Abstract
Tomato yellow leaf curl virus (TYLCV) causes serious yield reductions in China. The use of certain resistance genes in tomato varieties has alleviated the impact of the virus to a certain extent. Recently, varieties with the Ty-1, Ty-2, or Ty-3 genes lost their resistance to TYLCV in some areas in China. New genes should be introduced into tomato to maintain the resistance to TYLCV. Tomato line AVTO1227 has excellent resistance to disease due to the resistance gene ty-5. In this study, we screened different types of markers in a tomato F2 population to compare their accuracy and efficiency. The sequencing analysis results were consistent with the high resolution melting (HRM) marker genotype and field identification results. The result confirmed that the functional marker of ty-5 was accurate and reliable. The single nucleotide polymorphism-based HRM genotyping method established in this study can be used for the selection of breeding parent material, gene correlation analysis, and molecular marker-assisted breeding.
1. Introduction
Tomato (Solanum lycopersicum) belongs to the Solanaceae family, originating from South America. Due to its adaptability, easy cultivation, and high yield, it has become one of the most important cultivated vegetables in the world. Tomato yellow leaf curl virus (TYLCV) is a significant virus causing quality and yield reductions in tomato [1]. Susceptible tomato lines infected by TYLCV typically exhibit severe curling and yellowing of the top leaves, in addition to slow and stunted growth [2]. If the plants are attacked at the juvenile stage, it can lead to significant yield losses or even no harvest [3]. TYLCV is transmitted by the whitefly (Bemisia tabaci) [4]. With the increased frequency of international transport, tourism, trade, and other human activities, whitefly and TYLCV have spread all over the world. Disease management for TYLCV is difficult, as the whitefly has high reproductive capacity. Physical, chemical, and biological control methods can reduce the occurrence of TYLCV to a certain degree. However, these preventions are imperfect, uneconomical, and laborious, and may have negative consequences for the environment. Breeding TYLCV-resistant tomato cultivars offers an attractive method for controlling this disease and could fundamentally resolve the devastation caused by TYLCV [5,6,7,8].
Thus far, six TYLCV resistance genes, namely Ty-1, Ty-2, Ty-3, Ty-4, and Ty-6 and the recessive gene ty-5, have been identified from wild tomato species [9,10,11,12,13,14]. Molecular markers with close linkage with these genes could be used to improve tomato breeding efficiency. Ty-1, Ty-2, and Ty-3 have been introduced into commercial varieties by molecular marker-assisted selection (MAS) technology. The Ty-1 and Ty-3 alleles are located on chromosome 6 and code for an RNA-dependent RNA polymerase [3]. A mixed infection of TYLCV with cucumber mosaic virus compromised the resistance of Ty-1 and Ty-3 [15]. Ty-2 was introgressed into tomato via Solanum habrochaites strain B6013 and was found to be located on chromosome 11. This gene encodes a nucleotide-binding leucine-rich repeat protein [16,17]. TYLCV variation and recombination can overcome the resistance of typical R-genes. We found that some varieties with the Ty-2 gene have lost their resistance to TYLCV in the Shouguang and Cuxiong regions of China. Tomato varieties with Ty-1, Ty-2, and Ty-3 were severely challenged by TYLCV, and thus the introduction of other resistance genes into tomato varieties is critical for tomato production. The Ty-4 gene has not been widely introduced into cultivated tomato due to its weak effect on TYLCV resistance [12]. Ty-6 is a novel resistance gene that is located on chromosome 10 and is derived from Solanum chilense strain LA2779 [18].
The recessive gene ty-5 was mapped on chromosome 4 and identified from breeding line TY172 derived from Solanum peruvianum [19,20]. This gene encodes a messenger RNA surveillance factor Pelota [13,21]. We introduced a cultivated tomato AVTO1227 containing the ty-5 gene from the Asian Vegetable Research and Development Centre. TYLCV-inoculation of AVTO1227 exhibited a high level of resistance. In this study, Insertion and Deletion (InDel) markers, single nucleotide polymorphism (SNP) markers, PCR fragments sequencing, and high resolution melting (HRM) techniques were used to analyze the tomato genotypes containing the TYLCV-resistance ty-5 gene. The objectives of this study included to establish a MAS breeding platform, enrich the collection of resistant tomato varieties, speed up breeding selection, and provide a basis for clarifying the resistance mechanism of ty-5 resistance-related genes.
