Identification and Characterization of Two Putative Citrus Phosphomannose Isomerase (CsPMI) Genes as Selectable Markers for Mature Citrus Transformation
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Constructs
2.2. Plant Materials and Agrobacterium Transformation
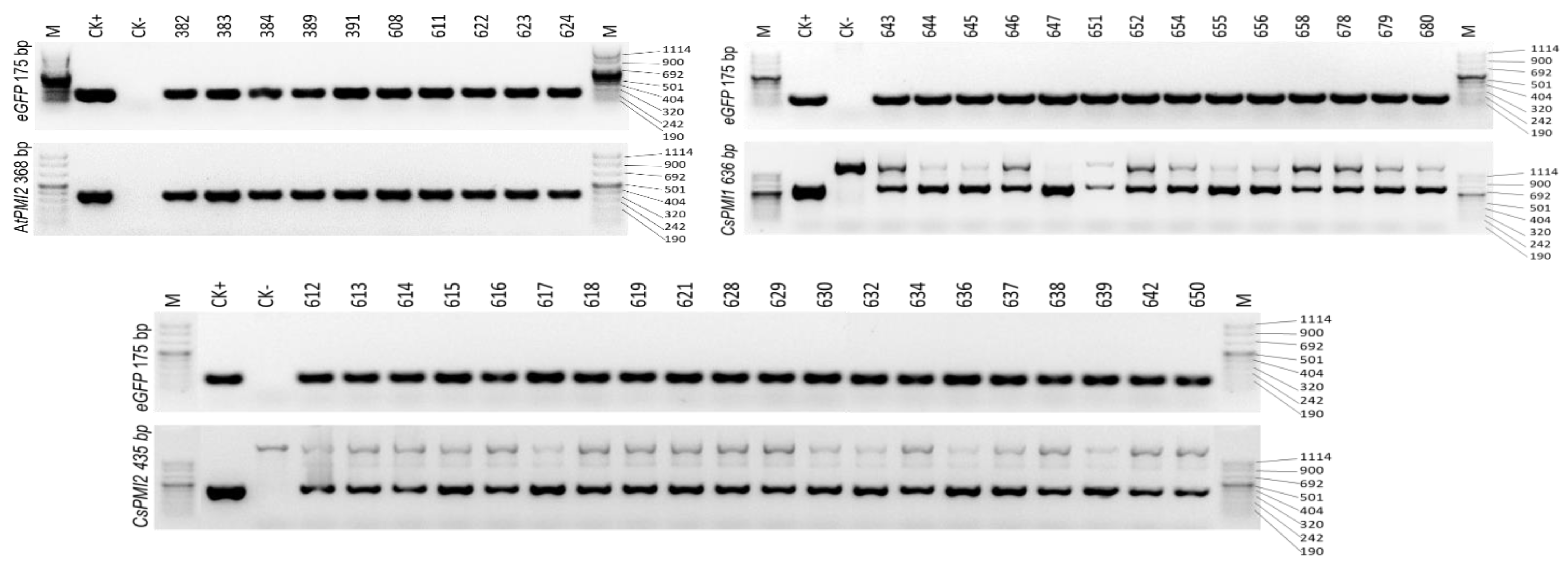
2.3. PCR Confirmation of Transgenics
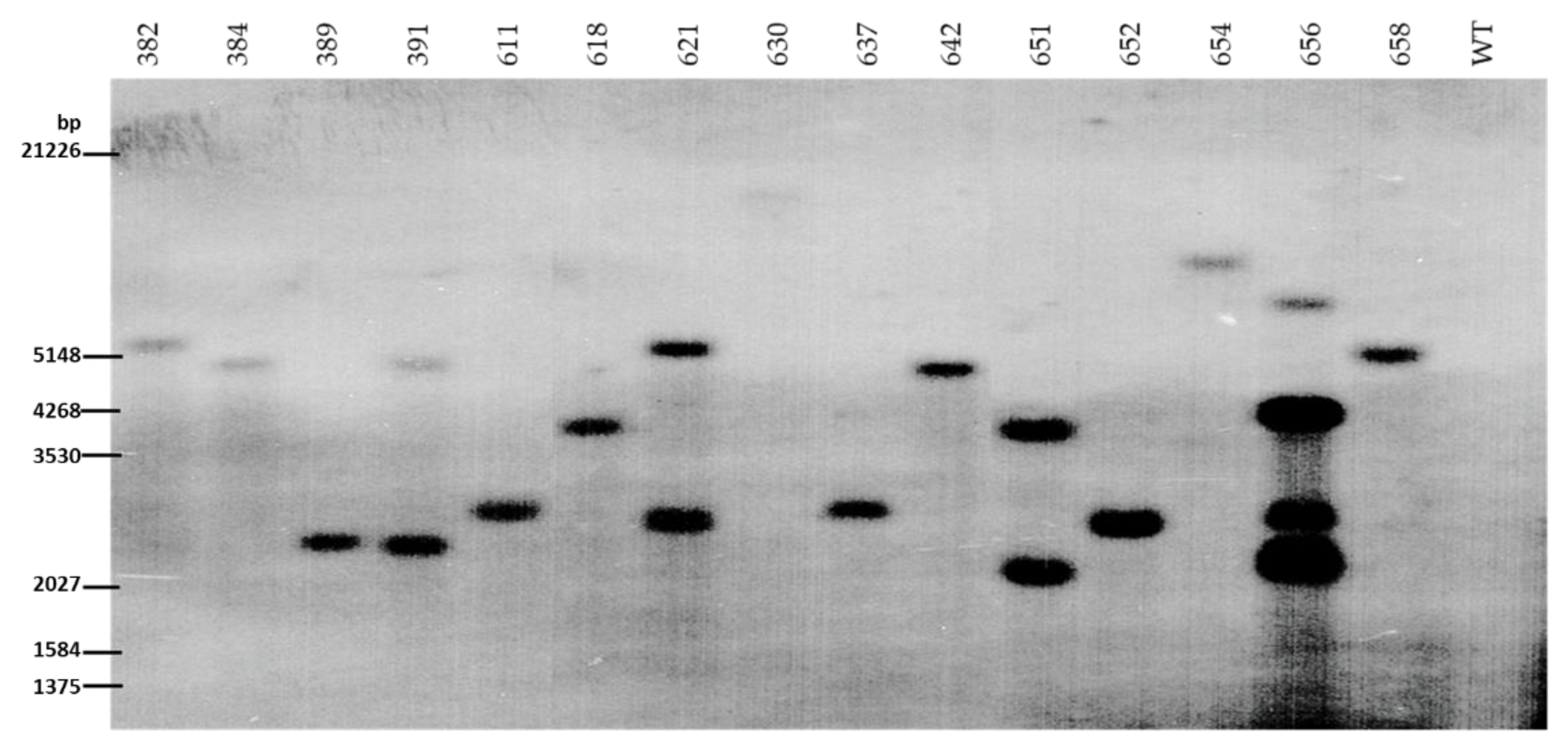
2.4. Southern Blot Analysis
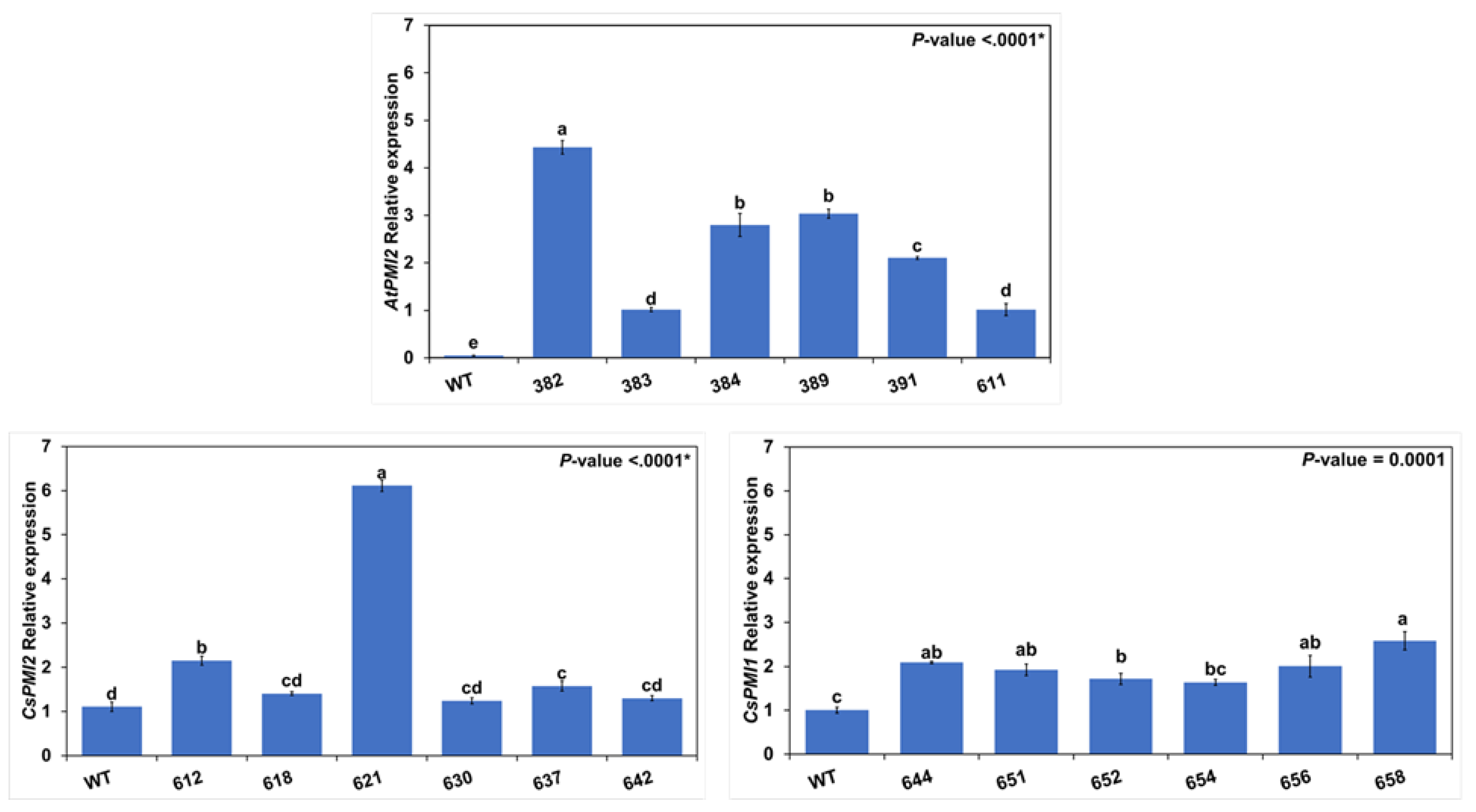
2.5. Quantitative Real-Time Reverse Transcriptase-PCR (qRT-PCR)
2.6. PMI In Vivo Assay
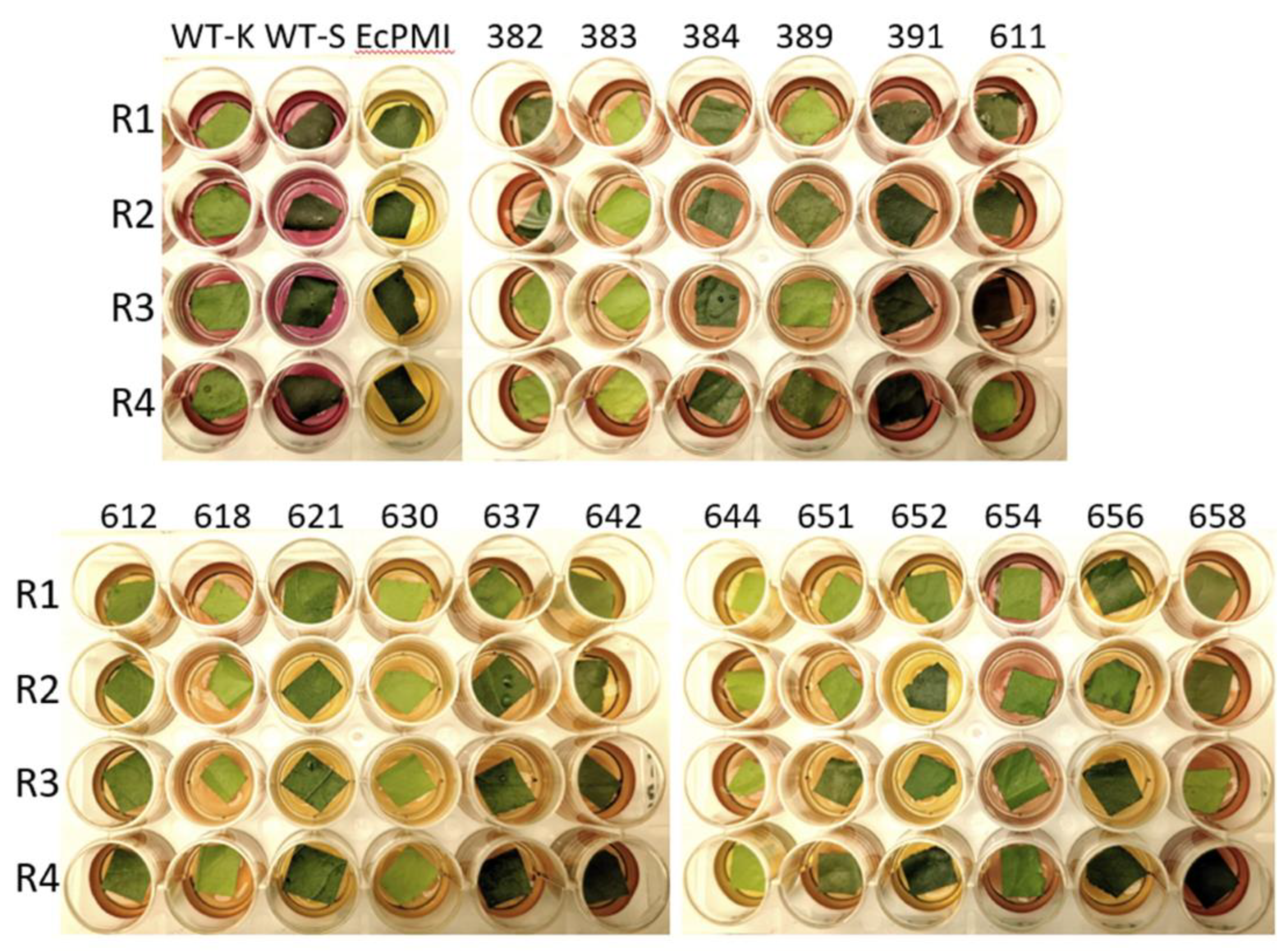
2.7. Chlorophenol Red Assay
2.8. Statistical Analysis of Transgenic Production
3. Results
3.1. CsPMI1 and CsPMI2 Are Evolutionarily Distantly Related
3.2. Generation of Transgenics with Phosphomannose Isomerase Selectable Markers
3.3. Molecular Analysis of the Transgenics
3.4. PMI In Vivo Enzyme Assay
3.5. Confirmation of PMI Expression Using Chlorophenol Red Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Schouten, H.J.; Krens, F.A.; Jacobsen, E. Cisgenic plants are similar to traditionally bred plants: International regulations for genetically modified organisms should be altered to exempt cisgenesis. EMBO Rep. 2006, 7, 750–753. [Google Scholar] [CrossRef]
- Espinoza, C.; Schlechter, R.; Herrera, D.; Torres, E.; Serrano, A.; Medina, C.; Arce-Johnson, P. Cisgenesis and intragenesis: New tools for improving crops. Biol. Res. 2013, 46, 323–331. [Google Scholar] [CrossRef]
- Nielsen, K.M. Transgenic organisms—Time for conceptual diversification? Nat. Biotechnol. 2003, 21, 227–228. [Google Scholar] [CrossRef]
- Rommens, C.M.; Humara, J.M.; Ye, J.; Yan, H.; Richael, C.; Zhang, L.; Perry, R.; Swords, K. Crop improvement through modification of the plant’s own genome. Plant Physiol. 2004, 135, 421–431. [Google Scholar] [CrossRef]
