Transcriptomic Analysis of Salt Stress Response in Pleurotus ostreatus
Abstract
1. Introduction
2. Materials and Methods
2.1. Pleurotus Ostreatus (PO) Mycelia
2.2. Mycelium Total RNA Extraction
2.3. cDNA (Complementary DNA) Library Production
2.4. Transcriptome Analysis
2.5. DEG (Difference Expressed Gene) Analysis
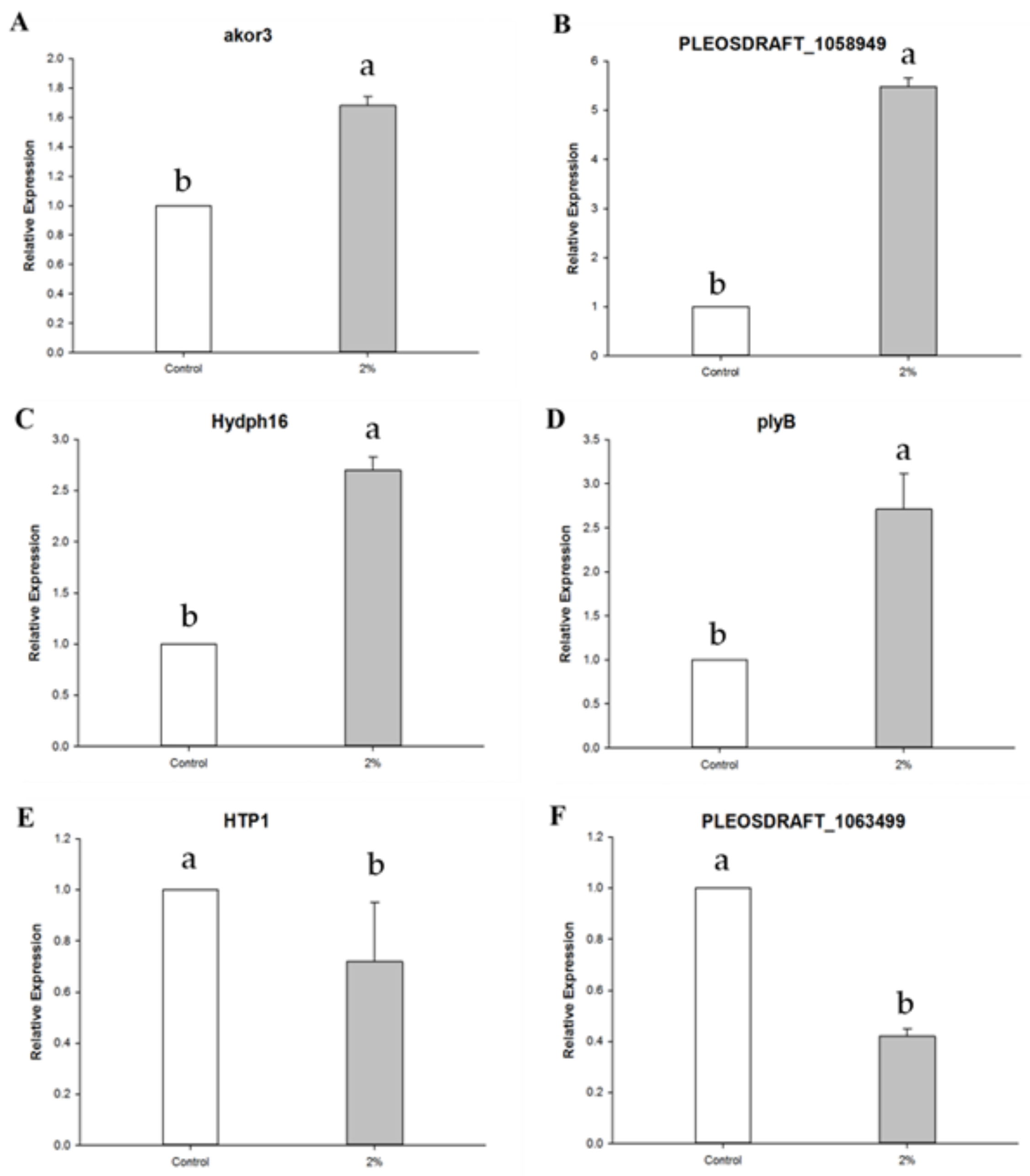
2.6. qRT-PCR (Quantitative Real Time Polymerase Chain Reaction)
3. Results
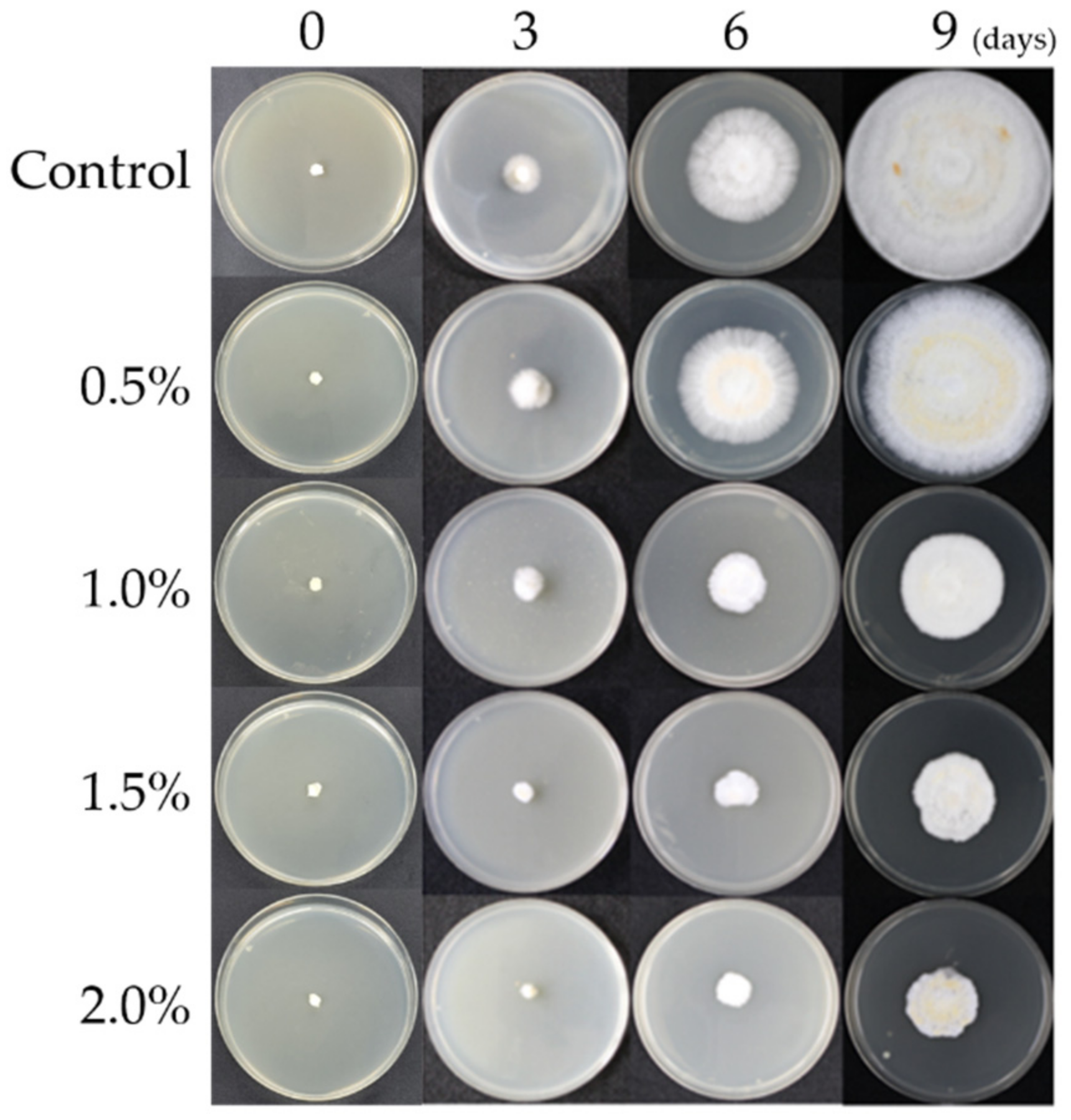
3.1. Mycelial Growth Characteristics According to the NaCl Concentration
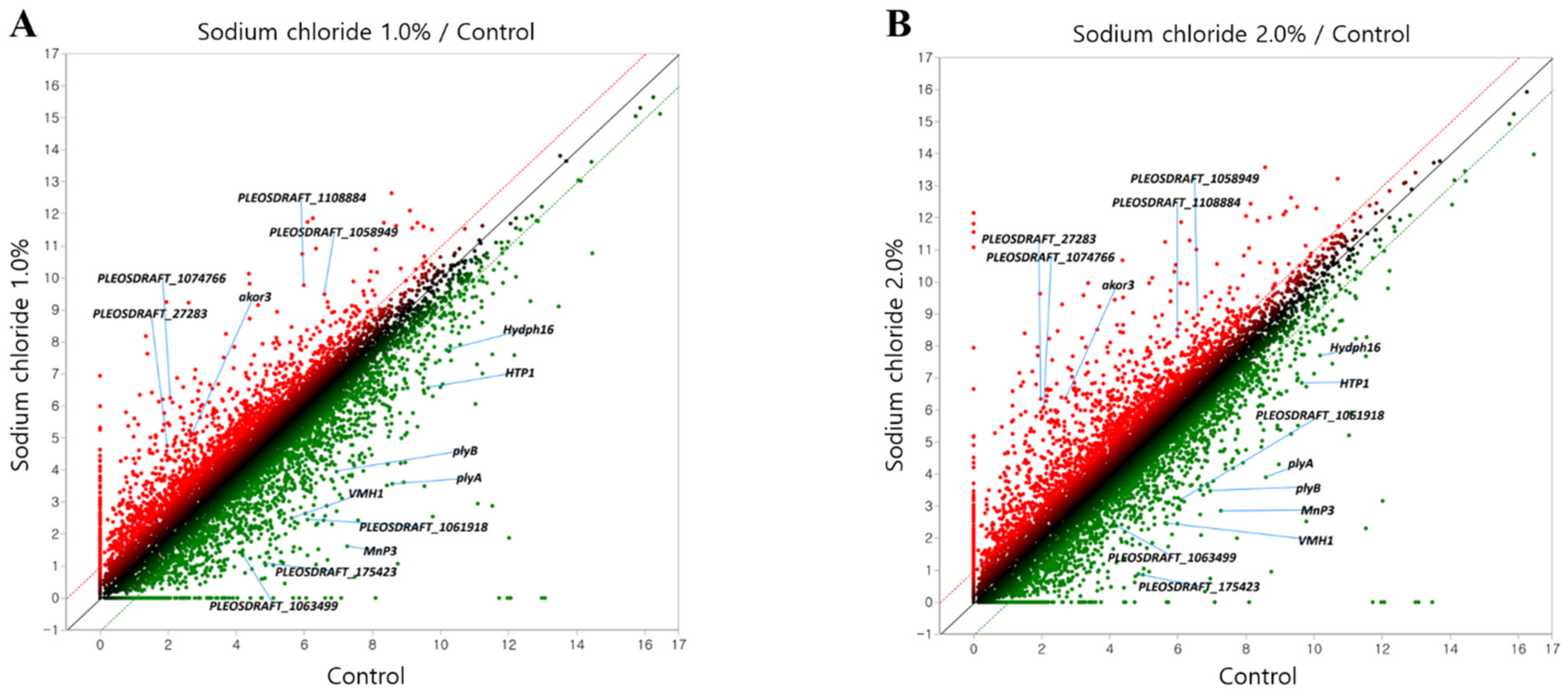
3.2. Gene Expression Pattern by NaCl Treatment
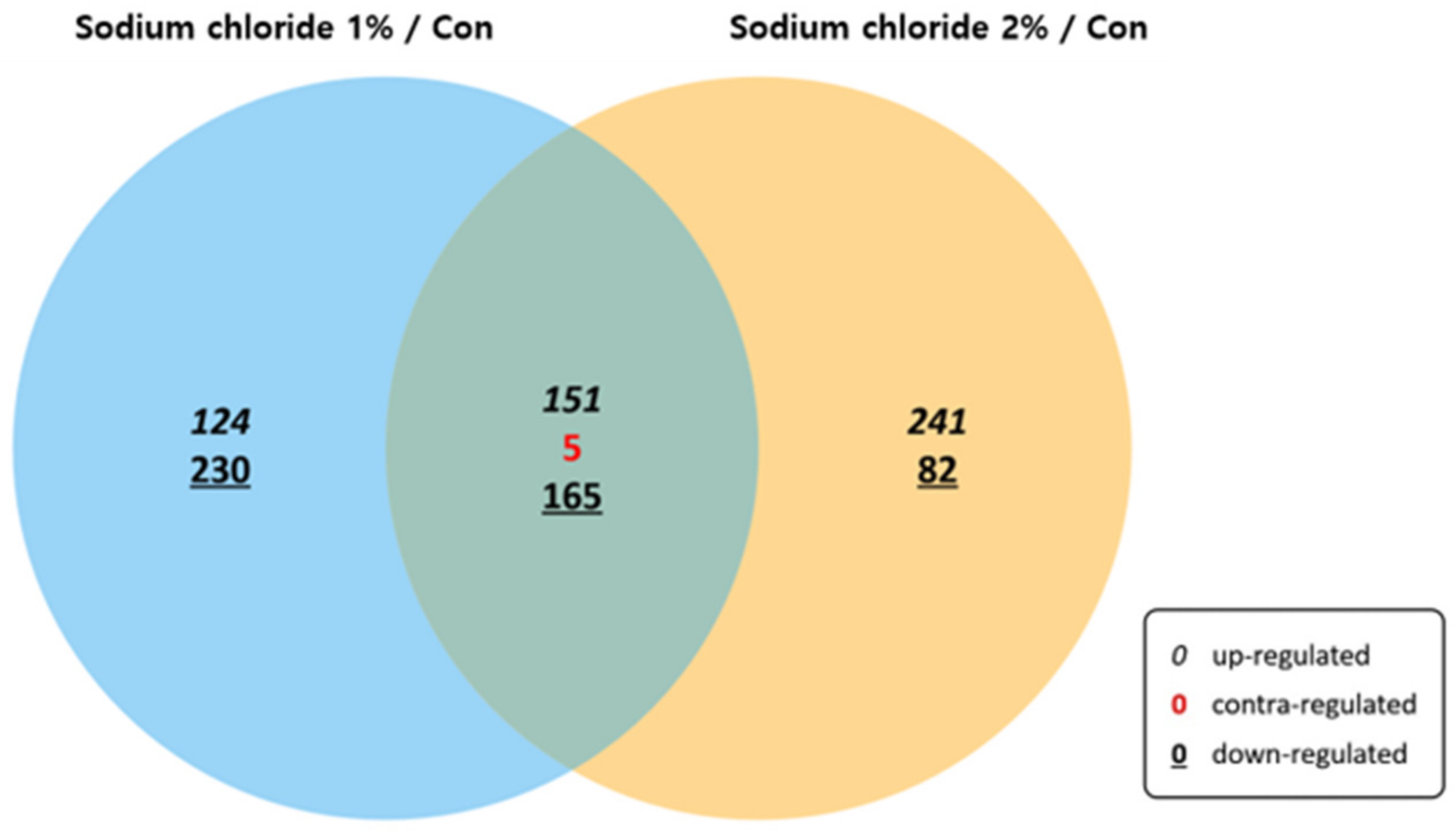
3.3. Comparison of Gene Expression Profiles
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gull, A.; Lone, A.A.; Wani, N.U.I. Biotic and Abiotic Stresses in Plants; IntechOpen: London, UK, 2019; pp. 1–19. [Google Scholar] [CrossRef]
- Dhanya Thomas, T.T.; Jos, T.P. UV radiation priming: A means of amplifying the inherent potential for abiotic stress tolerance in crop plants. Environ. Exp. Bot. 2017, 138, 57–66. [Google Scholar] [CrossRef]
- Jang, M.J.; Lee, Y.H.; Cho, Y.K.; Koo, H.M.; Oh, T.S. Growth properties of Neolentinus lepideus according to the light environment. J. Mushroom 2015, 13, 125–128. [Google Scholar] [CrossRef][Green Version]
- Won, S.Y.; Jang, M.J.; Ju, Y.C.; Lee, Y.B. Optimum CO2 Concentration for Fruit-body Formation and Yieldof Pleurotus ferulae Mushroom in the Growing Facilitiyfor Bottle Cultivation. J. Bio-Environ. Control 2010, 19, 77–81. [Google Scholar]
