Genetic Diversity and Genome-Wide Association Study of Architectural Traits of Spray Cut Chrysanthemum Varieties
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Phenotypic Evaluation of Architectural Traits
2.3. Phenotypic Data Analysis
2.4. GWAS and Candidate Gene Annotation
3. Results
3.1. Analysis of Significantly Different Architectural Traits of Spray Cut Chrysanthemum between EN2019 and EN2020
3.2. Descriptive Statistics of the Architectural Characteristics of Spray Cut Chrysanthemum in EN2019 and EN2020
3.3. Correlation Analysis of Architectural Traits in EN2019 and EN2020
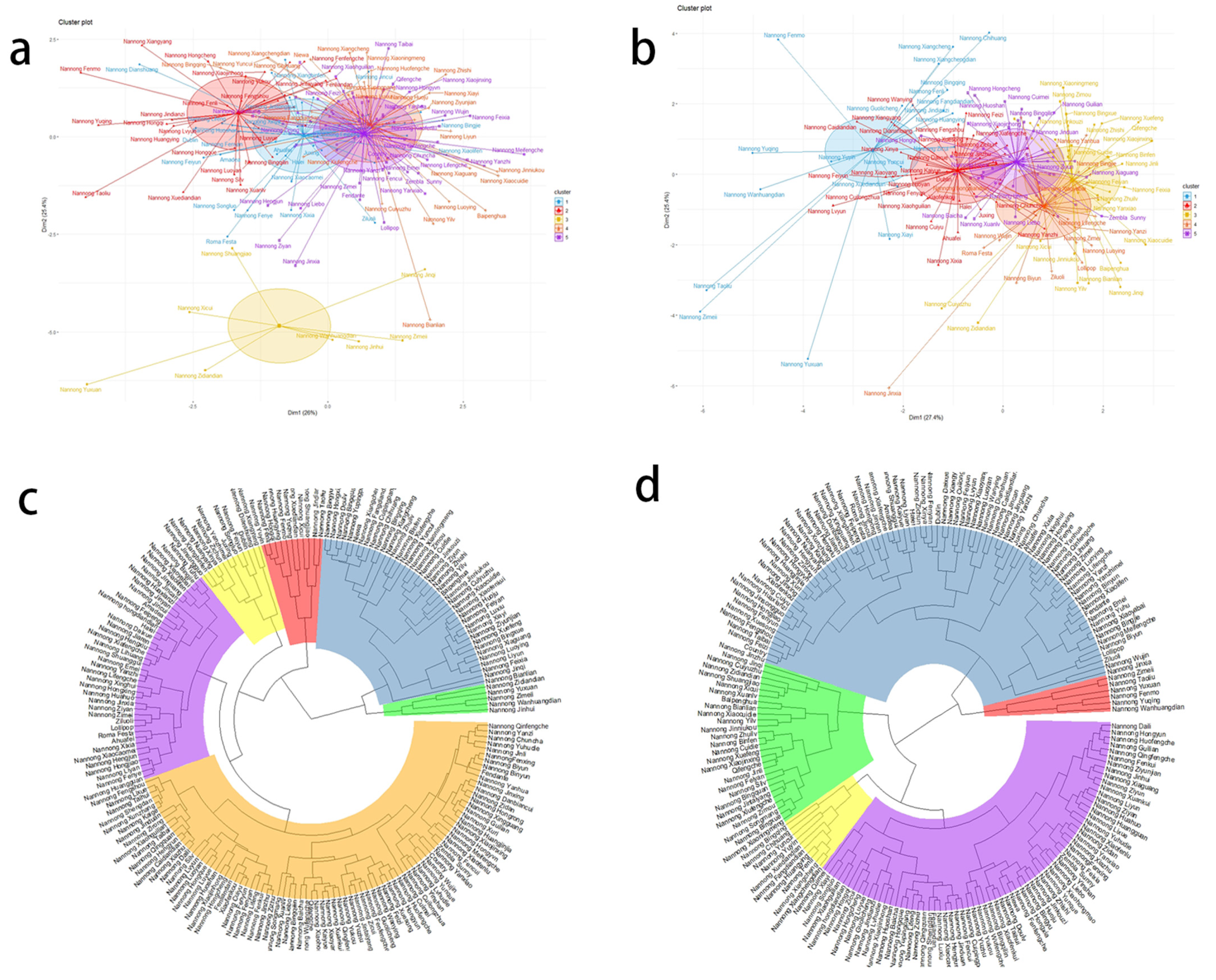
3.4. Cluster Analysis of the Architectural Traits of 195 Spray Cut Chrysanthemum Species
3.5. GWAS and Mining of Genes Controlling Plant Architecture
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Su, J.; Jiang, J.; Zhang, F.; Liu, Y.; Ding, L.; Chen, S.; Chen, F. Current achievements and future prospects in the genetic breeding of chrysanthemum: A review. Hortic. Res. 2019, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhou, X.; Xi, L.; Li, J.; Zhao, R.; Ma, N.; Zhao, L. Roles of DgBRC1 in regulation of lateral branching in chrysanthemum (Dendranthema × grandiflora cv. Jinba). PLoS ONE 2013, 8, e61717. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Jeong, B.R. Side lighting enhances morphophysiology by inducing more branching and flowering in chrysanthemum grown in controlled environment. Int. J. Mol. Sci. 2021, 22, 12019. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhang, L.; Yu, Q.; Zhang, J.; Li, P.; Zhang, Y.; Xing, X.; Ding, L.; Fang, W.; Chen, F.; et al. Integrated Signals of Jasmonates, Sugars, Cytokinins and Auxin Influence the Initial Growth of the Second Buds of Chrysanthemum after Decapitation. Biology 2021, 10, 440. [Google Scholar] [CrossRef]
- Barbier, F.F.; Dun, E.A.; Kerr, S.C.; Chabikwa, T.G.; Beveridge, C.A. An update on the signals controlling shoot branching. Trends Plant Sci. 2019, 24, 220–236. [Google Scholar] [CrossRef]
- Rameau, C.; Bertheloot, J.; Leduc, N.; Andrieu, B.; Foucher, F.; Sakr, S. Multiple pathways regulate shoot branching. Front. Plant Sci. 2015, 5, 741. [Google Scholar] [CrossRef]
- Nordström, A.; Tarkowski, P.; Tarkowska, D.; Norbaek, R.; Åstot, C.; Dolezal, K.; Sandberg, G. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: A factor of potential importance for auxin–cytokinin-regulated development. Proc. Natl. Acad. Sci. USA 2004, 101, 8039–8044. [Google Scholar] [CrossRef]
- Tanaka, M.; Takei, K.; Kojima, M.; Sakakibara, H.; Mori, H. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 2006, 45, 1028–1036. [Google Scholar] [CrossRef]
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 2012, 335, 1348–1351. [Google Scholar] [CrossRef]