2. Materials and Methods
2.1. Plant Materials
The resistant tomato line ‘AVTO1227’ containing the ty-5 gene was crossed to the susceptible line Moneymaker to produce F1 generation. The F1 plants were then selfed to develop a segregating F2 population. Part of the F2 plants and recombinants were subjected to disease resistance evaluation and marker validation.
2.2. TYLCV Inoculation
Whiteflies carrying the yellow leaf curl virus were maintained in Moneymaker plants in our lab. Three-leaf-stage tomato seedlings were transferred to a phytotron with viruliferous whiteflies for 2 weeks, following which the trays containing the seedlings were moved into another phytotron without whiteflies. The whiteflies on the seedlings were killed by imidacloprid immediately after moving into the phytotron. Imidacloprid was used again three days later to make sure all the whiteflies were killed. The seedlings were then planted into plant pots and used for scoring [11].
2.3. Disease Assessment
Forty days after inoculation, the plants were assessed for TYLCV infection severity. The TYLCV symptoms were visually evaluated using a 0–4 disease severity index (DSI) described by Wang et al. [1], where 0 = no visible symptoms, with inoculated plants appearing similar to non-inoculated plants; 4 = severe stunting, yellowing, and curling, with plant growth having ceased. Intermediate scores (e.g., 0.5 and 1.5) represent intermediate disease morphologies based on the above scale and previously described methods. For recombinant screening, plants with a DSI ≤ 1 were considered resistant, whereas plants with a DSI > 1 were rated as susceptible.
2.4. DNA Extraction
Genomic DNA was extracted from young leaves using the CTAB method. The DNA quality was assessed by 1% agarose gels stained with ethidium bromide and visualized under ultraviolet light. The purity (OD260/280 = 1.8–2.0) and concentration of the sample DNA were determined using an Eppendorf BioSpectrometer® UV/Vis spectrophotometer, following which the DNA was adjusted to a final concentration of 10 ng·μL−1 and stored at −20 °C.
2.5. Marker Analysis
Primers were designed using Primer Premier 5 software (Version 5.00). The primers used in this study are listed in Table 1. SlNAC1 was adopted from Hutton et al. [20]. The InDel marker ty5-17 was previously designed by our lab. HRM-ty5-1 was designed according to the SNP in the first exon of ty-5, and HRM-ty5-2 was designed according to the SNP in the promoter of ty-5 [1].

Table 1.
Primers and reaction conditions for the developed markers.
2.6. Marker Analysis
PCR amplification for the SlNAC1 marker was performed in a total volume of 20 μL containing 2 μL genomic DNA, 10 μL Master Mix (Tsingke Biotechnology Co., Ltd. Nanjing, China), and 0.2 μL of each 10 μM primer. The PCR conditions were as follows: initial denaturation for 4 min at 94 °C followed by 38 cycles of 45 s at 94 °C, 45 s at 48 °C, 45 s at 72 °C, and a final extension of 10 min at 72 °C. The PCR products were digested by the TaqI restriction enzyme at 65 °C for 3 h. The digested products were tested on 1% agarose gels.
The PCR amplification for the ty5-17 marker was performed following the method described above. The PCR products were tested on 8% polyacrylamide gel (PAGE).
2.7. HRM Marker Genotype Analysis
The PCR amplification and HRM analysis were performed on a LightCycler®480 II instrument (Roche, Basel, Switzerland) using a 96-well reaction module. The LightCycler®480 II High Resolution Melting Master kit was purchased from Roche. The PCR was performed in a 20 μL volume with 10 ng DNA, 1 × Master Mix (Tsingke Biotechnology Co., Ltd., Nanjing, China), 2.0 m M MgCl2, and 0.2 μM of each primer.
The initial PCR reaction conditions included incubation at 95 °C for 10 min, followed by 45 cycles of 95 °C for 15 s, 58 °C for 15 s, and 72 °C for 20 s. This was followed by melting curve reaction conditions of 95 °C for 1 min, 40 °C for 1 min, and then up to 65 °C for 1 s; following which the temperature was ramped to 95 °C for the fluorescence collection process (25 times/°C) and then finally cooled to 40 °C. Each sample had three repetitions. The results were analyzed using Gene Scanning software from LightCycler®480 II (Roche, Basel, Switzerland).
