Genome-Wide Identification of Sucrose Transporter Genes and Functional Analysis of RsSUC1b in Radish (Raphanus sativus L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Identification of SUC Gene Family in Radish
2.3. Sequence Characteristic and Phylogenetic Analysis
2.4. RNA Extraction and RT-qPCR Analysis
2.5. Subcellular Localization of RsSUC1b
2.6. Construction of Expression Vector and Genetic Transformation
2.7. Determination of Soluble Sugar Contents
2.8. Statistical Analysis
3. Results
3.1. Identification of the Radish SUC Gene Family
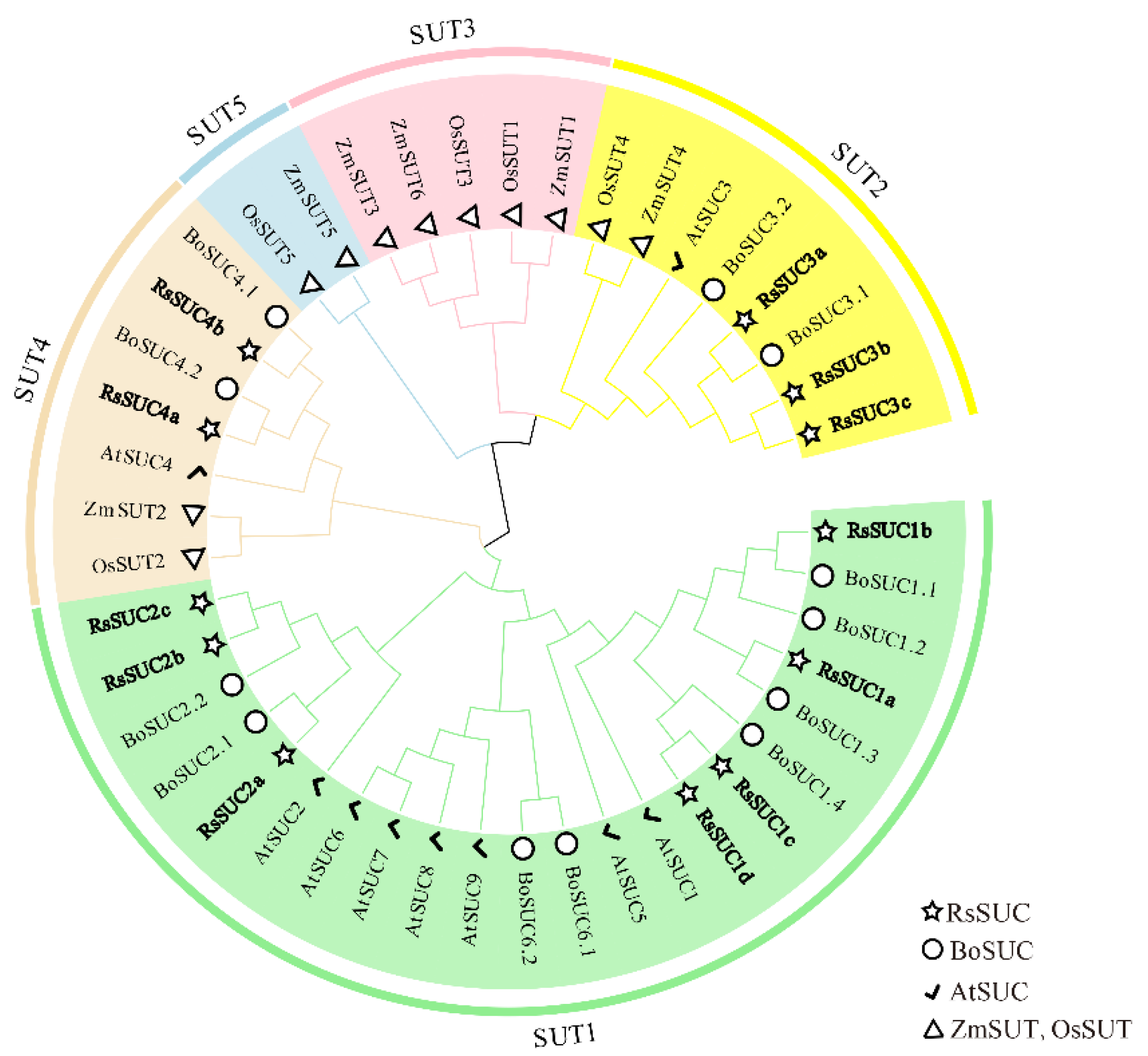
3.2. Phylogenetic Analysis of RsSUC Proteins
3.3. Gene Structure and Conserved Motifs Analysis of RsSUCs
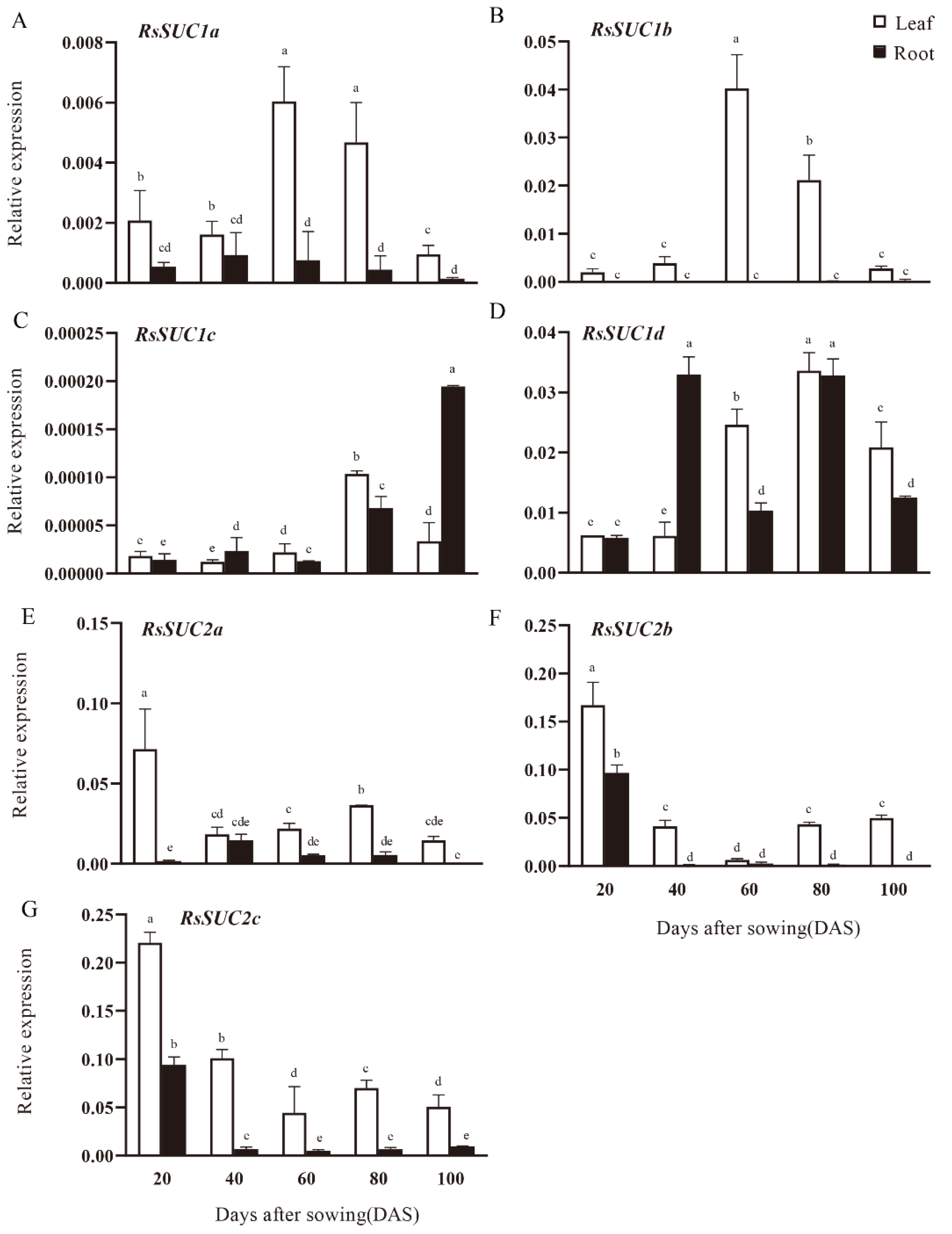
3.4. Expression Analysis of RsSUC Genes during Radish Taproot Development
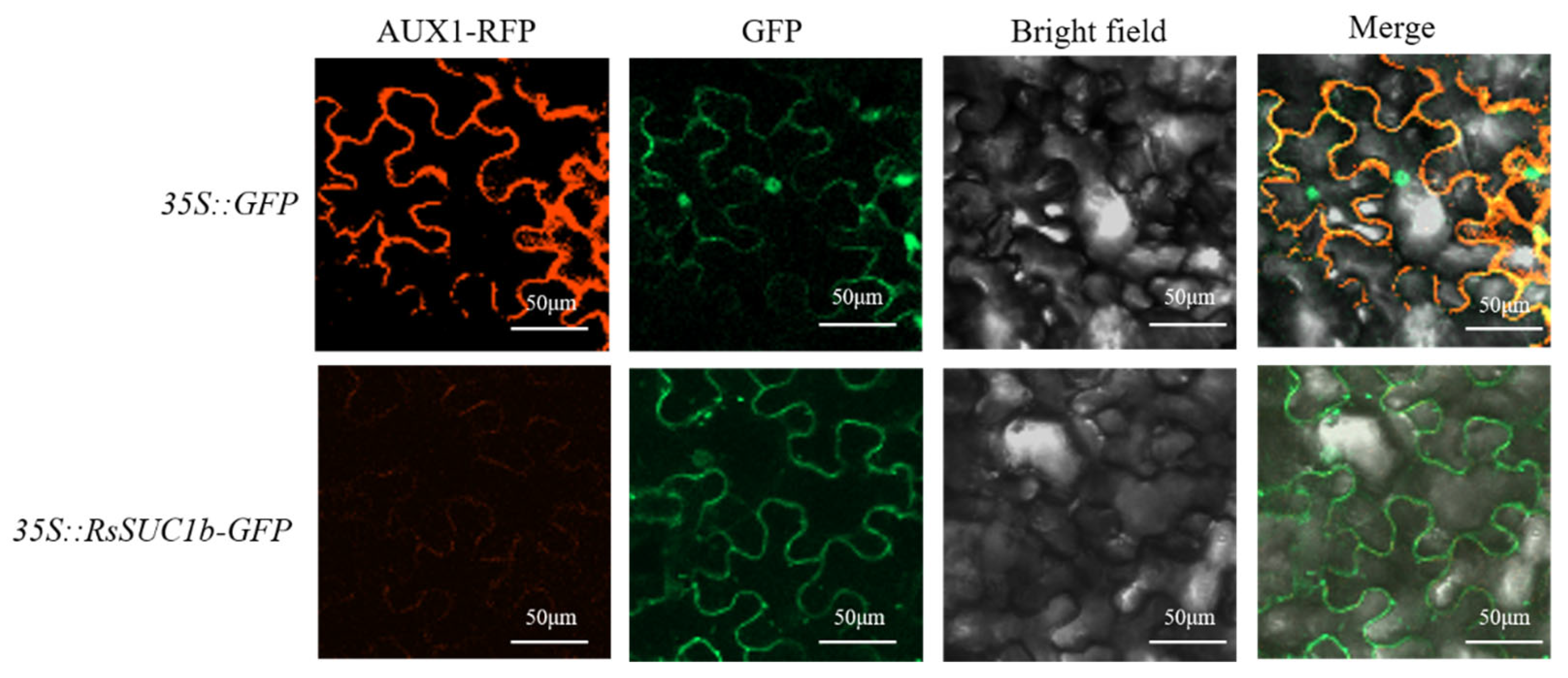
