Abstract
In order to best conserve, as well as utilize, traditional apple germplasm in Norway, an apple heritage cultivar collection was established in Ullensvang, western Norway, which aims to become the National Clonal Germplasm Repository. The establishment of the apple heritage cultivar collection was preceded by a molecular study that aimed to genotype a large number of apple accessions maintained in various ex situ sites in western and south-eastern Norway, using a rather small set of eight SSR markers. However limited, the marker set managed to identify synonyms, homonyms, and duplicates within and among the investigated collections. In this study, 171 apple accessions from the Ullensvang apple heritage cultivar collection were genotyped using a set of 20 different SSR markers. Approximately half of the accessions have been previously genotyped using eight SSR markers, enabling an assessment of whether the use of a larger marker set would yield a more accurate characterization. Based on the obtained molecular data, the apple heritage cultivar collection was determined to hold a key part of the overall genetic diversity of the Norwegian apple germplasm. Furthermore, the twelve additional SSR markers were able to differentiate several accessions groups originally thought to be synonyms, as well as to provide a more detailed insight into the genetic structure of this germplasm.
1. Introduction
Apple (Malus × domestica Borkh.) is economically and culturally the most important temperate fruit crop in the world and it is well adapted to the temperate climate zone []. The conservation and utilization of plant genetic resources (PGR) in agriculture has received increasing attention over the last few decades and is one of the foci in the development of sustainable food production. Existing fruit PGR are the result of centuries of adaptation, natural selection and breeding, traditionally by farmers but more recently by expert breeders. Availability and informative value of plant germplasms are becoming more and more important for the future preservation and sustainable use of genetic resources []. However, this task is particularly challenging for trees whose life cycles are very long []. The importance of the research and conservation of local, traditional cultivars is evident, as this genetically heterogeneous material represents a potential source of positive pomological traits and resistance to biotic (including pests and diseases) and abiotic stress []. Consequently, future breeding efforts must include sustainable use of these resources. The overall and long-term changes in the structure and methods in agriculture production significantly affect the state of genetic diversity in the agricultural sector. The majority of commercial farmers cultivate modern international cultivars under intensive production methods. This means that traditional varieties have been replaced and are scarce in commercial agriculture. It is important to note that besides the agricultural production aspect, biodiversity of agricultural species also represents an important part of biocultural identity and heritage. However, there is no sustainable conservation without utilization. In order to promote the utilization of PGR, the first step must consist of collecting as many relevant data about them as possible. The evaluation of important commercial characteristics and the genetic diversity of fruit accessions would enable pre-selection; a trend present in many leading PGR collections around the world.
Apple genetic resources in Norway are currently conserved within 19 local clonal archives. However, during establishment of these ex situ collections, primary focus was not on capturing as much of the diversity as possible, but instead on preserving cultivars of particular importance to specific fruit-growing areas. Although the origin of these individual cultivars varies greatly (Norway, Sweden, England, North America, Russia, etc.), they all are traditionally cultivated in Norway. To identify redundancies within the collection as well as to assess the genetic diversity and structure of apple germplasm currently being conserved in Norway, eight highly polymorphic SSR markers were used in the genetic characterization of 181 apple accessions in six ex situ collections []. After this mentioned study, an apple heritage cultivar collection was established in Ullensvang, Norway, which aims to become the National Clonal Germplasm Repository. This measure has been taken in order to ensure the long-term sustainability of the conservation process as well as to enhance the possibility of the utilization of the collected germplasm. The accessions selected for the apple heritage cultivar collection were considered a priority according to the national mandate list. It is important to note that some of the accessions determined to be synonyms or duplicates by Gasi et al. [] were included in the collection due to their phenotypic divergence assessed by curators of the local clonal archives.
Although the number of microsatellite loci analyzed in similar studies at the time of the publication by Gasi et al. [] was as high as 24 SSRs [], the use of a limited number of SSR markers was justified with the argument that molecular markers were employed to increase the financial sustainability of the conservation process (by identifying redundancies). Therefore, the high cost of using an ever-larger set of markers would represent an unnecessary financial burden to conservation efforts.
Since that time, numerous large scale genetic studies on apple accessions have been conducted using everything from 13 [,], 14 [], 15 [,], 16 [], and up to 19 different SSRs []. Although none of the mentioned studies surpass the Lassois et al. [] study in terms of the sheer number of microsatellite markers, there is a notable increase in the lower end number of SSRs employed in genetic studies on apple accession through the years. Namely, Gasi et al. [] mentioned that early on, and at that time, more recent studies relied on the use of as few as eight SSRs in their investigations [,]. This could be due to technological as well as methodological progress, such as the development of the SSR genotyping one-tube reaction kit [], which has reduced the necessary time and labor, as well as the overall cost associated with SSR genotyping.