2.8. Sequencing Analysis
The PCR amplifications were carried out in 20 μL of total reaction volume containing 2 μL genomic DNA, 10 μL Master Mix (Tsingke Biotechnology Co., Ltd.), and 0.2 μL of each 10-μM primer. The PCR conditions were as follows: initial denaturation for 4 min at 94 °C followed by 38 cycles of 45 s at 94 °C, 45 s at 58 °C, 45 s at 72 °C, and a final extension of 10 min at 72 °C. The PCR amplification products were sequenced by Kingsley Biological Technology Co., Ltd. (Nanjing, China). The results of the sequence were analyzed by Chormas software (Version 1.62).
3. Results
3.1. CAPS Marker Analysis
The SlNACI marker exhibited codominant characteristics. Samples of 10 plants from the F2 population were analyzed, and the parent samples were used as the control. The electrophoresis results are indicated in Figure 1. Lanes 1–5 represent the homozygous susceptible genotype, with a 161 bp band. Lane 6 contained two bands of 206 bp and 161 bp, representing the heterozygous susceptible genotype. Lanes 7–10 only had a band of 206 bp representing the homozygous resistant plants.
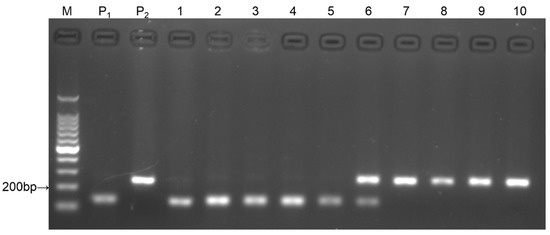
Figure 1.
Gel image of the parents and F2 individuals amplified by primer SlNACI. Note: M: 100 bp marker; P1: Moneymaker; P2: AVTO1227; lanes 1–10: F2 individuals.
3.2. InDel Marker Analysis
The ty5-17 marker possessed codominant characteristics. The PCR amplification bands were 172 bp and 187 bp, with a difference of only 9 bp between the two DNA bands. Samples of 10 plants (different from the plants used in Section 3.1) from the F2 population were analyzed. The polyacrylamide gel results (Figure 2) showed that lanes 1–5 represent the homozygous resistant genotype, with a 172 bp band. Lanes 6 and 7 are the heterozygous susceptible genotype, containing two bands of 172 bp and 187 bp. Lanes 8–10 represent the homozygous susceptible genotype, with a band of 187 bp.
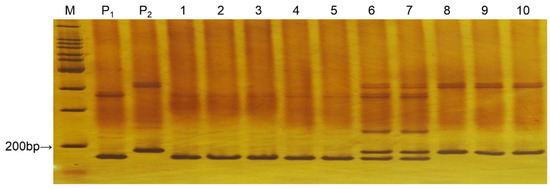
Figure 2.
Gel image of the parents and F2 individuals amplified by primer ty5-17. Note: M: 100 bp marker; P1: AVTO1227; P2: Moneymaker; lanes 1–10: F2 individuals.
3.3. HRM Analysis
Two transversions within the promoter region and a SNP in the exon were detected on ty-5. The exon site was used to design HRM-ty5-1 and the promoter SNP site was used for the design of HRM-ty5-2. Both could discriminate the three genotypes (homozygous resistant, heterozygous susceptible, and homozygous susceptible) based on different melting curves. AVTO1227, Moneymaker, and one F1 plant were used as the standard samples of the homozygous resistant, homozygous susceptible, and heterozygous susceptible genotypes, respectively. The samples of the 12 randomly selected plants from the F2 population were analyzed by HRM-ty5-1 and HRM-ty5-2, with three repeats tested. The results are shown in Figure 3. The curves of different genotypes showed different shapes.
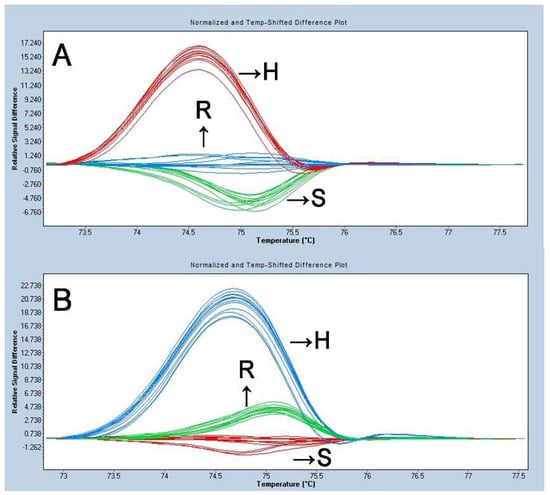
Figure 3.