- Qin, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011, 52, 1569–1582. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N. Abscisic acid and abiotic stress signaling. Plant Signal. Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef]
- Wu, X.; Yuan, J.; Luo, A.; Chen, Y.; Fan, Y. Drought stress and re-watering increase secondary metabolites and enzyme activity in dendrobium moniliforme. Ind. Crops Prod. 2016, 94, 385–393. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef]
- Ma, L.; Li, Y.; Yu, C.; Wang, Y.; Li, X.; Li, N.; Bu, N. Alleviation of exogenous oligochitosan on wheat seedlings growth under salt stress. Protoplasma 2012, 249, 393–399. [Google Scholar] [CrossRef]
- Rivero, R.M.; Mestre, T.C.; Mittler, R.O.N.; Rubio, F.; Garcia-Sanchez, F.R.A.N.C.I.S.C.O.; Martinez, V. The combined effect of salinity and heat reveals a specific physiological, biochemical and molecular response in tomato plants. Plant Cell Environ. 2014, 37, 1059–1073. [Google Scholar] [CrossRef]
- Chokshi, K.; Pancha, I.; Ghosh, A.; Mishra, S. Salinity induced oxidative stress alters the physiological responses and improves the biofuel potential of green microalgae Acutodesmus dimorphus. Bioresour. Technol. 2017, 244, 1376–1383. [Google Scholar] [CrossRef]
- Tresner, H.D.; Hayes, J.A. Sodium chloride tolerance of terrestrial fungi. Appl. Microbiol. 1971, 22, 210–213. [Google Scholar] [CrossRef]
- Jhune, C.S.; Sul, H.J.; Kong, W.S.; You, Y.B.; Kim, J.H.; Cheong, J.C.; Lee, C.J. Effects of NaCl Concentration on Mycelial Growth and Fruiting Body Yields of Oyster Mushrooms, Pleurotus spp. J. Mushroom 2009, 7, 173–181. [Google Scholar]