- Wu, C.; Trieu, A.; Radhakrishnan, P.; Kwok, S.F.; Harris, S.; Zhang, K.; Wang, J.; Wan, J.; Zhai, H.; Takatsuto, S.; et al. Brassinosteroids regulate grain filling in rice. Plant Cell 2008, 20, 2130–2145. [Google Scholar] [CrossRef]
- Xia, X.; Dong, H.; Yin, Y.; Song, X.; Gu, X.; Sang, K.; Zhou, J.; Shi, K.; Zhou, Y.; Foyer, C.H.; et al. Brassinosteroid signaling integrates multiple pathways to release apical dominance in tomato. Proc. Natl. Acad. Sci. USA 2021, 118, e2004384118. [Google Scholar] [CrossRef]
- Mason, M.G.; Ross, J.J.; Babst, B.A.; Wienclaw, B.N.; Beveridge, C.A. Sugar demand, not auxin, is the initial regulator of apical dominance. Proc. Natl. Acad. Sci. USA 2014, 111, 6092–6097. [Google Scholar] [CrossRef]
- Wang, B.; Smith, S.M.; Li, J. Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 2018, 69, 437–468. [Google Scholar] [CrossRef]
- Adams, H.D.; Collins, A.D.; Briggs, S.P.; Vennetier, M.; Dickman, L.T.; Sevanto, S.A.; Garcia-Forner, N.; Powers, H.H.; McDowell, N.G. Experimental drought and heat can delay phenological development and reduce foliar and shoot growth in semiarid trees. Glob. Chang. Biol. 2015, 21, 4210–4220. [Google Scholar] [CrossRef]
- Fleisher, D.H.; Timlin, D.J.; Reddy, V.R. Temperature influence on potato leaf and branch distribution and on canopy photosynthetic rate. Agron. J. 2006, 98, 1442–1452. [Google Scholar] [CrossRef]
- Leduc, N.; Roman, H.; Barbier, F.; Péron, T.; Huché-Thélier, L.; Lothier, J.; Demotes-Mainard, S.; Sakr, S. Light signaling in bud outgrowth and branching in plants. Plants 2014, 3, 223–250. [Google Scholar] [CrossRef]
- Bahmani, I.; Hazard, L.; Varlet-Grancher, C.; Betin, M.; Lemaire, G.; Matthew, C.; Thom, E. Differences in tillering of long- and short-leaved perennial ryegrass genetic lines under full light and shade treatments. Crop Sci. 2000, 40, 1095–1102. [Google Scholar] [CrossRef]
- Evers, J.B.; Vos, J.; Andrieu, B.; Struik, P.C. Cessation of tillering in spring wheat in relation to light interception and red: Far-red ratio. Ann. Bot. 2006, 97, 649–658. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, Y.; Xu, G. How does nitrogen shape plant architecture? J. Exp. Bot. 2020, 71, 4415–4427. [Google Scholar] [CrossRef]
- He, X.; Qu, B.; Li, W.; Zhao, X.; Teng, W.; Ma, W.; Ren, Y.; Li, B.; Li, Z.; Tong, Y. The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol. 2015, 169, 1991–2005. [Google Scholar] [CrossRef]
- Tian, D.; Wang, P.; Tang, B.; Teng, X.; Li, C.; Liu, X.; Zou, D.; Song, S.; Zhang, Z. GWAS Atlas: A curated resource of genome-wide variant-trait associations in plants and animals. Nucleic Acids Res. 2020, 48, D927–D932. [Google Scholar] [CrossRef] [PubMed]