Additional developments in genetic diversity studies on apple germplasm has been the use of SNP arrays, either in combination with microsatellites [] or as an exclusive method for the genotyping of apple accessions [,]. Other high-throughput approaches, such as the use of Diversity Arrays Technology (DArT) markers, have also been employed in genetic studies on apples []. However, regardless of the new genomic approaches, microsatellites remain an effective and cost-efficient marker []. The cost-effectiveness of this marker system is perhaps still the most alluring aspect, especially for national gene banks struggling with the financial sustainability of their conservation efforts. Additionally, there is an existing set of SSR markers recommended by the European Cooperation Program for Plant Genetic Resources (ECPGR) which allows data comparison between different studies []. In this study, 171 apple accessions from the Ullensvang apple heritage cultivar collection were genotyped using 20 different SSR markers to obtain SSR profiles for each accession within this candidate collection for the Norwegian National Clonal Germplasm Repository. Among the genotyped accessions, approximately half (93 accessions) have been previously genotyped using eight SSR markers by Gasi et al. []. The use of additional markers on these accessions had the added benefit of answering the question of whether the use of a larger marker set would yield a more accurate characterization of the mentioned genetic resource. In order to quantify this accuracy, the concordance in terms of synonyms, homonyms, and duplicates between the present and 2016 study was primarily examined. Furthermore, the obtained molecular data on all 171 accessions were used to investigate the genetic diversity and structure of the analyzed apple germplasm.
2. Materials and Methods
2.1. Plant Material and Managements
The apple accessions archive was planted in 2018 and 2019 at the experimental farm of NIBIO Ullensvang (60.318655, 6.652948). The soil is a sandy loam with approximately 4% organic matter, being very uniform in morphological and physical characteristics (color and structure). All accessions were grafted on M9 rootstocks spaced 1 × 3.5 m apart, three trees of each accession and all trees were trained as spindle trees, and pruned to a maximum height of about 2.5–3 m. The selected trees are homogeneous in terms of flower set, vigor, and health status. The weeds under the trees were removed by a 1 m wide herbicide strip using glyfosat (trade name Roundup with 360 g/L of glyphosate, Monsanto Crop Sciences) which was maintained in each season, together with frequent grass mowing in the inter-rows. Pest managements were conducted according to integrated protocols, spraying against major pests (insects and diseases) when needed []. When water deficits occurred, trees were irrigated by drip irrigation. All trees received the same number of fertilizers based on soil and leaf analysis. Hand thinning was carried out at the end of June in order to achieve optimum crop loads of good fruit quality (15 cm apart between fruitlets).
2.2. Genetic Analyses
During the late spring of 2021, young leaf tissues (approximately 20 mg/sample) were collected from 171 apple accessions maintained at the apple heritage cultivar collection in Ullensvang, Norway (Table 1). The tissue was silica-dried before being reduced to a fine powder using a Qiagen Tissue Lyser device (QIAGEN, Hilden, Germany). DNA extraction was carried out using the commercially available NucleoSpin Plant II, a Mini kit for DNA from plants (Macherey-Nagel, Dueren, Germany), following the manufacturer’s recommendation. Twenty SSR markers were chosen based on their polymorphism reported in previous studies on apples [,,,]. It is important to note that more than half of the SSRs recommended by ECPGR have been included in this study and eight of them have previously been used by Gasi et al. []. Forward primers were labeled with three different fluorescent dyes (6-FAM, HEX, and TAMRA). Based on a size range and annealing temperature, eighteen primer pairs were combined into six multiplex reactions (MIX1-CH02B10, CH05E03, CH02C02a; MIX2-CH03D12, CH02C11, CH01D03; MIX3-CH01F07a, CH04E03, CH01D09; MIX4-CH01H01, CH01H02, CH01H10; MIX5-CH02C02a, CH02C02b, CH04E02; and MIX6-EMPC117, CH02C09, GD12), and the remaining two primer pairs (CH05e04 and CH02C06) were single amplified. All PCR reactions were conducted in a total volume of 15 µL, containing 2 mM of MgCl2, 1 × PCR buffer, 0.2 mM of dNTPs, 0.05 U/µL of TaqNovaHS DNA Polymerase (Blirt, Gdańsk, Poland), and 10–50 ng of template DNA. All the primer pairs were amplified as described in Gianfranceschi et al. [], Liebhard et al. [], and Gasi et al. [], with minor modifications. Diluted PCR products were mixed with Hi-Di formamide (Applied Biosystems) and a prepared size standard. An ABI PRISM 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) was used for the electrophoretic separation of PCR products. Alleles were sized relative to the internal size standard LIZ 500 (Applied Biosystems, Warrington, UK). GeneMapper v. 5 (Applied Biosystems) was used for allele scoring. In case of any ambiguity (samples differing in only a few bp on one or few loci or displaying uncharacteristic peak morphology) samples were rerun on the Genetic Analyzer.

Table 1.
A total of 171 apple accessions conserved in Ullensvang, western Norway, analyzed using 20 SSR markers, as well as assignment of each non-admixed genotype to a genetic cluster (GC) (K = 4) defined by Structure [] (probability of membership qI > 80%).
2.3. Biostatistical Analysis
Allele frequencies, gene diversity [], and the number of effective alleles for each of the 20 microsatellite loci were calculated in the population genetics software SPAGeDI 1.4 []. Calculated allele frequencies enabled the identification of rare alleles (alleles with a frequency lower than 0.05%). In order to calculate the sufficient number of SSRs needed to differentiate individual accessions, an accumulation curve approach of the “poppr package” was used in R []. A Bayesian model-based cluster procedure within Structure ver. 2.2.3 [] was employed in order to conduct a Structure analysis. Genetic clusters (K), reconstructed under panmictic population (RPP) assumptions, were computed on individuals testing K (log-likelihood) = 1–10 for all accessions, based on the assumption that the sampled cultivars were from an unknown origin. For each K, ten independent runs were conducted. Tests were based on the admixture model, where allelic frequencies were correlated, and which assumes different Fst values for specific sub-clusters in a burn-in period of 200,000 and 500,000 iterations. Structure harvester ver. 0.6.1 application [], which implements the Evanno method [], was employed to estimate the most probable K value. After determining the K value, the individuals were assigned to specific clusters via the run with the maximum likelihood []. The genotypes were assigned to the groups according to their highest membership coefficient, with the assigning probability (qI) of 80% according to similar studies [,,,]. All accessions with the probability of membership to the Genetic clusters below 80% were deemed admixed. An analysis of Molecular Variance (AMOVA) [], based on the stepwise mutation model [], was conducted within GenoType software [GenoType/GenoDive package [] with 1000 permutations.