- Muthangya, M.; Hashim, S.O.; Amana, J.M.; Mshandete, A.M.; Kivaisi, A.K. Optimization of Pleurotus mushroom cultivation on saline sisal solid waste. World Appl. Sci. J. 2013, 23, 1146–1150. [Google Scholar]
- Boumaaza, B.; Benkhelifa, M.; Belkhoudja, M. Effects of two salts compounds on mycelial growth, sporulation, and spore germination of six isolates of Botrytis cinerea in the western north of Algeria. Int. J. Microbiol. 2015, 2015, 572626. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2011, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.D.; Kim, Y.G.; Kim, H.G.; Han, G.H.; Moon, C.S.; Hur, I.B. Bottle Cultivation of Pleurotus ostreatus and Agrocybe aegerita using agricultural by-product. Korean J. Mycol. 1998, 26, 47–50. [Google Scholar]
- Chang, H.Y.; Park, H.S.; Yoon, J.S. Substitute cheap supplements development for Pleurotus ostreatus cultivation usiug food by-product dried wastes. J. Mushroom 2008, 6, 126–130. [Google Scholar]
- Chang, Q.; Griest, T.A.; Harter, T.M.; Petrash, J.M. Functional studies of aldo-keto reductases in Saccharomyces cerevisiae. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2007, 1773, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Petrash, J.M.; Murthy BS, N.; Young, M.; Morris, K.; Rikimaru, L.; Griest, T.A.; Harter, T. Functional genomic studies of aldo–keto reductases. Chem. Biol. Interact. 2001, 130, 673–683. [Google Scholar] [CrossRef]