- Kost, T.D.; Jänsch, M.; Gessler, C.; Patocchi, A.; Broggini, G.; Flachowsky, H. Generation of a cisgenic apple line of cultivar ‘Gala’ with increased fire blight resistance. In Proceedings of the XIV EUCARPIA Symposium on Fruit Breeding and Genetics 1172, Bologna, Italy, 17 October 2017; pp. 79–84. [Google Scholar]
- Krens, F.A.; Schaart, J.G.; Van der Burgh, A.M.; Tinnenbroek-Capel, I.E.; Groenwold, R.; Kodde, L.P.; Broggini, G.A.; Gessler, C.; Schouten, H.J. Cisgenic apple trees; development, characterization, and performance. Front. Plant Sci. 2015, 6, 286. [Google Scholar] [CrossRef][Green Version]
- Vanblaere, T.; Flachowsky, H.; Gessler, C.; Broggini, G.A. Molecular characterization of cisgenic lines of apple ‘Gala’ carrying the Rvi6 scab resistance gene. Plant Biotechnol. J. 2014, 12, 2–9. [Google Scholar] [CrossRef]
- Würdig, J.; Flachowsky, H.; Saß, A.; Peil, A.; Hanke, M.-V. Improving resistance of different apple cultivars using the Rvi6 scab resistance gene in a cisgenic approach based on the Flp/FRT recombinase system. Mol. Breed. 2015, 35, 1–18. [Google Scholar] [CrossRef]
- Krens, F.; Salentijn, E.; Schaart, J.; Schouten, H.; Jacobsen, E. Current progress in trans-and cisgenic apple and strawberry breeding. Acta Hortic. 2012, 941, 37–48. [Google Scholar] [CrossRef]
- Haverkort, A.; Jacobsen, E.; Visser, R.; Boonekamp, P.; Vossen, J.; Kessel, G.; Hutten, R.; Franke, A.; Lotz, L. Sustainable resistance against phytophthora in potato through cisgenic. In Proceedings of the Eleventh EuroBlight Workshop, Lelystad, The Netherlands, 28–31 October 2008; pp. 259–260. [Google Scholar]
- An, C.; Orbovic, V.; Mou, Z. An efficient intragenic vector for generating intragenic and cisgenic plants in citrus. Am. J. Plant Sci. 2013, 4, 2131. [Google Scholar] [CrossRef]
- Merritt, B.A.; Zhang, X.; Triplett, E.W.; Mou, Z.; Orbović, V. Selection of transgenic citrus plants based on glyphosate tolerance conferred by a citrus 5-enolpyruvylshikimate-3-phosphate synthase variant. Plant Cell Rep. 2021, 40, 1947–1956. [Google Scholar] [CrossRef]