- Cano-Gamez, E.; Trynka, G. From GWAS to function: Using functional genomics to identify the mechanisms underlying complex diseases. Front. Genet. 2020, 11, 424. [Google Scholar] [CrossRef]
- Su, J.; Zhang, F.; Li, P.; Guan, Z.; Fang, W.; Chen, F. Genetic variation and association mapping of waterlogging tolerance in chrysanthemum. Planta 2016, 244, 1241–1252. [Google Scholar] [CrossRef]
- Su, J.; Zhang, F.; Chong, X.; Song, A.; Guan, Z.; Fang, W.; Chen, F. Genome-wide association study identifies favorable SNP alleles and candidate genes for waterlogging tolerance in chrysanthemums. Hortic. Res. 2019, 6, 21. [Google Scholar] [CrossRef]
- Sumitomo, K.; Shirasawa, K.; Isobe, S.; Hirakawa, H.; Hisamatsu, T.; Nakano, Y.; Yagi, M.; Ohmiya, A. Genome-wide association study overcomes the genome complexity in autohexaploid chrysanthemum and tags SNP markers onto the flower color genes. Sci. Rep. 2019, 9, 13947. [Google Scholar] [CrossRef]
- He, Y.; Wu, D.; Wei, D.; Fu, Y.; Cui, Y.; Dong, H.; Tan, C.; Qian, W. GWAS, QTL mapping and gene expression analyses in Brassica napus reveal genetic control of branching morphogenesis. Sci. Rep. 2017, 7, 15971. [Google Scholar] [CrossRef]
- Li, F.; Chen, B.; Xu, K.; Gao, G.; Yan, G.; Qiao, J.; Li, J.; Li, H.; Li, L.; Xiao, X.; et al. A genome-wide association study of plant height and primary branch number in rapeseed (Brassica napus). Plant Sci. 2016, 242, 169–177. [Google Scholar] [CrossRef]
- Liu, H.; Wang, J.; Zhang, B.; Yang, X.; Hammond, J.P.; Ding, G.; Wang, S.; Cai, H.; Wang, C.; Xu, F. Genome-wide association study dissects the genetic control of plant height and branch number in response to low-phosphorus stress in Brassica napus. Ann. Bot. 2021, 128, 919–930. [Google Scholar] [CrossRef]
- Zheng, M.; Peng, C.; Liu, H.; Tang, M.; Yang, H.; Li, X.; Liu, J.; Sun, X.; Wang, X.; Xu, J.; et al. Genome-wide association study reveals candidate genes for control of plant height, branch initiation height and branch number in rapeseed (Brassica napus L.). Front. Plant Sci. 2017, 8, 1246. [Google Scholar] [CrossRef] [PubMed]
- Yano, K.; Morinaka, Y.; Wang, F.; Huang, P.; Takehara, S.; Hirai, T.; Ito, A.; Koketsu, E.; Kawamura, M.; Kotake, K.; et al. GWAS with principal component analysis identifies a gene comprehensively controlling rice architecture. Proc. Natl. Acad. Sci. USA 2019, 116, 21262–21267. [Google Scholar] [CrossRef] [PubMed]
- Chong, X.; Zhang, F.; Wu, Y.; Yang, X.; Zhao, N.; Wang, H.; Guan, Z.; Fang, W.; Chen, F. A SNP-enabled assessment of genetic diversity, evolutionary relationships and the identification of candidate genes in chrysanthemum. Genome Biol. Evol. 2016, 8, 3661–3671. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Luo, F.; Pei, Z.; Zhao, X.; Liu, H.; Jiang, Y.; Sun, S. Genome-wide association study for plant architecture and bioenergy traits in diverse sorghum and sudangrass germplasm. Agronomy 2020, 10, 1602. [Google Scholar] [CrossRef]