- Yu, J.; Sun, H.; Zhang, J.; Hou, Y.; Zhang, T.; Kang, J.; Long, R. Analysis of Aldo-Keto Reductase Gene Family and Their Responses to Salt, Drought, and Abscisic Acid Stresses in Medicago truncatula. Int. J. Mol. Sci. 2020, 21, 754. [Google Scholar] [CrossRef]
- Chen, S.; Liu, J.; Pei, H.; Li, J.; Zhou, J.; Xiang, H. Molecular investigation of a novel thermostable glucan phosphorylase from Thermoanaerobacter tengcongensis. Enzym. Microb. Technol. 2007, 41, 390–396. [Google Scholar] [CrossRef]
- Schinzel, R.; Nidetzky, B. Bacterial α-glucan phosphorylases. FEMS Microbiol. Lett. 1999, 171, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wang, S.; Lin, Y.; Xu, W.; Ye, D.; Xiong, Y.; Guan, K.L. Acetylation negatively regulates glycogen phosphorylase by recruiting protein phosphatase 1. Cell Metab. 2012, 15, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Hofrichter, M.; Kellner, H.; Pecyna, M.J.; Ullrich, R. Fungal unspecific peroxygenases: Heme-thiolate proteins that combine peroxidase and cytochrome P450 properties. Monooxygenase Peroxidase Peroxygenase Prop. Mech. Cytochrome 2015, 450, 341–368. [Google Scholar]
- Alonso-Gil, S.; Parkan, K.; Kaminský, J.; Pohl, R.; Miyazaki, T. Unlocking the Hydrolytic Mechanism of GH92 α-1, 2-Mannosidases: Computation Inspires the use of C-Glycosides as Michaelis Complex Mimics. Chem. A Eur. J. 2022, 28, e202200148. [Google Scholar] [CrossRef] [PubMed]
- Ahn, Y.O.; Zheng, M.; Bevan, D.R.; Esen, A.; Shiu, S.H.; Benson, J.; Shih, M.C. Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 35. Phytochemistry 2007, 68, 1510–1520. [Google Scholar] [CrossRef]