- de Vetten, N.; Wolters, A.-M.; Raemakers, K.; van der Meer, I.; ter Stege, R.; Heeres, E.; Heeres, P.; Visser, R. A transformation method for obtaining marker-free plants of a cross-pollinating and vegetatively propagated crop. Nat. Biotechnol. 2003, 21, 439–442. [Google Scholar] [CrossRef]
- Qin, J.; Wang, Y.; Zhu, C. Biolistic transformation of wheat using the HMW-GS 1Dx5 gene without selectable markers. Genet. Mol. Res. 2014, 13, 4361–4371. [Google Scholar] [CrossRef]
- Hu, L.; Li, H.; Qin, R.; Xu, R.; Li, J.; Li, L.; Wei, P.; Yang, J. Plant phosphomannose isomerase as a selectable marker for rice transformation. Sci. Rep. 2016, 6, 25921. [Google Scholar] [CrossRef]
- Peña, L.; Cervera, M.; Fagoaga, C.; Romero, J.; Ballester, A.; Soler, N.; Pons, E.; Rodríguez, A.; Peris, J.; Juárez, J. Citrus. Comp. Transgenic Crop Plants 2009, 1–62. [Google Scholar] [CrossRef]
- Srivastava, V.; Ow, D.W. Marker-free site-specific gene integration in plants. Trends Biotechnol. 2004, 22, 627–629. [Google Scholar] [CrossRef]
- Privalle, L.S.; Wright, M.; Reed, J.; Hansen, G.; Dawson, J.; Dunder, E.M.; Chang, Y.-F.; Powell, M.L.; Meghji, M. Phosphomannose isomerase–a novel system for plant selection. Can. Wheat Board Agrium Foragen 2000, 171. [Google Scholar] [CrossRef]
- Duan, Y.; Zhai, C.; Li, H.; Li, J.; Mei, W.; Gui, H.; Ni, D.; Song, F.; Li, L.; Zhang, W. An efficient and high-throughput protocol for Agrobacterium-mediated transformation based on phosphomannose isomerase positive selection in Japonica rice (Oryza sativa L.). Plant Cell Rep. 2012, 31, 1611–1624. [Google Scholar] [CrossRef]
- Gui, H.; Li, X.; Liu, Y.; Han, K.; Li, X. The relationship between PMI (manA) gene expression and optimal selection pressure in Indica rice transformation. Plant Cell Rep. 2014, 33, 1081–1090. [Google Scholar] [CrossRef]
- Hoa, T.T.C.; Al-Babili, S.; Schaub, P.; Potrykus, I.; Beyer, P. Golden Indica and Japonica rice lines amenable to deregulation. Plant Physiol. 2003, 133, 161–169. [Google Scholar] [CrossRef]