- Su, J.; Li, L.; Zhang, C.; Wang, C.; Gu, L.; Wang, H.; Wei, H.; Liu, Q.; Huang, L.; Yu, S. Genome-wide association study identified genetic variations and candidate genes for plant architecture component traits in Chinese upland cotton. Theor. Appl. Genet. 2018, 131, 1299–1314. [Google Scholar] [CrossRef]
- Reinhardt, D.; Kuhlemeier, C. Plant architecture. EMBO Rep. 2002, 3, 846–851. [Google Scholar] [CrossRef]
- Atkin, O.K.; Loveys, B.; Atkinson, L.J.; Pons, T.J. Phenotypic plasticity and growth temperature: Understanding interspecific variability. J. Exp. Bot. 2006, 57, 267–281. [Google Scholar] [CrossRef]
- Li, P.; Chen, J.; Li, Y.; Zhang, K.; Wang, H. Possible mechanisms of control of Fusarium wilt of cut chrysanthemum by Phanerochaete chrysosporium in continuous cropping fields: A case study. Sci. Rep. 2017, 7, 15994. [Google Scholar] [CrossRef]
- Mortensen, L.M. Effects of air humidity on growth, flowering, keeping quality and water relations of four short-day greenhouse species. Sci. Hortic. 2000, 86, 299–310. [Google Scholar] [CrossRef]
- Körner, O.; Challa, H. Temperature integration and process-based humidity control in chrysanthemum. Comput. Electron. Agric. 2004, 43, 1–21. [Google Scholar] [CrossRef]
- Casal, J.J. Shade avoidance. Arab. Book/Am. Soc. Plant Biol. 2012, 10, e0157. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Abernathy, S.D.; White, R.H.; Finlayson, S.A. Photosynthetic photon flux density and phytochrome B interact to regulate branching in Arabidopsis. Plant Cell Environ. 2011, 34, 1986–1998. [Google Scholar] [CrossRef]
- Holalu, S.V.; Reddy, S.K.; Blackman, B.K.; Finlayson, S.A. Phytochrome interacting factors 4 and 5 regulate axillary branching via bud abscisic acid and stem auxin signalling. Plant Cell Environ. 2020, 43, 2224–2238. [Google Scholar] [CrossRef]
- Kebrom, T.H.; Burson, B.L.; Finlayson, S.A. Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol. 2006, 140, 1109–1117. [Google Scholar] [CrossRef]
- Kebrom, T.H.; Mullet, J.E. Transcriptome profiling of tiller buds provides new insights into PhyB regulation of tillering and indeterminate growth in sorghum. Plant Physiol. 2016, 170, 2232–2250. [Google Scholar] [CrossRef]
- Wang, X.; Chen, E.; Ge, X.; Gong, Q.; Butt, H.; Zhang, C.; Yang, Z.; Li, F.; Zhang, X. Overexpressed BRH1, a RING finger gene, alters rosette leaf shape in Arabidopsis thaliana. Sci. China Life Sci. 2018, 61, 79–87. [Google Scholar] [CrossRef]