A factorial correspondence analysis (FCA), based on a matrix of binary microsatellite allele presence/absence data, was performed using the “dudi.coa” routine in R 4.1.3 [], as suggested by Muller and McCusker []. The graphical display of the FCA results was conducted with the rgl package [], also in R 4.1.3.
Similarity among all 171 accessions was determined using an Unweighted Pair Group Method with Arithmetic mean (UPGMA) cluster analysis based on a matrix with pairwise comparisons using Jaccards similarity coefficient. Calculations were carried out with the use of R packages philentropy [] and usedist []. The dendrogram was constructed in MEGA 6 software (Molecular Evolutionary Genetics Analysis) [].
All input data for statistical software was prepared with the MADC v. 2.0 computer program [].
3. Results and Discussion
3.1. Allele Polymorphism
In this study twenty primer pairs amplified 288 distinct alleles, or an average of 14.3 alleles per locus (Table 2). These values are higher than the ones reported from the previous study on 181 apple accessions from Norway (11.9) []. A similar value for this parameter has been reported by Marconi et al. [] (14.6), who investigated 175 mostly Italian apple accessions provided by 10 apple collections, using 19 SSR markers. Higher values for an average number of alleles per SSR locus such as 18.5 reported by van Treuren et al. [], 16.7 by Urrestarazu et al. [], 16.8 by Liang et al. [], 19.5 by Lassois et al. [], 17.7 by Pereira-Lorenzo et al. [] and up to 23.06 by Urrestarazu et al. []. The differences in the reported values are probably a product of a vast difference in the number of analyzed accessions between the studies. The effective number of alleles per locus was 5.43 (Table 2), higher than reported by Gasi et al. (2016) [] (4.53) and similar to two studies on Italian apple germplasm by Liang et al. [] (5.64) and Marconi et al. [] (5.94). A higher value for the effective number of alleles per locus has been obtained in probably the most extensive apple germplasm study to this day, which included almost 2500 apple accessions [] (6.59) from collections located in nine European countries (Norwegian collections were not included in the research).

Table 2.
Number of detected alleles, number of rare alleles (frequency < 0.05), number of effective alleles, allele size range, gene diversity calculated for 171 apple accessions maintained in the apple heritage cultivar collection Ullensvang, Norway.
The difference between the average allele number and average effective allele number obtained in this study can be attributed to the relatively high presence of rare alleles (177 alleles or 61.8%). A high presence of rare alleles can indicate that a large portion of the investigated germplasm has not been extensively used in breeding programs. This is particularly the case with traditional Norwegian cultivars. The obtained values are, however, lower compared to the 73.4% of rare alleles detected for apple accessions from three broad European geographic regions (North + East, West and South) []. The calculated gene diversity for all loci was 0.76, ranging between 0.47 for EMPC117 and 0.92 for CH02C06 (Table 2). The calculated value is very similar to that previously reported on the Norwegian apple germplasm by Gaši et al. [] (0.75). Somewhat higher values for gene diversity have been reported by Urrestarazu et al. [] (0.82), Liang et al. [] (0.83), Lassois et al. [] (0.82), Urrestarazu et al. [] (0.83), Marconi et al. [] (0.81) and Pereira-Lorenzo et al. [] (0.815).
The detection of more than two different alleles per locus, which was taken as an indication of a triploid state (none of the primer pairs displayed the ability to amplify more than one locus), was detected in 22 (12.86%) apple accessions, which is in line with the data previously reported, where this state was present in 12% of the apple accessions from Norway []. It is important to note that only those accessions displaying more than two alleles on at least three loci, after the rerun of the sample on the Genetic Analyzer, were deemed as triploids. Although this approach is not a substitute for flow cytometry, a complete correlation between triploids detected with SSRs and flow cytometry has been reported by Pereira-Lorenzo et al. [].
A higher percentage of genotypes with three detected alleles per locus have been reported by studies conducted on Spanish [] (24%), Bosnian [,] (27%), Spanish and Portuguese [] (24%), as well as Turkish (18.9%) [] apple germplasm. A higher frequency of triploid apple accession among germplasm from southern Europe compared to northern Europe has been noted by Kanlic et al. [], who proposed that this was due to the effect of climatic conditions on the pollination rates and differences in the traditional end-use of the fruit. Namely, the authors argued that pollination issues due to unequal segregation of the chromosomes during meiosis, prevalent among triploids, might have been the bases for negative selection pressure in a Northern climate, which is less conducive for pollination.