- Lucca, P.; Ye, X.; Potrykus, I. Effective selection and regeneration of transgenic rice plants with mannose as selective agent. Mol. Breed. 2001, 7, 43–49. [Google Scholar] [CrossRef]
- Gadaleta, A.; Giancaspro, A.; Blechl, A.; Blanco, A. Phosphomannose isomerase, pmi, as a selectable marker gene for durum wheat transformation. J. Cereal Sci. 2006, 43, 31–37. [Google Scholar] [CrossRef]
- Reed, J.; Privalle, L.; Powell, M.L.; Meghji, M.; Dawson, J.; Dunder, E.; Sutthe, J.; Wenck, A.; Launis, K.; Kramer, C. Phosphomannose isomerase: An efficient selectable marker for plant transformation. Vitr. Cell. Dev. Biol. Plant 2001, 37, 127–132. [Google Scholar] [CrossRef]
- Wright, M.; Dawson, J.; Dunder, E.; Suttie, J.; Reed, J.; Kramer, C.; Chang, Y.; Novitzky, R.; Wang, H.; Artim-Moore, L. Efficient biolistic transformation of maize (Zea mays L.) and wheat (Triticum aestivum L.) using the phosphomannose isomerase gene, pmi, as the selectable marker. Plant Cell Rep. 2001, 20, 429–436. [Google Scholar] [CrossRef]
- Hur, S.-H.; Min, B.-W. Efficient development of transgenic Cabbage with jasmonic acid carboxyl methyltransferase (JMT) gene based on PMI/mannose selection system. Plant Breed Biotechnol. 2015, 3, 226–237. [Google Scholar] [CrossRef]
- Ku, J.J.; Park, Y.H.; Park, Y.D. A non-antibiotic selection system uses the phosphomannose-isomerase (PMI) gene for Agrobacterium-mediated transformation of Chinese cabbage. J. Plant Biol. 2006, 49, 115–122. [Google Scholar] [CrossRef]
- Min, B.W.; Cho, Y.N.; Song, M.J.; Noh, T.K.; Kim, B.K.; Chae, W.K.; Park, Y.S.; Choi, Y.D.; Harn, C.H. Successful genetic transformation of Chinese cabbage using phosphomannose isomerase as a selection marker. Plant Cell Rep. 2007, 26, 337–344. [Google Scholar] [CrossRef]
- Ballester, A.; Cervera, M.; Pena, L. Evaluation of selection strategies alternative to nptII in genetic transformation of citrus. Plant Cell Rep. 2008, 27, 1005–1015. [Google Scholar] [CrossRef]