- Yan, S.; Tang, Z.; Su, W.; Sun, W. Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics 2005, 5, 235–244. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651. [Google Scholar] [CrossRef]
- Marschner, H. Minernal Nutirition of Higher Plant; Academic Press: London, UK, 1995; pp. 662–663. [Google Scholar]
- Lee, B.M.; Shim, S.I.; Lee, S.G.; Kang, B.H.; Chung, I.M.; Kim, K.H. Physiological response on saline tolerance between halophytes and glycophytes. Korean J. Environ. Agric. 1999, 18, 61–65. [Google Scholar]
- Choi, S.C.; Kim, J.G.; Choo, Y.S. Effects of Salt Stress on Inorganic Ions and Glycine Betaine Contents in Leaves of Beta vulgaris var. cicla L. Korean J. Ecol. Environ. 2013, 46, 388–394. [Google Scholar] [CrossRef]
- Lee, S.B.; Cho, K.M.; Shin, P.; Yang, C.H.; Back, N.H.; Lee, K.B.; Chung, D.Y. Effect of soil salinity on growth, yield and nutrients uptake of whole crop barley in newly reclaimed land. Korean J. Environ. Agric. 2013, 32, 332–337. [Google Scholar] [CrossRef][Green Version]
- Park, S.C.; Kim, M.D.; Kim, S.H.; Kim, Y.H.; Jeong, J.C.; Lee, H.S.; Kwak, S.S. Enhanced drought and oxidative stress tolerance in transgenic sweetpotato expressing a codA gene. J. Plant Biotechnol. 2015, 42, 19–24. [Google Scholar] [CrossRef][Green Version]
- Phae, C.G.; Chu, Y.S.; Park, J.S. Investigation of Affect on Composting process and plant growth of Salt concetration in food waste. J. Korea Org. Resour. Recycl. Assoc. 2002, 10, 103–111. [Google Scholar]
| Gene Symbol | Sequence | Tm (°C) | Product Size |
|---|---|---|---|