3.2. Genetic Identity and Relationships
Among the 171 accessions, the following four clear cases of duplicates (accessions with identical names and identical SSR profiles on all 20 analyzed loci) were detected: “Stor Granat”, “Gravenstein”, “Katja”, and “Filipa” (Figure 1). Any phenotypic differences between the duplicates, which led to their inclusion in the Ullensvang apple heritage cultivar collection, are either caused by genetic factors, but not detectable with 20 SSRs used, or a consequence of environmental factors.
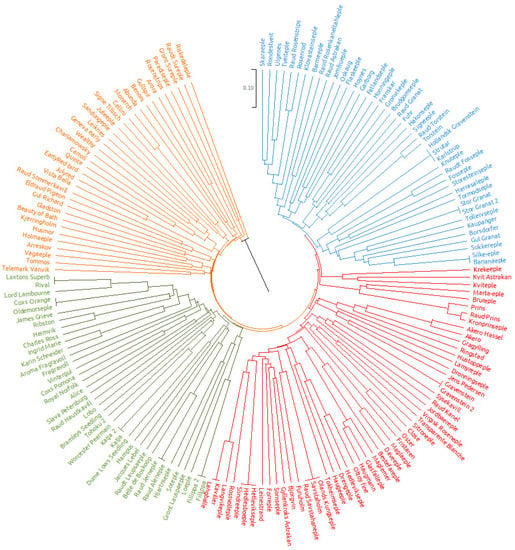
Figure 1.
UPGMA cluster analysis based on polymorphisms of SSR data for 171 apple accessions using Jaccard’s similarity coefficient.
Overall, 8 groups of synonyms (2 or more accessions with different names but identical SSR profiles on all 20 analyzed loci) were found among the 171 apple accessions collected in Norway. Namely, the accession “Rival” had an identical SSR profile to “Laxtons Superb”, “Langballe” as “Kaviller”, “Oldemorseple” as “Cox’s Orange”, “Hollandsk Gravenstein” as both “Strutar” and “Karlstrup”, “RaudÅleneple” as “RaudJerneple”, “Silke-eple” as “Bananeple”, “Sävstaholm” as both “Raud Sävstahaneple” and “Ølands Kungseple” and “Hedlesbøeple” as both “Hedlevikseple” and “Slindreeple”. The percentage of redundancies in the analyzed collection was 8.8%, which is lower compared to the previous study on Norwegian apple collections [] (13%), as well what has been reported for apple collections in the Netherlands [] (32%), Spain [] (47%), Italy [] (34%) and France (34%) []. Higher values for redundancies have also been reported by Urrestarazu et al. [] (16%) among and within 14 large European apple collections. The lower redundancies reported in this study are probably due to the fact that prior to the formation of the apple heritage cultivar collection in Ullensvang, a molecular study conducted by Gasi et al. [] already identified several duplicates and synonyms.
Considering the overall number of analyzed accessions, the number of mislabeling through synonyms and homonyms is rather low in the Ullensvang apple heritage cultivar collection. The previous genetic study conducted on six Norwegian ex situ apple collections by Gasi et al. [] certainly aided in avoiding mislabeling and redundancies; however, it is worth considering that a significant portion of accessions presented here have not previously been genotyped.
UPGMA cluster analysis grouped all 171 apple accessions into a dendrogram, with duplicates occurring on the same line (Figure 1). The cluster analysis was able to group all genotypes into four large clusters, with the first one (counterclockwise) containing several old English cultivars (“Cellini”, “Gladstone” and “Beauty of Bath”), as well as numerous cultivars developed through breeding programs in the USA (“Benoni”, “Monarch”, “Geneva early”, “EarlyRed bird”, “Julyred” and “VistaBella”). Furthermore, within this cluster, several winter hardy cultivars (“Wealthy”, “Charlamowsky”, “Carroll” and “Quinte”) were grouped next to each other. The second major cluster was also dominated by old English cultivars such as “Cox’s Orange” and “Cox’s Pomona”, as well as their presumed progenitor “Ribston”. The Swedish cultivar “Katja”, a cross between “James Grieve” (itself a seedling of “Cox’s Orange”) and “Worcester Pearmain” also grouped in this cluster together with both of its parents. Additionally included in this cluster was the old Danish cultivar “Filippa”. The two remaining major clusters consisted of traditional Norwegian apple cultivars and represented very diverse groups with different levels of genetic relatedness between individual genotypes.