SNP genotyping analysis for TYLCV resistance in tomato with HRM-ty-5-1 (A) and HRM-ty-5-2 (B). Note: R: homozygous resistant; S: homozygous susceptible; H: heterozygous.
3.4. Sequencing Results
The results presented by the HRM analysis were further investigated and confirmed by PCR sequencing. The PCR amplification products of the 12 samples analyzed with HRM-ty5-1 were sequenced, and the results of the genotype were consistent with the HRM analysis. The sequencing results (Figure 4) demonstrates that at the same site, the homozygous resistant genotype contained a G base, the homozygous susceptible contained a T base, and the heterozygous susceptible contained both the G and T bases.
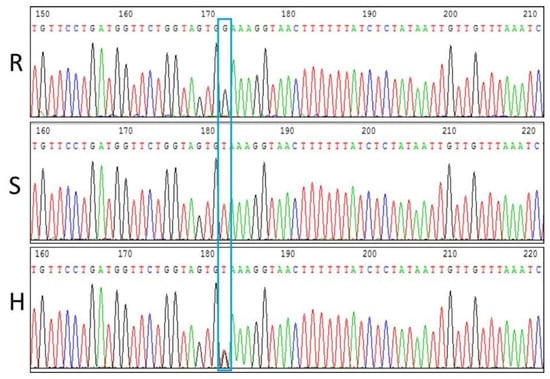
Figure 4.
Sequencing results of tomato genotypes with different resistance levels to TYLCV. Note: R: homozygous resistant; S: homozygous susceptible; H: heterozygous. The blue box indicates the SNP site.
3.5. Comparison of the Genotypes of Different Markers
Molecular markers SlNAC1 and ty5-17 were used to analyze 1500 plants of the F2 population. A total of 19 recombinant plants were detected between these two markers. HRM marker analysis and the sequencing method were used to analyze the genotypes of these 19 recombinants, and the results indicated that the phenotype of all recombinants was consistent with the HRM marker and sequencing results. The phenotypes of plants 1, 2, 13, and 18 were consistent with SlNAC1, and those of plants 3, 5, 10, 12, 14, 16, and 17 were consistent with the genotype of ty5-17. The accuracy of the HRM-ty5-1 and HRM-ty5-2 marker was 100% (Table 2).

Table 2.
Comparative analysis of tomato reaction to TYLCV with different types of markers.
4. Discussion
With the development of biotechnology, a variety of genetic markers have been developed based on DNA polymorphisms. Molecular marker technology has been widely used in the identification, detection, and assisted selection of parent materials and varieties with breeding applications. MAS is an effective tool for the breeding of tomato resistance. However, the accuracy of linkage marker selection depends on the degree of linkage between the marker and the resistance gene. Even closely linked molecular markers can result in false positives due to the recombination between the resistance genes and markers in separated progeny, reducing the selection efficiency. A functional marker (gene internal marker) of the resistance gene, which is a specific marker with a selection accuracy rate of 100% [22], can overcome the shortcomings of linkage markers. Comparing the test results and application analysis between the different markers, genotyping with HRM-ty5-1, HRM-ty5-2 and sequencing have 100% accuracy. In this study, the SNP marker represented a functional marker within the gene, there was no issue with false positives, the amplification was stable, the genotype was clear, and the disease resistance gene could be effectively identified. The HRM technique can thus provide a detection platform for disease resistance breeding and improve the selection efficiency of disease-resistant materials.
