W.H.; Chung, H.; Jeong, S.; Lim, K.B.; Hwang, Y.J.; Kim, G.B.; et al. Elucidating the triplicated ancestral genome structure of radish based on chromosome-level comparison with the Brassica genomes. Theor. Appl. Genet. 2016, 129, 1357–1372. [Google Scholar] [CrossRef]
- Li, Y.; Shan, X.; Jiang, Z.; Zhao, L.; Jin, F. Genome-wide identification and expression analysis of the GA2ox gene family in maize (Zea mays L.) under various abiotic stress conditions. Plant Physiol. Biochem. 2021, 166, 621–633. [Google Scholar] [CrossRef]
- Wang, Y.; Ying, J.; Zhang, Y.; Xu, L.; Zhang, W.; Ni, M.; Zhu, Y.; Liu, L. Genome-Wide Identification and Functional Characterization of the Cation Proton Antiporter (CPA) Family Related to Salt Stress Response in Radish (Raphanus sativus L.). Int. J. Mol. Sci. 2020, 4, 8262. [Google Scholar] [CrossRef]
- Mahesh, K.; Balaraju, P.; Ramakrishna, B.; Rao, S.S.R. Effect of Brassinosteroids on Germination and Seedling Growth of Radish (Raphanus sativus L.) under PEG-6000 Induced Water Stress. Am. J. Plant Sci. 2013, 4, 2305–2313. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Tang, Y.; Gao, S.; Su, S.; Hong, L.; Wang, W.; Fang, Z.; Li, X.; Ma, J.; Quan, W. Comprehensive analyses of the annexin gene family in wheat. BMC Genom. 2016, 17, 415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; He, X.; Chang, P.; Jiang, H.; Gong, D.; Sun, Q. Genome-wide identification and characterization of TCP family genes in Brassica juncea var. tumida. Peer. J. 2020, 8, e9130. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, 302–305. [Google Scholar] [CrossRef]
- Fan, L.; Xu, L.; Wang, Y.; Tang, M.; Liu, L. Genome- and Transcriptome-Wide Characterization of bZIP Gene Family Identifies Potential Members Involved in Abiotic Stress Response and Anthocyanin Biosynthesis in Radish (Raphanus sativus L.). Int. J. Mol. Sci. 2019, 20, 6334. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Ying, J.; Tang, M.; Wang, Y.; Xu, L.; Liu, M.; Liu, L. Genome-wide identification of AUX/IAA in radish and functional characterization of RsIAA33 gene during taproot thickening. Gene 2021, 795, 145782. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattike, R.A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [Green Version]
- Shen, F.; Ying, J.; Xu, L.; Sun, X.; Wang, J.; Wang, Y.; Mei, Y.; Zhu, Y.; Liu, L. Characterization of Annexin gene family and functional analysis of RsANN1a involved in heat tolerance in radish (Raphanus sativus L.). Physiol. Mol. Biol. Plants 2021, 27, 2027–2041. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Machanick, P.; Bailey, T.L. MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 2011, 27, 1696–1697. [Google Scholar] [CrossRef] [Green Version]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef] [Green Version]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genom. Biol. 2019, 20, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Mitsui, Y.; Shimomura, M.; Komatsu, K.; Namiki, N.; Shibata-Hatta, M.; Imai, M.; Katayose, Y.; Mukai, Y.; Kanamori, H.; Kurita, K.; et al. The radish genome and comprehensive gene expression profile of tuberous root formation and development. Sci. Rep. 2015, 5, 10835. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar]
- Bailey, T.L.; Gribskov, M. Combining evidence using p-values: Application to sequence homology searches. Bioinformatics 1998, 14, 48–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, M.; Xu, L.; Wang, Y.; Cheng, W.; Luo, X.; Xie, Y.; Fan, L.; Liu, L. Genome-wide characterization and evolutionary analysis of heat shock transcription factors (HSFs) to reveal their potential role under abiotic stresses in radish (Raphanus sativus L.). BMC Genom. 2019, 20, 772. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Su, N.; Jia, L.; Tian, J.; Li, H.; Huang, L.; Shen, Z.; Cui, J. Transcriptome analysis of radish sprouts hypocotyls reveals the regulatory role of hydrogen-rich water in anthocyanin biosynthesis under UV-A. BMC Plant Biol. 2018, 18, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Liu, W.; Shen, H.; Zhu, X.; Zhai, L.; Xu, L.; Wang, R.; Gong, Y.; Limera, C.; Liu, L. Identification of Radish (Raphanus sativus L.) miRNAs and Their Target Genes to Explore miRNA-Mediated Regulatory Networks in Lead (Pb) Stress Responses by High-Throughput Sequencing and Degradome Analysis. Plant Mol. Biol. Rep. 2015, 33, 358–376. [Google Scholar] [CrossRef]