3.3. Accuracy of a Larger Marker Set
In order to quantify a potential increase in genetic identity accuracy, achieved through the use of additional 12 SSR markers, the concordance in terms of synonyms, homonyms, and duplicates between the present and 2016 study was examined. The accessions “Åkerø” and “Åkerø Hassel”, although possessing very similar names, differed in four SSR loci. However, in the 2016 study, “Åkerø Hassel”, and “Åkerø” had identical genetic profiles on all eight of the analyzed loci. The accessions “Fosseple” from Njøs and “Raudt Fosseple” were determined to be genetically identical in the 2016 study, while the use of additional SSR markers found a difference between the two accessions in three SSR loci. However, these differences were only in terms of 2 bps. As “Raudt Fosseple” (eng. “Red Fosseple”) and “Fosseple” are genetically very similar and their names vary only in the prefix raudt or red, it is reasonable to assume that “Raudt Fosseple” probably is a red mutant. The accessions “Raud Torstein” and “Torstein” differed in only 2 bps on one of the 20 SSR loci, while these accessions were indistinguishable in the 2016 study, due to the use of a limited number of SSR loci. The accessions “Laxtons Superb” and “Lord Lambourne” were determined to be synonyms in the 2016 study, whereas in the 2021 study, the use of additional SSR markers managed to show a clear difference in SSR profiles in five of the additional loci analyzed here. The accessions “Prins” and “Kronprinseple”, were determined to be synonyms in the 2016 study. However, these two, as well as a third accession carrying the name “Prins” (“Raud Prins”), slightly differentiated with the use of a much larger set of SSR loci. Namely, “Raud Prins” and “Kronprinseple” differed in one locus, “Raud Prins” and “Prins” differed in two loci, while “Prins” and “Kronprinseple’ differed in three of the 20 loci. It is important to note that these differences were only in terms of 2 to 4 bps, still making these accessions genetically highly similar. Overall, the use of a larger set of SSR markers gave more accurate insight into the genetic identity of the apple germplasm maintained in Norwegian ex situ collections. However, it is hard to quantify if this deeper insight justified the extra cost of employing an additional 12 SSR markers. Namely, using an accumulation curve approach (Figure 2), it was determined that only six SSRs, of the 20 analyzed in the study, were sufficient to differentiate all accessions with a differing SSR profile. However, it is worth noting that diversity studies rarely rely on markers exclusively for the differentiation of individual accessions, but also for investigating genetic relationships and underlying the genetic structure, where the use of additional markers can be beneficial. Aside from this, the collection would benefit greatly from a thorough pomological characterization, which would add a new dimension to the obtained molecular data. Joint molecular and phenotypic data should serve as a sound basis for the exclusion of any redundancies identified in this study. The benefits of joint the evaluation of genetic and phenotypic characteristics were recently reported by Wiehle et al. [].
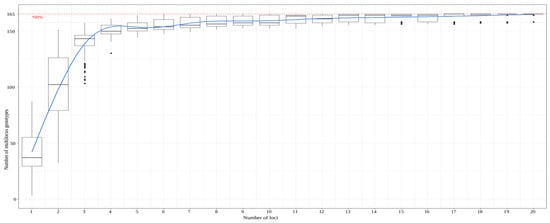
Figure 2.
The minimum number of microsatellite markers needed to differentiate distinct accessions, determined using an accumulation curve calculated in the poppr package within R.
3.4. Genetic Structure
A Bayesian analysis was implemented on 171 accessions in order to investigate the underlying structure of the analyzed collection. The subsequent ΔK analyses [] revealed a maximum value for K = 4. After assigning individual accessions to genetic clusters (GC), to which they displayed a probability of membership above 80%, 25 accessions grouped in GC1, 25 in GC2, 43 in GC3, and 16 in GC4 (Table 1). A total of 62 accessions were determined to be admixed for K = 4 (Table 1). The total percentage of admixed accessions was 36%, which is higher than in the previous study on apple germplasm from Norway (28.86%) []. This is to be expected due to the use of a much greater number of SSRs. The grouping into the GCs reflected in many ways the results of the previous UPGMA cluster analysis, with GC4 containing numerous old English cultivars (“Charles Ross”, “Cox’s Orange”, “Ribston”, “Worcester Pearmain”, etc.), as well as some progeny of these cultivars (“Ingrid Marie” and “Katja”). The GC3 contained a mix of old English, as well as cultivars developed in North American breeding programs. Additionally, in this GC, winter hardy cultivars (“Wealthy”, “Charlamowsky”, “Carroll” and “Quinte”) were classified with a probability of membership above 80%. As with the cluster analyses based on Jaccard’s similarity coefficient, the final two GCs (GC1 and GC2) held numerous traditional Norwegian apple cultivars. Unlike other GCs, containing many foreign apple cultivars, traditional Norwegian accession found in GC1 and GC2, developed under specific environmental conditions present in Norway, as well as under certain selection pressure exerted by the local farmer population with their specific customs and food preferences.
In spite of the differences in the two statistical approaches, there is a high degree of concordance between the results of the UPGMA cluster analyses (Figure 1) and the Bayesian analyses (Table 1). This serves to strengthen the conclusions on genetic relationships between certain segments of the analyzed germplasm, presented within this study.
An analysis of molecular variance (AMOVA) carried out on the previously described GCs revealed that a significant part of the variance (14%; p < 0.01) was ascribed to differences among the analyzed four GCs, indicating a significant genetic differentiation between these groups. A factorial correspondence analysis (FCA) (Figure 3) displayed an overlap between the GCs containing old English varieties (GC3 and GC4), while the remaining two GCs clearly separated from each, as well from the GCs containing foreign cultivars. In general, the analyses of the genetic structure conducted on the accessions maintained at the apple heritage cultivar collection in Ullensvang, indicate a more complex structure compared to the results of the previous study on Norwegian apple germplasm. Namely, Gasi et al. [] reported that all analyzed apple accessions maintained in six ex situ collections were classified into two main GCs. The first one containing traditional Scandinavian cultivars, while the second GC including international cultivars, as well as numerous old apple cultivars introduced to Norway from western Europe and North America. The added sub-structuring obtained in this study could be a consequence of more detailed genetic information gathered through the use of an additional 12 SSR markers.