In this research, we compared the efficiency of different marker types linked with ty-5. Cleaved amplified polymorphism sequences (CAPS) are based on restriction enzymes, with the restriction enzyme digesting the PCR amplification sequence to produce DNA polymorphism. SlNAC1 markers are codominant CAPS markers with specific enzyme digestion sites. This marker is located far from the target gene and can easily recombine during resistance identification, leading to inaccurate detection results. The SNP sites in the first exon and the promoter of ty-5 cannot be converted to CAPS marker directly. A dCAPS (derived-cleaved amplified polymorphic sequences) marker 14IY5 based on the SNP located at the first exon was developed [21]. Although 14IY5 and HRM-ty5-1 target the same SNP, HRM-ty5-1 is more accurate than 14IY5, as HRM markers do not require introduction of mismatches in the primer. Besides that, HRM marker do not need additional time for restriction enzyme digestion and gel electrophoresis. The detection results can been read on the computer directly when the qPCR is completed. HRM-ty5-1 is more efficient and suitable for testing large numbers of samples than 14IY5.
InDels are detected at the genomic level, with differently-sized nucleotide fragment insertions or deletions producing DNA polymorphisms. According to the insertion or deletion sites, specific primers are designed to amplify the DNA sequence. The ty5-17 marker is an InDel marker that also has the characteristics of being codominant, but the PCR amplification products need to be tested using polyacrylamide gel, the operation steps of which are complex and time consuming, and thus it is also difficult to use these for testing large numbers of samples.
SNPs are detected at the genomic level and are the result of single base mutations (substitutions, insertions, or deletions) producing DNA polymorphisms. Compared with other molecular markers, SNPs have the advantages of high distribution density and genetic stability and are widely distributed in the tomato genome. SNPs are typically tested using direct sequencing, TaqMan probe, CAPS, HRM, and other methods [23,24].
HRM is a post-PCR technique that, through small increments in temperature (0.01–0.2 °C/s), measures the rate of DNA chain dissociation [25,26]. The PCR amplification products are combined with double-stranded DNA binding dye during the heating process, and when melted, each PCR product will exhibit a specific characteristic melting or disassociation behavior [27,28]. Different SNP sites or heterozygous and homozygous alleles can form different shapes in the melting curve. Thus, HRM can be used for genotyping.
Direct sequencing refers to the sequencing of PCR amplification products followed by sequence comparison in order to determine a single nucleotide difference in the study sequence. Its testing accuracy can reach 100%, and it can be used to map the specific SNP location and base mutation type. However, direct sequencing is more costly, and long sequencing cycles require more time and thus a longer wait for the results. Direct sequencing is also difficult to use for testing large numbers of samples.
In contrast to the different markers discussed above, the HRM technique does not require either enzyme digestion or the separation of products by gel electrophoresis. Additionally, the PCR amplification and melting curves are completed at the same time in the same system, resulting in a closed operation and thus reduced contamination. In addition, the HRM reaction can analyze 96 or 348 samples on the LightCycler®480 II analyzer at once, which only requires 90–100 min from the beginning of the reaction to data generation. HRM has the advantages of simple operation, intuitive results, short analysis time, high specificity, and high sensitivity, and even a single base pair difference can be detected [28,29]. Due to the high throughput characteristic of HRM, it is suitable for the analysis of large numbers of samples, and thus HRM markers can be used to screen large populations [30]. For higher throughput, HRM-ty5 also can be converted into KASP (Kompetitive Allele-Specific PCR) markers.