- Boscariol, R.; Almeida, W.; Derbyshire, M.; Mourao Filho, F.; Mendes, B. The use of the PMI/mannose selection system to recover transgenic sweet orange plants (Citrus sinensis L. Osbeck). Plant Cell Rep. 2003, 22, 122–128. [Google Scholar] [CrossRef]
- Dutt, M.; Lee, D.H.; Grosser, J.W. Bifunctional selection–reporter systems for genetic transformation of citrus: Mannose-and kanamycin-based systems. Vitr. Cell. Dev. Biol. Plant 2010, 46, 467–476. [Google Scholar] [CrossRef]
- Wu, H.; Acanda, Y.; Canton, M.; Zale, J. Efficient biolistic transformation of immature citrus rootstocks using phosphomannose-isomerase selection. Plants 2019, 8, 390. [Google Scholar] [CrossRef]
- Lin, Y.; Huang, J. Characterization of an algal phosphomannose isomerase gene and its application as a selectable marker for genetic manipulation of tomato. Plant Diver. 2021, 43, 63–70. [Google Scholar] [CrossRef]
- Wang, Y.; Geng, L.; Gui, H.; Liu, Y.; Wang, Y.; Zhang, X.; Li, X. Soybean PMI genes as a selectable marker for corn and rice transformation. Maize Genet. Coop. News Lett. 2015, 89, 1–3. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Wu, H.; Acanda, Y.; Shankar, A.; Peeples, M.; Hubbard, C.; Orbović, V.; Zale, J. Genetic transformation of commercially important mature citrus scions. Crop Sci. 2015, 55, 2786–2797. [Google Scholar] [CrossRef]
- Murashige, T.; Tucker, D.P.H. Growth factor requirements of citrus tissue cultre. Intl. Citrus Symp. 1969, 11, 1155–1161. [Google Scholar]
- Cervera, M.; Juarez, J.; Navarro, L.; Pena, L. Genetic transformation of mature citrus plants. In Methods in Molecular Biology; Pena, L., Ed.; Humana Press: Totowa, NJ, USA, 2005; Volume 86, pp. 177–187. [Google Scholar]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pena, L.; Perez, R.M.; Cervera, M.; Juarez, J.A.; Navarro, L. Early events in Agrobacterium-mediated genetic transformation of citrus explants. Ann. Bot. 2004, 94, 67–74. [Google Scholar] [CrossRef]