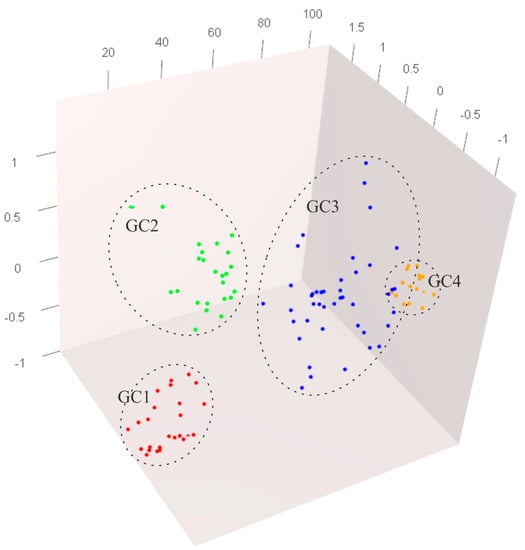
Figure 3.
Factorial Correspondence Analysis (FCA) of SSR data for four defined genetic clusters calculated using Structure [] (only genotypes with the likelihood of membership to individual GC above 80% are included in the analyses).
4. Conclusions
Based on the presented results, it can be concluded that the apple heritage cultivar collection established in Ullensvang contains the key part of the overall genetic diversity of Norwegian apple germplasm, which, until recently, has been mostly maintained at various different collections. With that in mind, the apple heritage cultivar collection represents a serious contender for the role of the National Clonal Germplasm Repository for apples in Norway. Although the use of twelve additional SSR markers was instrumental in differentiating between several accession groups originally thought to be synonyms, the added value of the use of a larger set of markers can also be seen through a better insight into the genetic structure of germplasm.
Author Contributions
Conceptualization, F.G. and M.M.; methodology and formal analysis, N.P., A.K., L.L. and O.F.; writing—original draft preparation F.G.; writing—review and editing M.F.A., M.M. and F.G.; project administration M.M., and funding acquisition; M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by Norwegian Agriculture Agency (project No. 2021/3847, Agros 143861).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
SSR data is avalaible from NIBIO Ullensvang at request.
Acknowledgments
The processing fees for publishing this paper were covered by the Mirsad Kurtovic memorial fund.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fotiric Akšić, M.D.; Dabić Zagorac, U.; Gašić, T.; Tosti, M.; Natić, M.; Meland, M. Analysis of apple fruit (Malus × domestica Borkh.) quality attributes obtained from organic and integrated production systems. Sustainability 2022, 14, 5300. [Google Scholar] [CrossRef]
- Lacis, G.; Trajkovski, V.; Rashal, I. Phenotypical Variability and Genetic Diversity within Accessions of the Swedish Sour Cherry (Prunus cerasus L.) Genetic Resources Collection. Biologija 2010, 56, 1–8. [Google Scholar] [CrossRef]
- Jolivet, C.; Höltken, A.M.; Liesebach, H.; Steiner, W.; Degen, B. Spatial Genetic Structure in Wild Cherry (Prunus avium L.): I. Variation among Natural Populations of Different Density. Tree Genet. Genomes 2011, 7, 271–283. [Google Scholar] [CrossRef]
- Kellerhals, M.; Bertschinger, L.; Gessler, S. Use of genetic resources in apple breeding and for sustainable fruit production. J. Fruit Ornam. Plant Res. 2004, 12, 53–62. [Google Scholar]
- Gaši, F.; Kanlić, K.; Stroil, B.K.; Pojskić, N.; Asdal, Å.; Rasmussen, M.; Kaiser, C.; Meland, M. Redundancies and Genetic Structure among ex situ Apple Collections in Norway Examined with Microsatellite Markers. HortScience 2016, 51, 1458–1462. [Google Scholar] [CrossRef] [Green Version]
- Lassois, L.; Denancé, C.; Ravon, E.; Guyader, A.; Guisnel, R.; Hibrand-Saint-Oyant, L.; Poncet, C.; Lasserre-Zuber, P.; Feugey, L.; Durel, C.E. Genetic diversity, population structure, parentage analysis, and construction of core collections in the French apple germplasm based on SSR markers. Plant Mol. Biol. Report. 2016, 34, 827–844. [Google Scholar] [CrossRef] [Green Version]
- Pereira-Lorenzo, S.; Ramos-Cabrer, A.M.; Ferreira, V.; Díaz-Hernández, M.B.; Carnide, V.; Pinto-Carnide, O.; Rodrigues, R.; Velázquez Barrera, M.E.; Rios-Mesa, D.; Ascasíbar-Errasti, J.; et al. Genetic diversity and core collection of Malus × domestica in northwestern Spain, Portugal and the Canary Islands by SSRs. Sci. Hortic. 2018, 240, 49–56. [Google Scholar] [CrossRef]
- Bakɪr, M.; Dumanoglu, H.; Aygun, A.; Erdogan, V.; Efe Dost, S.; Gülsen, O.; Serdar, U.; Kalkisim, O.; Bastas, K. Genetic diversity and population structure of apple germplasm from Eastern Black Sea region of Turkey by SSRs. Sci. Hortic. 2022, 294, 110793. [Google Scholar] [CrossRef]
- Baric, S.; Storti, A.; Hofer, M.; Guerra, W.; Dalla Via, J. Molecular Genetic Identification of Apple Cultivars Based on Microsatellite DNA Analysis. I. The Database of 600 Validated Profiles. Erwerbs-Obstbau 2020, 62, 117–154. [Google Scholar] [CrossRef]
- Larsen, B.; Toldam-Andersen, T.B.; Pedersen, C.; Ørgaard, M. Unravelling genetic diversity and cultivar parentage in the Danish apple gene bank collection. Tree Genet. Genomes 2017, 13, 1–12. [Google Scholar] [CrossRef]