Author Contributions
Conceptualization, Y.W. and L.S.; formal analysis, Y.W.; funding acquisition, Y.W., L.S. and T.Z.; project administration, L.Z.; resources, T.Z.; supervision, T.Z.; validation, W.Y. and L.S.; writing—original draft, Y.W. and L.S.; writing—review & editing, Y.W. and T.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31972424), the National Natural Science Foundation of China (31902025), the Seed Industry Revitalization Project of Jiangsu Province (JBGS [2021] 066), the fifth “333” high-level personnel training project of Jiangsu (BRA2020369), and the Natural Science Foundation of Youth in Jiangsu Province (BK20180306).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We gratefully acknowledge Duanyue Huang for his assistance during the experiments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, Y.; Jiang, J.; Zhao, L.; Zhou, R.; Yu, W.; Zhao, T. Application of Whole Genome Resequencing in Mapping of a Tomato Yellow Leaf Curl Virus Resistance Gene. Sci. Rep. 2018, 8, 9592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanssen, I.M.; Lapidot, M.; Thomma, B.P.H.J. Emerging Viral Diseases of Tomato Crops. Mol. Plant-Microbe Interact. 2010, 23, 539–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verlaan, M.G.; Hutton, S.F.; Ibrahem, R.M.; Kormelink, R.; Visser, R.G.F.; Scott, J.W.; Edwards, J.; Bai, Y. The Tomato Yellow Leaf Curl Virus Resistance Genes Ty-1 and Ty-3 Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases. PLoS Genet. 2013, 9, e1003399. [Google Scholar] [CrossRef] [Green Version]
- Cohen, S.; Harpaz, I. Periodic, Rather Than Continual Acquisition of a New Tomato Virus by Its Vector, The Tobacco Whitefly (Bemisia Tabaci Gennadius). Èntomol. Exp. Appl. 1964, 7, 155–166. [Google Scholar] [CrossRef]
- Scholthof, K.-B.G.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef]
- Pan, H.; Chu, D.; Yan, W.Q.; Su, Q.; Liu, B.M.; Wang, S.L.; Wu, Q.J.; Xie, W.; Jiao, X.G.; Li, R.M.; et al. Rapid Spread of Tomato Yellow Leaf Curl Virus in China Is Aided Differentially by Two Invasive Whiteflies. PLoS ONE 2012, 7, e34817. [Google Scholar] [CrossRef] [Green Version]
- Polston, J.E.; Lapidot, M. Management of Tomato yellow leaf curl virus: US and Israel Perspectives. In Tomato Yellow Leaf Curl Virus Disease; Spirnger: Dordrecht, The Netherlands, 2007; pp. 251–262. [Google Scholar] [CrossRef]
- Lapidot, M.; Legg, J.P.; Wintermantel, W.M.; Polston, J.E. Management of whitefly-transmitted viruses in open-field production systems. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2014; Volume 90, pp. 147–206. [Google Scholar] [CrossRef]
- Zamir, D.; Ekstein-Michelson, I.; Zakay, Y.; Navot, N.; Zeidan, M.; Sarfatti, M.; Eshed, Y.; Harel, E.; Pleban, T.; Van-Oss, H.; et al. Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, TY-1. Theor. Appl. Genet. 1994, 88, 141–146. [Google Scholar] [CrossRef]
- Yang, X.; Caro, M.; Hutton, S.F.; Scott, J.W.; Guo, Y.; Wang, X.; Rashid, H.; Szinay, D.; De Jong, H.; Visser, R.G.F.; et al. Fine mapping of the tomato yellow leaf curl virus resistance gene Ty-2 on chromosome 11 of tomato. Mol. Breed. 2014, 34, 749–760. [Google Scholar] [CrossRef] [Green Version]
- Ji, Y.; Schuster, D.J.; Scott, J.W. Ty-3, a begomovirus resistance locus near the Tomato yellow leaf curl virus resistance locus Ty-1 on chromosome 6 of tomato. Mol. Breed. 2007, 20, 271–284. [Google Scholar] [CrossRef]
- Ji, Y.; Scott, J.W.; Schuster, D.J.; Maxwell, D.P. Molecular Mapping of Ty-4, a New Tomato Yellow Leaf Curl Virus Resistance Locus on Chromosome 3 of Tomato. J. Am. Soc. Hortic. Sci. 2009, 134, 281–288. [Google Scholar] [CrossRef] [Green Version]
- Lapidot, M.; Karniel, U.; Gelbart, D.; Fogel, D.; Evenor, D.; Kutsher, Y.; Makhbash, Z.; Nahon, S.; Shlomo, H.; Chen, L.; et al. A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor Pelota. PLoS Genet. 2015, 11, e1005538. [Google Scholar] [CrossRef] [PubMed]
- Hutton, S.F.; Scott, J.W. Ty-6, a major begomovirus resistance gene located on chromosome 10. Rept. Tomato Genet. Coop. 2014, 64, 4–18. [Google Scholar]