- Cervera, M.; Juarez, J.; Navarro, A.; Pina, J.A.; Duran-Vila, N.; Navarro, L.; Pena, L. Genetic transformation and regeneration of mature tissues of woody fruit plants bypassing the juvenile stage. Transgenic Res. 1998, 7, 51–59. [Google Scholar] [CrossRef]
- Cervera, M.; Navarro, A.; Navarro, L.; Pena, L. Production of transgenic adult plants from clementine mandarin by enhancing cell competence for transformation and regeneration. Tree Physiol. 2008, 28, 55–66. [Google Scholar] [CrossRef]
- Chassy, B.M. Food safety risks and consumer health. New Biotechnol. 2010, 27, 534–544. [Google Scholar] [CrossRef]
- Erpen, L.; Tavano, E.; Harakava, R.; Dutt, M.; Grosser, J.; Piedade, S.; Mendes, B.; Mourão Filho, F. Isolation, characterization, and evaluation of three Citrus sinensis-derived constitutive gene promoters. Plant Cell Rep. 2018, 37, 1113–1125. [Google Scholar] [CrossRef]
- Dasgupta, K.; Hotton, S.; Belknap, W.; Syed, Y.; Dardick, C.; Thilmony, R.; Thomson, J.G. Isolation of novel citrus and plum fruit promoters and their functional characterization for fruit biotechnology. BMC Biotechnol. 2020, 20, 1–15. [Google Scholar] [CrossRef]
- Dutt, M.; Zambon, F.T.; Erpen, L.; Soriano, L.; Grosser, J. Embryo-specific expression of a visual reporter gene as a selection system for citrus transformation. PLoS ONE 2018, 13, e0190413. [Google Scholar] [CrossRef]


| Vector | Mannose + Sucrose (g L−1) | No. of Explants | No. of Shoots | Transgenic Shoots w/EGFP |
|---|---|---|---|---|
| AtPMI2 | 0/30 | 360 | 760 | 12 |
| 1.9/28.1 | 360 | 522 | 2 | |
| 3.8/26.2 | 360 | 482 | 7 | |
| 7.5/22.5 | 360 | 293 | 6 | |
| 15/15 | 360 | 289 | 7 | |
| 30/0 | 360 | 116 | 0 | |
| Sum | - | 2160 | 2463 | 34 |
| CsPMI1 | 0/30 | 400 | 684 | 8 |
| 1.9/28.1 | 400 | 570 | 6 | |
| 3.8/26.2 | 400 | 504 | 10 | |
| 7.5/22.5 | 400 | 334 | 9 | |
| 15/15 | 400 | 236 | 5 | |
| 30/0 | 400 | 72 | 1 | |
| Sum | - | 2400 | 2400 | 39 |
| CsPMI2 | 0/30 | 400 | 651 | 8 |
| 1.9/28.1 | 400 | 568 | 6 | |
| 3.8/26.2 | 400 | 442 | 9 | |
| 7.5/22.5 | 400 | 392 | 17 | |
| 15/15 | 400 | 296 | 9 | |