- Yang, Z.; Li, W.; Su, X.; Ge, P.; Ge, P.; Zhou, Y.; Hao, Y.; Shu, H.; Gao, C.; Cheng, S.; et al. Early Response of Radish to Heat Stress by Strand-Specific Transcriptome and miRNA Analysis. Int. J. Mol. Sci. 2019, 20, 3321. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Xu, L.; Wang, Y.; Shen, H.; Zhu, X.; Zhang, K.; Chen, Y.; Yu, R.; Limera, C.; Liu, L. Transcriptome-wide analysis of chromium-stress responsive microRNAs to explore miRNA-mediated regulatory networks in radish (Raphanus sativus L.). Sci. Rep. 2015, 5, 14024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Wang, Q.; Sun, R.; Xie, F.; Jones, D.C.; Zhang, B. Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci. Rep. 2014, 4, 6645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Chen, Y.Q.; Ding, A.M.; Chen, H.; Xia, F.; Wang, W.F.; Sun, Y.H. Genome-wide analysis of TCP family in tobacco. Genet. Mol. Res. 2016, 15, gmr7728. [Google Scholar] [CrossRef]
- Airoldi, C.A.; Davies, B. Gene duplication and the evolution of plant MADS-box transcription factors. J. Genet. Genom. 2012, 39, 157–165. [Google Scholar] [CrossRef]
- De Grassi, A.; Lanave, C.; Saccone, C. Genome duplication and gene-family evolution: The case of three OXPHOS gene families. Gene 2008, 421, 1–6. [Google Scholar] [CrossRef]
- Chen, P.; Li, J.; Ye, X.; Tan, B.; Zheng, X.; Cheng, J.; Wang, W.; Wang, H.; Gu, L.; Feng, J. Genome-wide identification of ziziphus jujuba TCP transcription factors and their expression in response to infection with jujube witches’ broom phytoplasma. Acta Physiol. Plant 2019, 41, 86. [Google Scholar] [CrossRef]
- Zheng, A.; Sun, F.; Cheng, T.; Wang, Y.; Xie, K.; Zhang, C.; Xi, Y. Genome-wide identification of members of the TCP gene family in switchgrass (Panicum virgatum L.) and analysis of their expression. Gene 2019, 702, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Ohashi, Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 2002, 30, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell. 2010, 22, 3574–3588. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Xu, X.; Mo, X.; Zhong, L.; Zhang, J.; Mo, B.; Kuai, B. Overexpression of TCP8 delays Arabidopsis flowering through a FLOWERING LOCUS C-dependent pathway. BMC Plant Biol. 2019, 19, 534. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, R.E.; Schommer, C.; Palatnik, J.F. Control of cell proliferation by microRNAs in plants. Curr. Opin. Plant Biol. 2016, 34, 68–76. [Google Scholar] [CrossRef]
- Shi, G.; Fu, J.; Rong, L.; Zhang, P.; Guo, C.; Xiao, K. TaMIR1119, a miRNA family member of wheat (Triticum aestivum), is essential in the regulation of plant drought tolerance. J. Integr. Agric. 2018, 17, 2369–2378. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.K.; Prasad, A.; Maurya, J.; Prasad, M. Regulation of small RNA-mediated high temperature stress responses in crop plants. Plant. Cell Rep. 2022, 41, 765–773. [Google Scholar] [CrossRef]
- Ali, M.; Javaid, A.; Naqvi, S.H.; Batcho, A.; Kayani, W.K.; Lal, A.; Sajid, I.A.; Nwogwugwu, J.O. Biotic stress triggered small RNA and RNAi defense response in plants. Mol. Biol. Rep. 2020, 47, 5511–5522. [Google Scholar] [CrossRef]
- Zhang, L.; Song, C.; Guo, D.; Guo, L.; Hou, X.; Wang, H. Identification of differentially expressed miRNAs and their target genes in response to brassinolide treatment on flowering of tree peony (Paeonia ostii). Plant Signal Behav. 2022, 17, 2056364. [Google Scholar] [CrossRef]