| akor3 | F: TTACGGCTCTCGACAGAGGA R: TTACCACCCAAGGACCTCCA | F: 59.3 R: 59.3 | 92 bp |
| PLEOSDRAFT _1058949 | F: TCATGGGCAGAACCCTTGAC R: ACCACCATTACCAAGAGCGG | F: 59.3 R: 59.3 | 137 bp |
| Hydph16 | F: ACATCCGTCATTGCTCTCGT R: GCTTCAACACCAGATGCACG | F: 57.3 R: 59.3 | 131 bp |
| plyB | F: GGAGGCCGTTCTCTCAAGAC R: TTACCACCCAAGGACCTCCA | F: 61.4 R: 59.3 | 91 bp |
| HTP1 | F: TCACCACCACCAAGTTCCAC R: ATGGTGCTCTGATTCGGGAC | F: 59.3 R: 59.3 | 147 bp |
| PLEOSDRAFT _1063499 | F: CTCGCCAGAACTACGTCTCC R: CTATAACCTGCCCGCTCTCG | F: 61.4 R: 61.4 | 114 bp |
| 18s rRNA | F: GCATGTGCACGCTTCACTAG R: CGTAGTCACACCGAGACGTT | F: 59.3 R: 59.3 | 117 bp |
| Step | Time | Temperature (°C) |
|---|---|---|
| Denaturation | 10 s | 95 |
| Combined Annealing/extension | 30 s | 54–56 |
| Number of cycles | 39 cycles | - |
| Treatments | Incubation Period (Days) | ||
|---|---|---|---|
| 3 | 6 | 9 | |
| Control | 14.8 ± 0.8 a | 40.7 ± 1.5 a | 80.0 ± 1.8 a |
| NaCl 0.5% | 14.7 ± 0.5 a | 40.3 ± 0.8 a | 78.7 ± 0.9 a |
| NaCl 1.0% | 8.5 ± 0.4 b | 22.7 ± 1.0 b | 45.4 ± 1.2 b |
| NaCl 1.5% | 7.6 ± 0.5 b | 18.2 ± 1.8 c | 32.3 ± 2.4 c |
| NaCl 2.0% | 6.3 ± 0.8 c | 8.2 ± 1.2 d | 25.5 ± 2.1 d |
| Gene Symbol | 4-Fold Change * | Annotation | ||
|---|---|---|---|---|
| Control | 1.0%/Con | 2.0%/Con | Product | |
| PLEOSDRAFT_1074766 | 10 | 25.34 | 21.30 | Glycoside hydrolase family 43 protein |
| PLEOSDRAFT_1108884 | 168 | 13.61 | 4.77 | Hypothetical protein |
| akor3 | 4 | 10.23 | 22.52 | Putative aldo-keto reductase |
| PLEOSDRAFT_27283 | 10 | 8.45 | 24.12 | Chitin deacetylase |