| 30/0 | 400 | 0 | 0 | |
| Sum | - | 2400 | 2442 | 49 |
| Variables a | Treatment Mannose + Sucrose (g L−1) b | Mean c | ±SE d |
|---|---|---|---|
| SL > 2 | 0/30 | 126.7 a | 15.3 |
| SL > 2 | 1.9/28.1 | 87.0 a | 2.1 |
| SL > 2 | 3.8/26.2 | 80.3 | 2.1 |
| SL > 2 | 7.5/22.5 | 48.9 | 2.5 |
| SL > 2 | 15/15 | 48.2 | 3.9 |
| SL > 2 | 30/0 | 19.4 | 8.5 |
| PS | 0/30 | 2.0 a | 0.5 |
| PS | 1.9/28.1 | 0.3 | 0.2 |
| PS | 3.8/26.2 | 1.1 a | 0.3 |
| PS | 7.5/22.5 | 1.0 a | 0.3 |
| PS | 15/15 | 1.2 a | 0.2 |
| PS | 30/0 | 0.0 | 0.0 |
| TES | 0/30 | 1.4 a | 0.2 |
| TES | 1.9/28.1 | 0.4 a | 0.2 |
| TES | 3.8/26.2 | 1.3 a | 0.3 |
| TES | 7.5/22.5 | 2.0 a | 0.5 |
| TES | 15/15 | 2.4 a | 0.5 |
| TES | 30/0 | 0.0 | 0.0 |
| TEE | 0/30 | 3.6 a | 1.1 |
| TEE | 1.9/28.1 | 0.7 | 0.4 |
| TEE | 3.8/26.2 | 1.8 a | 0.4 |
| TEE | 7.5/22.5 | 1.0 | 0.4 |
| TEE | 15/15 | 2.2 a | 0.6 |
| TEE | 30/0 | 0.0 | 0.0 |
| Variables a | Treatment Mannose + Sucrose (g L−1) b | Mean c | ±SE d |
|---|---|---|---|
| SL > 2 | 0/30 | 114.0 a | 8.1 |
| SL > 2 | 1.9/28.1 | 95.0 a | 10.4 |
| SL > 2 | 3.8/26.2 | 84.0 | 10.1 |
| SL > 2 | 7.5/22.5 | 55.7 | 2.2 |
| SL > 2 | 15/15 | 39.4 | 3.4 |
| SL > 2 | 30/0 | 12.0 | 1.6 |
| PS | 0/30 | 1.3 a | 0.4 |
| PS | 1.9/28.1 | 1.0 a | 0.4 |
| PS | 3.8/26.2 | 1.7 a | 0.3 |
| PS | 7.5/22.5 | 1.5 a | 0.2 |
| PS | 15/15 | 0.7 a | 0.1 |
| PS | 30/0 | 0.2 | 0.2 |
| TES | 0/30 | 1.2 a | 0.4 |
| TES | 1.9/28.1 | 1.3 a | 0.5 |
| TES | 3.8/26.2 | 2.3 a | 0.6 |
| TES | 7.5/22.5 | 2.6 a | 0.4 |
| TES | 15/15 | 2.0 a | 0.4 |
| TES | 30/0 | 1.0 a | 1.0 |
| TEE | 0/30 | 2.1 a | 0.7 |
| TEE | 1.9/28.1 | 1.6 a | 0.7 |
| TEE | 3.8/26.2 | 2.5 a | 0.3 |
| TEE | 7.5/22.5 | 2.5 a | 0.7 |
| TEE | 15/15 | 1.1 a | 0.2 |
| TEE | 30/0 | 0.2 a | 0.2 |
| Variable a | Treatment Mannose + Sucrose (g L−1) b | Mean c | ±SE d |
|---|---|---|---|
| SL > 2 | 0/30 | 108.5 a | 20.5 |
| SL > 2 | 1.9/28.1 | 94.7 a | 23.8 |
| SL > 2 | 3.8/26.2 | 73.7 a | 3.3 |
| SL > 2 | 7.5/22.5 | 65.3 | 8.9 |