- Mao, Y.; Wu, F.; Yu, X.; Bai, J.; Zhong, W.; He, Y. MicroRNA319a-targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol. 2014, 164, 710–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Li, S.; Ma, Q.; Wen, J.; Yan, K.; Li, Q. The Acer palmatum TCP Transcription Factor ApTCP2 Controls Leaf Morphogenesis, Accelerates Senescence, and Affects Flowering via miR319 in Arabidopsis thaliana. J. Plant Growth Regul. 2021, 41, 244–256. [Google Scholar] [CrossRef]
- Schommer, C.; Debernardi, J.M.; Bresso, E.G.; Rodriguez, R.E.; Palatnik, J.F. Repression of cell proliferation by miR319-regulated TCP4. Mol. Plant 2014, 7, 1533–1544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bresso, E.G.; Chorostecki, U.; Rodriguez, R.E.; Palatnik, J.F.; Schommer, C. Spatial Control of Gene Expression by miR319-Regulated TCP Transcription Factors in Leaf Development. Plant Physiol. 2018, 176, 1694–1708. [Google Scholar] [CrossRef] [PubMed]
- Es, S.W.; Silveira, S.R.; Rocha, D.I.; Bimbo, A.; Martinelli, A.P.; Dornelas, M.C.; Angenent, G.C.; Immink, R.G.H. Novel functions of the Arabidopsis transcription factor TCP5 in petal development and ethylene biosynthesis. Plant J. 2018, 94, 867–879. [Google Scholar]
- Mizoi, J.; Kanazawa, N.; Kidokoro, S.; Takahashi, F.; Qin, F.; Morimoto, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Heat-induced inhibition of phosphorylation of the stress-protective transcription factor DREB2A promotes thermotolerance of Arabidopsis thaliana. J. Biol. Chem. 2019, 294, 902–917. [Google Scholar] [CrossRef] [Green Version]

| Name | PKC | CK II | cAMP-cGMP | N-Myr | N-Glyc |
|---|---|---|---|---|---|
| RsTCP01 | 3 | 1 | 0 | 0 | 3 |
| RsTCP02 | 11 | 8 | 0 | 20 | 6 |
| RsTCP03 | 0 | 0 | 0 | 0 | 0 |
| RsTCP04 | 5 | 3 | 1 | 14 | 5 |
| RsTCP05 | 3 | 5 | 1 | 12 | 2 |
| RsTCP06 | 6 | 7 | 0 | 3 | 1 |
| RsTCP07 | 2 | 1 | 0 | 7 | 0 |
| RsTCP08 | 4 | 7 | 0 | 10 | 4 |
| RsTCP09 | 4 | 7 | 0 | 11 | 4 |
| RsTCP10 | 6 | 3 | 0 | 2 | 4 |
| RsTCP11 | 3 | 4 | 0 | 6 | 2 |
| RsTCP12 | 5 | 5 | 1 | 16 | 2 |
| RsTCP13 | 3 | 4 | 1 | 6 | 2 |
| RsTCP14 | 6 | 4 | 0 | 3 | 1 |
| RsTCP15 | 5 | 4 | 0 | 3 | 5 |
| RsTCP16 | 6 | 6 | 1 | 7 | 4 |
| RsTCP17 | 8 | 4 | 0 | 6 | 4 |
| RsTCP18 | 5 | 7 | 0 | 2 | 0 |
| RsTCP19 | 4 | 7 | 3 | 3 | 4 |
| RsTCP20 | 5 | 3 | 0 | 3 | 3 |
| RsTCP21 | 3 | 7 | 1 | 14 | 3 |
| RsTCP22 | 4 | 5 | 0 | 5 | 4 |
| RsTCP23 | 9 | 5 | 3 | 5 | 1 |
| RsTCP24 | 9 | 5 | 3 | 5 | 1 |
| RsTCP25 | 4 | 6 | 1 | 15 | 3 |
| RsTCP26 | 7 | 9 | 0 | 7 | 2 |
| RsTCP27 | 5 | 6 | 1 | 17 | 5 |
| RsTCP28 | 5 | 7 | 1 | 23 | 3 |
| RsTCP29 | 2 | 2 | 0 | 4 | 1 |
| RsTCP30 | 4 | 2 | 0 | 4 | 5 |
| RsTCP31 | 4 | 7 | 1 | 12 | 1 |
| RsTCP32 | 8 | 9 | 0 | 16 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mei, Y.; Liu, Z.; Zheng, J.; Wang, W.; Zu, Y.; Wu, Y.; Zhang, L.; Feng, R.; Shen, F. Genome-Wide Identification and Expression Pattern Analysis of the TCP Gene Family in Radish (Raphanus sativus L.). Horticulturae 2022, 8, 656. https://doi.org/10.3390/horticulturae8070656
Mei Y, Liu Z, Zheng J, Wang W, Zu Y, Wu Y, Zhang L, Feng R, Shen F. Genome-Wide Identification and Expression Pattern Analysis of the TCP Gene Family in Radish (Raphanus sativus L.). Horticulturae. 2022; 8(7):656. https://doi.org/10.3390/horticulturae8070656
Chicago/Turabian StyleMei, Yi, Zhe Liu, Jiaqiu Zheng, Weiwei Wang, Yanxia Zu, Yongcheng Wu, Lina Zhang, Ruchao Feng, and Feng Shen. 2022. "Genome-Wide Identification and Expression Pattern Analysis of the TCP Gene Family in Radish (Raphanus sativus L.)" Horticulturae 8, no. 7: 656. https://doi.org/10.3390/horticulturae8070656
APA StyleMei, Y., Liu, Z., Zheng, J., Wang, W., Zu, Y., Wu, Y., Zhang, L., Feng, R., & Shen, F. (2022). Genome-Wide Identification and Expression Pattern Analysis of the TCP Gene Family in Radish (Raphanus sativus L.). Horticulturae, 8(7), 656. https://doi.org/10.3390/horticulturae8070656