- Testolin, R.; Foria, S.; Baccichet, I.; Messina, R.; Danuso, F.; Losa, A.; Scarbolo, E.; Stocco, M.; Cipriani, G. Genotyping apple (Malus × domestica Borkh.) heirloom germplasm collected and maintained by the Regional Administration of Friuli Venezia Giulia (Italy). Sci. Hortic. 2019, 252, 229–237. [Google Scholar] [CrossRef]
- Urrestarazu, J.; Denancé, C.; Ravon, E.; Guyader, A.; Guisnel, R.; Feugey, L.; Poncet, C.; Lateur, M.; Houben, P.; Ordidge, M.; et al. Analysis of the genetic diversity and structure across a wide range of germplasm reveals prominent gene flow in apple at the European level. BMC Plant Biol. 2016, 16, 130. [Google Scholar] [CrossRef] [PubMed]
- Marconi, G.; Ferradini, N.; Russi, L.; Concezzi, L.; Veronesi, F.; Albertini, E. Genetic Characterization of the Apple Germplasm Collection in Central Italy: The Value of Local Varieties. Front. Plant Sci. 2018, 9, 1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hokanson, S.C.; Lamboy, W.F.; Szewc-McFadden, A.K.; McFerson, J.R. Microsatellite (SSR) variation in a collection of Malus (apple) species and hybrids. Euphytica 2001, 118, 281–294. [Google Scholar] [CrossRef]
- Garkava-Gustavsson, L.; Mujaju, C.; Sehic, J.; Zborowska, A.; Backes, G.M.; Hietaranta, T.; Antonius, K. Genetic diversity in Swedish and Finnish heirloom apple cultivars revealed with SSR markers. Sci. Hortic. 2013, 162, 43–48. [Google Scholar] [CrossRef]
- Cmejlova, J.; Rejlova, M.; Paprstein, F.; Cmejla, R. A new one-tube reaction kit for the SSR genotyping of apple (Malus × domestica Borkh.). Plant Sci. 2021, 303, 110768. [Google Scholar] [CrossRef]
- Skytte af Sätra, J.; Troggio, M.; Odilbekov, F.; Sehic, J.; Mattisson, H.; Hjalmarsson, I.; Ingvarsson, P.K.; Garkava-Gustavsson, L. Genetic Status of the Swedish Central collection of heirloom apple cultivars. Sci. Hortic. 2020, 272, 109599. [Google Scholar] [CrossRef]
- Leforestier, D.; Ravon, E.; Muranty, H.; Cornille, A.; Lemaire, C.; Giraud, T.; Durel, C.E.; Branca, A. Genomic basis of the differences between cider and dessert apple varieties. Evolut. Appl. 2015, 8, 650–661. [Google Scholar] [CrossRef]
- Muranty, H.; Denancé, C.; Feugey, L.; Crépin, J.L.; Barbier, Y.; Tartarini, S.; Ordidge, M.; Troggio, M.; Lateur, M.; Nybom, H.; et al. Using whole-genome SNP data to reconstruct a large multi-generation pedigree in apple germplasm. BMC Plant Biol. 2020, 20, 2. [Google Scholar] [CrossRef] [Green Version]
- Ordidge, M.; Kirdwichai, P.; Baksh, M.F.; Venison, E.P.; Gibbings, J.G.; Dunwell, J.M. Genetic analysis of a major international collection of cultivated apple varieties reveals previously unknown historic heteroploid and inbred relationships. PLoS ONE 2018, 13, e0202405. [Google Scholar] [CrossRef] [Green Version]
- Hodel, R.G.; Segovia-Salcedo, M.C.; Landis, J.B.; Crowl, A.A.; Sun, M.; Liu, X.; Gitzendanner, M.A.; Douglas, N.A.; Germain-Aubrey, C.C.; Chen, S.; et al. The report of my death was an exaggeration: A review for researchers using microsatellites in the 21st century. APPS 2016, 4, apps.1600025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, F.F. Common Set of ECPGR SSR Markers for Malus Characterization. In Report of a Working Group on Malus/Pyrus, Fourth Meeting, Weggis, Switzerland, 7–9 March 2012; Lateur, M., Ordidge, M., Engels, J., Lipman, E., Eds.; Bioversity International: Rome, Italy, 2013; pp. 26–27, (abstract of presentation). [Google Scholar]
- Akšić, M.F.; Lazarević, K.; Šegan, S.; Natić, M.; Tosti, T.; Ćirić, I.; Meland, M. Assessing the Fatty Acid, Carotenoid, and Tocopherol Compositions of Seeds from Apple Cultivars (Malus domestica Borkh.) Grown Norway. Foods 2021, 10, 1956. [Google Scholar] [CrossRef] [PubMed]
- Gianfranceschi, L.; Seglias, N.; Tarchini, R.; Komjanc, M.; Gessler, C. Simple sequence repeats for the genetic analysis of apple. Theor. Appl. Genet. 1998, 96, 1069–1076. [Google Scholar] [CrossRef]
- Hokanson, S.C.; Szewc-McFadden, A.K.; Lamboy, W.F.; McFerson, J.R. Microsatellite (SSR) markers reveal genetic identities, genetic diversity and relationships in a Malus × domestica borkh. core subset collection. Theor. Appl. Genet. 1998, 97, 671–683. [Google Scholar] [CrossRef]
- Liebhard, R.; Gianfranceschi, L.; Koller, B.; Ryder, C.D.; Tarchini, R.; Van de Weg, E.; Gessler, C. Development and characterisation of 140 new microsatellites in apple (Malus x domestica Borkh.). Mol. Breed. 2002, 10, 217–241. [Google Scholar] [CrossRef]