- Butterbach, P.; Verlaan, M.G.; Dullemans, A.; Lohuis, D.; Visser, R.G.F.; Bai, Y.; Kormelink, R. Tomato yellow leaf curl virus resistance by Ty-1 involves increased cytosine methylation of viral genomes and is compromised by cucumber mosaic virus infection. Proc. Natl. Acad. Sci. USA 2014, 111, 12942–12947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, X.; Yan, Z.; Wang, X.; Wang, Y.; Arens, M.; Du, Y.; Visser, R.G.F.; Kormelink, R.; Bai, Y.; Wolters, A.-M.A. The NLR Protein Encoded by the Resistance Gene Ty-2 Is Triggered by the Replication-Associated Protein Rep/C1 of Tomato Yellow Leaf Curl Virus. Front. Plant Sci. 2020, 11, 545306. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Ohnishi, J.; Saito, A.; Ohyama, A.; Nunome, T.; Miyatake, K.; Fukuoka, H. An NB-LRR gene, TYNBS1, is responsible for resistance mediated by the Ty-2 Begomovirus resistance locus of tomato. Theor. Appl. Genet. 2018, 131, 1345–1362. [Google Scholar] [CrossRef]
- Gill, U.; Scott, J.W.; Shekasteband, R.; Ogundiwin, E.; Schuit, C.; Francis, D.M.; Sim, S.-C.; Smith, H.; Hutton, S.F. Ty-6, a major begomovirus resistance gene on chromosome 10, is effective against Tomato yellow leaf curl virus and Tomato mottle virus. Theor. Appl. Genet. 2019, 132, 1543–1554. [Google Scholar] [CrossRef] [Green Version]
- Anbinder, I.; Reuveni, M.; Azari, R.; Paran, I.; Nahon, S.; Shlomo, H.; Chen, L.; Lapidot, M.; Levin, I. Molecular dissection of Tomato leaf curl virus resistance in tomato line TY172 derived from Solanum peruvianum. Theor. Appl. Genet. 2009, 119, 519–530. [Google Scholar] [CrossRef]
- Hutton, S.F.; Scott, J.W.; Schuster, D.J. Recessive Resistance to Tomato yellow leaf curl virus from the Tomato Cultivar Tyking Is Located in the Same Region as Ty-5 on Chromosome 4. HortScience 2012, 47, 324–327. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Chung, D.J.; Lee, J.M.; Yeam, I. Development and Application of Gene-Specific Markers for Tomato Yellow Leaf Curl Virus Resistance in Both Field and Artificial Infections. Plants 2021, 10, 9. [Google Scholar] [CrossRef]
- Arens, P.; Mansilla, C.; Deinum, D.; Cavellini, L.; Moretti, A.; Rolland, S.; van der Schoot, H.; Calvache, D.; Ponz, F.; Collonnier, C.; et al. Development and evaluation of robust molecular markers linked to disease resistance in tomato for distinctness, uniformity and stability testing. Theor. Appl. Genet. 2010, 120, 655–664. [Google Scholar] [CrossRef] [Green Version]
- Gut, I.G. Automation in genotyping of single nucleotide polymorphisms. Hum. Mutat. 2001, 17, 475–492. [Google Scholar] [CrossRef] [PubMed]
- Syvänen, A.-C. Accessing genetic variation: Genotyping single nucleotide polymorphisms. Nat. Rev. Genet. 2001, 2, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Druml, B.; Cichna-Markl, M. High resolution melting (HRM) analysis of DNA—Its role and potential in food analysis. Food Chem. 2014, 158, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Reed, G.H.; Kent, J.O.; Wittwer, C.T. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics 2007, 8, 597–608. [Google Scholar] [CrossRef] [Green Version]
- Montgomery, J.; Wittwer, C.T.; Palais, R.; Zhou, L. Simultaneous mutation scanning and genotyping by high-resolution DNA melting analysis. Nat. Protoc. 2007, 2, 59–66. [Google Scholar] [CrossRef]
- Li, Y.-D.; Chu, Z.-Z.; Liu, X.-G.; Jing, H.-C.; Liu, Y.-G.; Hao, D.-Y. A Cost-effective High-resolution Melting Approach using the EvaGreen Dye for DNA Polymorphism Detection and Genotyping in Plants. J. Integr. Plant Biol. 2010, 52, 1036–1042. [Google Scholar] [CrossRef]
- Reed, G.H.; Wittwer, C.T. Sensitivity and Specificity of Single-Nucleotide Polymorphism Scanning by High-Resolution Melting Analysis. Clin. Chem. 2004, 50, 1748–1754. [Google Scholar] [CrossRef] [Green Version]
- Vossen, R.H.; Aten, E.; Roos, A.; Den Dunnen, J.T. High-Resolution Melting Analysis (HRMA)-More than just sequence variant screening. Hum. Mutat. 2009, 30, 860–866. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).