- Tominaga-Wada, R.; Wada, T. CPC-ETC1 chimeric protein localization data in Arabidopsis root epidermis. Data Brief 2018, 18, 1773–1776. [Google Scholar] [CrossRef]
- Hsieh, W.-P.; Hsieh, H.-L.; Wu, S.-H. Arabidopsis bZIP16 transcription factor integrates light and hormone signaling pathways to regulate early seedling development. Plant Cell 2012, 24, 3997–4011. [Google Scholar] [CrossRef][Green Version]


| Trait | Environment | Max | Min | Rang | Mean | SD | CV/% | Skewness | Kurtosis | r |
|---|---|---|---|---|---|---|---|---|---|---|
| Plant height/cm | EN2019 | 153.67 | 36.10 | 117.57 | 101.84 | 21.20 | 20.82 | −0.22 | −0.16 | 0.794 ** |
| EN2020 | 141.42 | 42.98 | 98.43 | 94.67 | 19.89 | 21.01 | −0.15 | −0.55 | ||
| Number of leaf nodes | EN2019 | 73.33 | 19.17 | 54.17 | 46.51 | 8.62 | 18.52 | 0.24 | 0.93 | 0.701 ** |
| EN2020 | 79.50 | 27.17 | 52.33 | 47.32 | 9.82 | 20.75 | 0.63 | 0.41 | ||
| Total number of lateral buds | EN2019 | 73.33 | 19.17 | 54.17 | 45.40 | 8.94 | 19.68 | 0.20 | 0.81 | 0.649 ** |
| EN2020 | 79.17 | 26.33 | 52.83 | 46.60 | 9.77 | 20.96 | 0.57 | 0.43 | ||
| Number of upper primary branches | EN2019 | 13.00 | 2.00 | 11.00 | 5.04 | 1.60 | 31.75 | 1.56 | 4.16 | 0.593 ** |
| EN2020 | 15.33 | 2.67 | 12.67 | 5.95 | 2.13 | 35.82 | 1.26 | 2.16 | ||
| Number of lateral flower buds | EN2019 | 52.67 | 2.83 | 49.83 | 10.83 | 9.30 | 85.84 | 2.45 | 6.69 | 0.738 ** |
| EN2020 | 56.50 | 2.67 | 53.83 | 11.25 | 8.30 | 73.81 | 2.71 | 9.85 | ||
| Stem diameter/mm | EN2019 | 7.87 | 3.62 | 4.24 | 5.54 | 0.70 | 12.67 | 0.25 | 0.58 | 0.618 ** |
| EN2020 | 7.21 | 2.77 | 4.44 | 5.07 | 0.67 | 13.30 | −0.07 | 0.86 | ||
| Primary branch diameter/mm | EN2019 | 4.40 | 1.24 | 3.16 | 2.18 | 0.46 | 20.90 | 1.34 | 3.35 | 0.712 ** |
| EN2020 | 3.88 | 1.25 | 2.62 | 2.09 | 0.45 | 21.40 | 1.14 | 2.26 | ||
| Primary branch angle/° | EN2019 | 45.00 | 16.22 | 28.78 | 31.49 | 4.72 | 14.99 | 0.22 | 0.28 | 0.718 ** |
| EN2020 | 53.38 | 20.11 | 33.17 | 31.03 | 4.30 | 13.85 | 0.80 | 3.11 | ||
| Primary branch length/cm | EN2019 | 78.08 | 2.22 | 75.86 | 16.47 | 8.75 | 53.11 | 3.15 | 15.82 | 0.521 ** |
| EN2020 | 54.96 | 1.63 | 53.33 | 13.39 | 7.77 | 58.06 | 3.04 | 11.42 |
| EN2019 | Plant Height | Number of Leaf Nodes | Total Number of Lateral Buds | Number of Upper Primary Branches | Number of Lateral Flower Buds | Stem Diameter | Primary Branch Diameter | Primary Branch Angle | Primary Branch Length | |
|---|---|---|---|---|---|---|---|---|---|---|
| EN2020 | ||||||||||
| Plant height | 1 | 0.196 ** | 0.188 ** | −0.298 ** | −0.195 ** | 0.153 * | 0.027 | 0.112 | 0.142 * | |
| Number of leaf nodes | 0.250 ** | 1 | 0.913 ** | 0.197 ** | 0.079 | 0.388 ** | 0.012 | −0.154 * | −0.059 | |