- Fernandez-Fernandez, F.; Harvey, N.G.; James, C.M. Isolation and characterization of polymorphic microsatellite markers from european pear (Pyrus Communis L.). Mol. Ecol. 2006, 6, 1039–1041. [Google Scholar] [CrossRef]
- Gaši, F.; Žulj-Mihaljević, M.; Šimon, S.; Grahić, J.; Pojskić, N.; Kurtović, M.; Nikolić, D.; Pejić, I. Genetic structure of apple accessions maintained ex situ in Bosnia and Herzegovina examined by microsatellite markers. Genetika 2013, 45, 467–478. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef]
- Hardy, O.J.; Vekemans, X. A versatile computer program to analyse spatial genetic structure at the individual or population level. Mol. Ecol. 2002, 2, 618–620. [Google Scholar] [CrossRef] [Green Version]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; Von Holdt, B.M. Structure harvester: A Website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Vigouroux, Y.; Glaubitz, J.C.; Matsuoka, Y.; Goodman, M.M.; Sánchez, G.J.; Doebley, J. Population structure and genetic diversity of New World maize races assessed by DNA microsatellites. Am. J. Bot. 2008, 95, 1240–1253. [Google Scholar] [CrossRef] [PubMed]
- Urrestarazu, J.; Miranda, C.; Santesteban, L.G.; Royo, J.B. Genetic diversity and structure of local apple cultivars from northeastern Spain assessed by microsatellite markers. Tree Genet. Genomes 2012, 8, 1163–1180. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance interfered from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Ohta, T.; Kimura, M. A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet. Resour. 1973, 22, 201–204. [Google Scholar] [CrossRef]
- Meirmans, P.; Van Tienderen, P. Genotype and genodive: Two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes 2004, 4, 792–794. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: http://www.R-project.org/ (accessed on 15 March 2022).
- Muller, L.A.H.; McCusker, J.H. Microsatellite analysis of genetic diversity among clinical and nonclinical Saccharomyces cerevisiae isolates suggests heterozygote advantage in clinical environments. Mol. Ecol. 2009, 18, 2779–2786. [Google Scholar] [CrossRef] [Green Version]
- Adler, D.; Murdoch, D. RGL: 3D Visualization Device System (Open GL), R Package Version 0.93.945; 2013. Available online: http://rgl.neoscientists.org (accessed on 15 March 2022).
- Drost, H.G. Philentropy: Information Theory and Distance Quantification with R. J. Open Source Softw. 2018, 3, 765. [Google Scholar] [CrossRef]
- Bittinger, K. Usedist: Functions to Re-Arrange, Extract, and Work with Distances, R package version 0.4.0. 2020. Available online: https://CRAN.R-project.org/package=usedist (accessed on 15 March 2022).
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grahić, A.; Grahić, J. MADC—Marker Analysis Data Compiler User’s Manual. 2017. Available online: www.divisionagro.ba/apps/docs/madc-marker-analysis-data-compiler/usermanual (accessed on 15 March 2022).
- Van Treuren, R.; Kemp, H.; Ernsting, G.; Jongejans, B.; Houtman, H.; Visser, L. Microsatellite genotyping of apple (Malus · domestica Borkh.) genetic resources in the Netherlands: Application in collection management and variety identification. Genet. Resour. Crop Evolut. 2010, 57, 853–865. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.; Dondini, L.; De Franceschi, P.; Paris, R.; Sansavini, S.; Tartarini, S. Genetic diversity, population structure and construction of a core collection of apple cultivars from Italian germplasm. Plant Mol. Biol. Rep. 2015, 33, 458–473. [Google Scholar] [CrossRef]
- Pereira-Lorenzo, S.; Ramos-Cabrer, A.M.; Díaz-Hernández, M.B. Evaluation of genetic identity and variation of local apple cultivars (Malus × domestica Borkh.) from Spain using microsatellite markers. Genet. Resour. Crop. Evolut. 2007, 54, 405–420. [Google Scholar] [CrossRef]
- Gasi, F.; Simon, S.; Pojskic, N.; Kurtovic, M.; Pejic, I. Genetic assessment apple germplasm in Bosnia and Herzegovina using microsatellite and morphologic markers. Sci. Hortic. 2010, 126, 164–171. [Google Scholar] [CrossRef]
- Kanlić, K.; Kalamujic-Stroil, B.; Grahic, J.; Asdal, Å.; Meland, M.; Kurtovic, M.; Gasi, F. Influence of selection pressure on the frequency of triploid genotypes among different traditional apple germplasms. Work. Fac. Agric. Food Sci. Univ. Sarajevo 2016, 66, 287–290. [Google Scholar]
- Wiehle, M.; Nawaz, M.A.; Dahlem, R.; Alam, I.; Khan, A.A.; Gailing, O.; Mueller, M.; Buerkert, A. Pheno-genetic studies of apple varieties in northern Pakistan: A hidden pool of diversity. Sci. Hortic. 2021, 281, 109950. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).