| PLEOSDRAFT_1058949 | 764 | 7.70 | 5.31 | Alpha-1,4 glucan phosphorylase (EC 2.4.1.1) |
| Hydph16 | 943 | 0.195 | 0.172 | Hydrophobin |
| plyB | 626 | 0.127 | 0.094 | Pleurotolysin B |
| HTP1 | 2077 | 0.116 | 0.141 | Heme-thiolate peroxidase |
| VMH1 | 67 | 0.116 | 0.094 | Hydrophobin |
| PLEOSDRAFT_1063499 | 105 | 0.085 | 0.211 | Glycoside hydrolase family 92 protein |
| PLEOSDRAFT_1061918 | 205 | 0.071 | 0.135 | Pectinesterase |
| PLEOSDRAFT_175423 | 17 | 0.057 | 0.139 | Small secreted protein with six-cysteine repeat motif-containing protein |
| plyA | 922 | 0.036 | 0.042 | Ostreolysin-like protein |
| MnP3 | 788 | 0.016 | 0.041 | Peroxidase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pyeon, H.-Y.; Park, Y.-J.; Lee, S.-C.; Jang, M.-J. Transcriptomic Analysis of Salt Stress Response in Pleurotus ostreatus. Horticulturae 2022, 8, 1147. https://doi.org/10.3390/horticulturae8121147
Pyeon H-Y, Park Y-J, Lee S-C, Jang M-J. Transcriptomic Analysis of Salt Stress Response in Pleurotus ostreatus. Horticulturae. 2022; 8(12):1147. https://doi.org/10.3390/horticulturae8121147
Chicago/Turabian StylePyeon, Ha-Young, Youn-Jin Park, Sang-Chul Lee, and Myoung-Jun Jang. 2022. "Transcriptomic Analysis of Salt Stress Response in Pleurotus ostreatus" Horticulturae 8, no. 12: 1147. https://doi.org/10.3390/horticulturae8121147
APA StylePyeon, H.-Y., Park, Y.-J., Lee, S.-C., & Jang, M.-J. (2022). Transcriptomic Analysis of Salt Stress Response in Pleurotus ostreatus. Horticulturae, 8(12), 1147. https://doi.org/10.3390/horticulturae8121147