| Total number of lateral buds | 0.253 ** | 0.986 ** | 1 | 0.133 | 0.041 | 0.334 ** | −0.016 | −0.160 * | −0.138 | |
| Number of upper primary branches | −0.369 ** | 0.134 | 0.126 | 1 | 0.231 ** | 0.110 | −0.200 ** | −0.293 ** | −0.317 ** | |
| Number of lateral flower buds | −0.232 ** | 0.351 ** | 0.341 ** | 0.240 ** | 1 | 0.109 | 0.579 ** | −0.074 | 0.534 ** | |
| Stem diameter | 0.322 ** | 0.250 ** | 0.257 ** | −0.019 | −0.191 ** | 1 | 0.394 ** | −0.041 | 0.044 | |
| Primary branch diameter | 0.096 | 0.028 | 0.026 | −0.466 ** | 0.368 ** | 0.147 * | 1 | 0.147 * | 0.639 ** | |
| Primary branch angle | 0.063 | −0.215 ** | −0.242 ** | −0.253 ** | −0.108 | −0.031 | 0.264 ** | 1 | 0.148 * | |
| Primary branch length | 0.113 | 0.063 | 0.057 | −0.402 ** | 0.513 ** | −0.279 ** | 0.661 ** | 0.160 * | 1 | |
| SNP Site | Significantly Associated Traits | Candidate Genes | Homologs in Arabidopsis |
|---|---|---|---|
| 19__104723464 | Number of lateral flower buds/Number of upper primary branches | evm.model.scaffold_940.421 | AT2G18790.1 (phytochrome B, phyB) |
| 9__215337463 | Number of leaf nodes/Total number of lateral buds | evm.model.scaffold_11169.22 | AT3G61460.1 (brassinosteroid-responsive RING-H2, BRH1) |
| 17__280870788 | Total number of lateral buds | evm.model.scaffold_3682.68 | AT2G46410.1 (Homeodomain-like superfamily protein, CPC) |
| 21__242369101 | Total number of lateral buds | evm.model.scaffold_881.80 | AT2G35530.1 (basic region/leucine zipper transcription factor 16, bZIP16) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, D.; Zhang, L.; Su, J.; Yu, Q.; Zhang, J.; Fang, W.; Wang, H.; Guan, Z.; Chen, F.; Song, A. Genetic Diversity and Genome-Wide Association Study of Architectural Traits of Spray Cut Chrysanthemum Varieties. Horticulturae 2022, 8, 458. https://doi.org/10.3390/horticulturae8050458
Sun D, Zhang L, Su J, Yu Q, Zhang J, Fang W, Wang H, Guan Z, Chen F, Song A. Genetic Diversity and Genome-Wide Association Study of Architectural Traits of Spray Cut Chrysanthemum Varieties. Horticulturae. 2022; 8(5):458. https://doi.org/10.3390/horticulturae8050458
Chicago/Turabian StyleSun, Daojin, Luyao Zhang, Jiangshuo Su, Qi Yu, Jiali Zhang, Weimin Fang, Haibin Wang, Zhiyong Guan, Fadi Chen, and Aiping Song. 2022. "Genetic Diversity and Genome-Wide Association Study of Architectural Traits of Spray Cut Chrysanthemum Varieties" Horticulturae 8, no. 5: 458. https://doi.org/10.3390/horticulturae8050458
APA StyleSun, D., Zhang, L., Su, J., Yu, Q., Zhang, J., Fang, W., Wang, H., Guan, Z., Chen, F., & Song, A. (2022). Genetic Diversity and Genome-Wide Association Study of Architectural Traits of Spray Cut Chrysanthemum Varieties. Horticulturae, 8(5), 458. https://doi.org/10.3390/horticulturae8050458