| SL > 2 | 15/15 | 49.3 | 6.9 |
| SL > 2 | 30/0 | 15.5 | 2.6 |
| PS | 0/30 | 1.4 a | 0.2 |
| PS | 1.9/28.1 | 1.0 a | 0.1 |
| PS | 3.8/26.2 | 1.4 a | 0.2 |
| PS | 7.5/22.5 | 2.7 | 0.4 |
| PS | 15/15 | 1.6 a | 0.3 |
| PS | 30/0 | 0.0 | 0.0 |
| TES | 0/30 | 1.7 a | 0.5 |
| TES | 1.9/28.1 | 1.6 a | 0.6 |
| TES | 3.8/26.2 | 2.0 a | 0.3 |
| TES | 7.5/22.5 | 4.4 | 0.7 |
| TES | 15/15 | 3.7 | 0.9 |
| TES | 30/0 | 0.0 a | 0.0 |
| TEE | 0/30 | 2.0 a | 0.2 |
| TEE | 1.9/28.1 | 1.5 a | 0.2 |
| TEE | 3.8/26.2 | 2.1 a | 0.3 |
| TEE | 7.5/22.5 | 4.2 | 0.7 |
| TEE | 15/15 | 2.4 a | 0.4 |
| TEE | 30/0 | 0.0 | 0.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, H.; Canton, M.; Mahmoud, L.M.; Weber, K.R.; Michalczyk, G.Z.; Dutt, M.; Zale, J.M. Identification and Characterization of Two Putative Citrus Phosphomannose Isomerase (CsPMI) Genes as Selectable Markers for Mature Citrus Transformation. Horticulturae 2022, 8, 204. https://doi.org/10.3390/horticulturae8030204
Wu H, Canton M, Mahmoud LM, Weber KR, Michalczyk GZ, Dutt M, Zale JM. Identification and Characterization of Two Putative Citrus Phosphomannose Isomerase (CsPMI) Genes as Selectable Markers for Mature Citrus Transformation. Horticulturae. 2022; 8(3):204. https://doi.org/10.3390/horticulturae8030204
Chicago/Turabian StyleWu, Hao, Michel Canton, Lamiaa M. Mahmoud, Katherine R. Weber, Gillian Z. Michalczyk, Manjul Dutt, and Janice M. Zale. 2022. "Identification and Characterization of Two Putative Citrus Phosphomannose Isomerase (CsPMI) Genes as Selectable Markers for Mature Citrus Transformation" Horticulturae 8, no. 3: 204. https://doi.org/10.3390/horticulturae8030204
APA StyleWu, H., Canton, M., Mahmoud, L. M., Weber, K. R., Michalczyk, G. Z., Dutt, M., & Zale, J. M. (2022). Identification and Characterization of Two Putative Citrus Phosphomannose Isomerase (CsPMI) Genes as Selectable Markers for Mature Citrus Transformation. Horticulturae, 8(3), 204. https://doi.org/10.3390/horticulturae8030204