3.5. Subcellular Localization of RsSUC1b
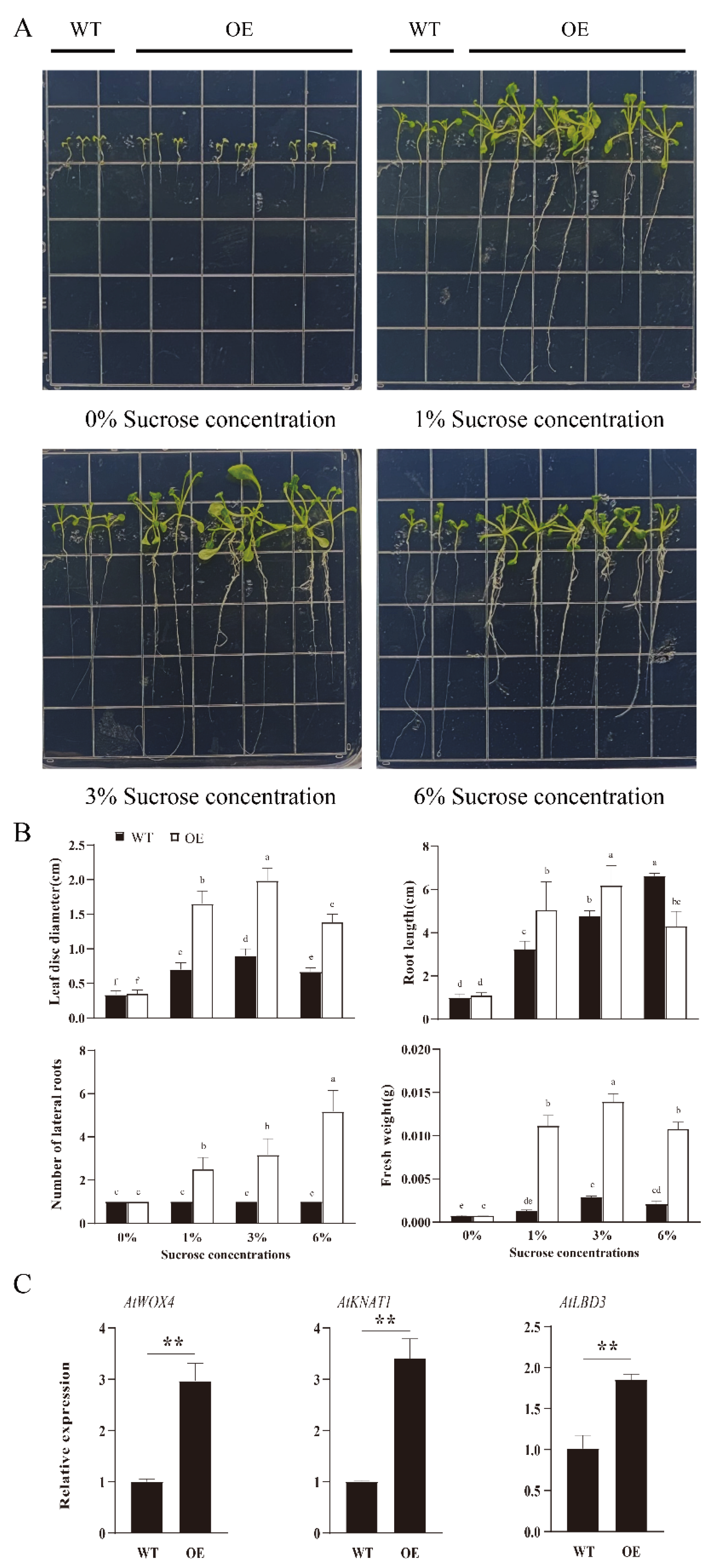
3.6. Effects of RsSUC1b Overexpression on Seedling Growth under Different Sucrose Treatments
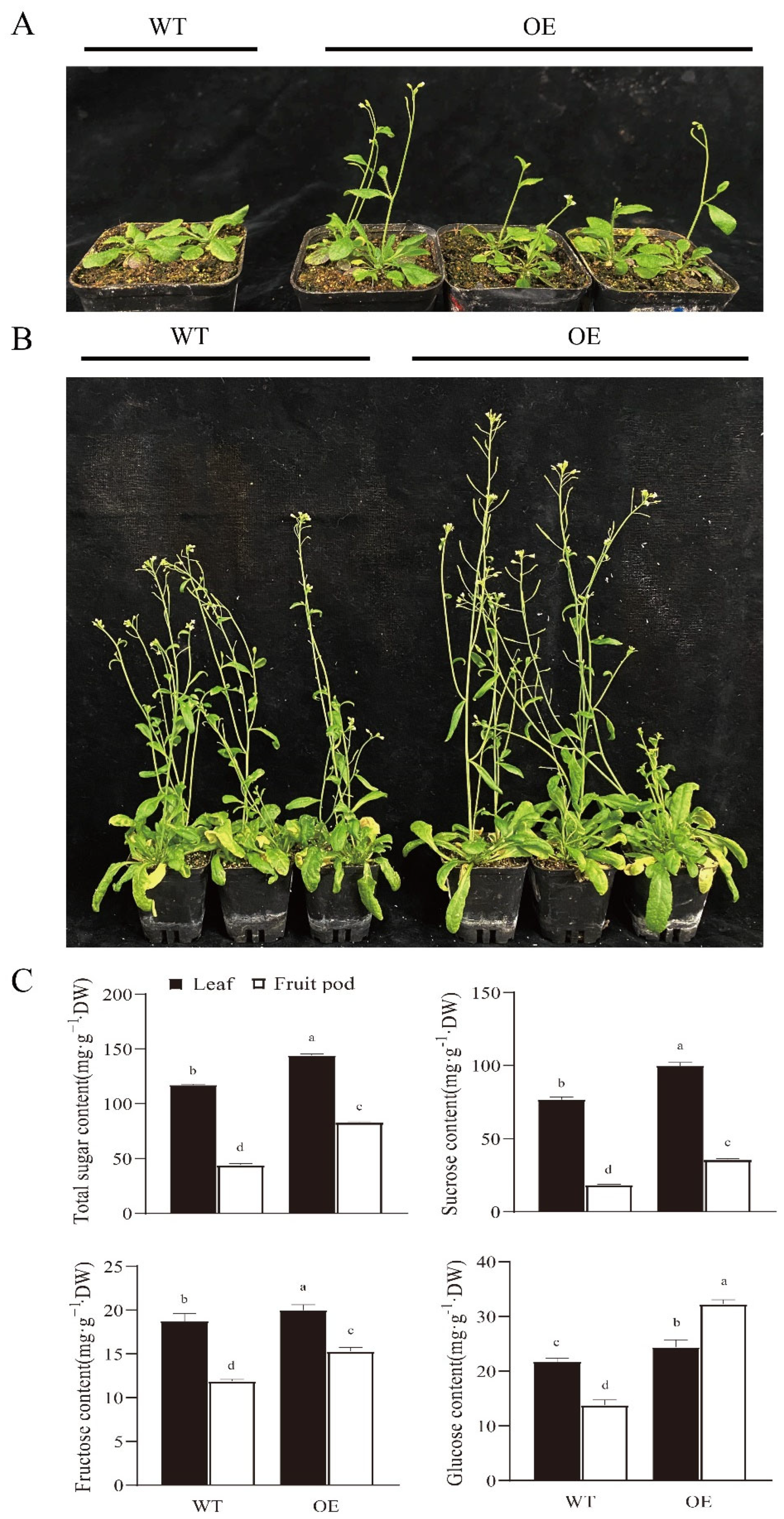
3.7. Overexpression of RsSUC1b in Arabidopsis Resulted in Early Flowering, Increased Height, and Higher SSCs of Plants
4. Discussion
4.1. Characterization of SUC Gene Family Members in Radish
4.2. Expression Patterns and Functional Diversity of RsSUC Genes
4.3. Sucrose Transporters Play Fundamental Roles in Plant Growth and Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fan, L.X.; Xu, L.; Wang, Y.; Tang, M.J.; Liu, L.W. Genome-and transcriptome-wide characterization of bZIP gene family identifies potential members involved in abiotic stress response and anthocyanin biosynthesis in radish (Raphanus sativus L.). Int. J. Mol. Sci. 2019, 16, 6334. [Google Scholar] [CrossRef] [PubMed]
- Masakazu, H.; Daiki, T.; Tatsuo, A.; Ikuo, T. Variations in the soluble sugar and organic acid contents in radish (Raphanus sativus L.) cultivars. Int. J. Food Sci. Technol. 2011, 46, 2387–2392. [Google Scholar]
- Kang, J.N.; Kim, J.S.; Lee, S.M.; Won, S.Y.; Seo, M.S.; Kwon, S.J. Analysis of phenotypic characteristics and sucrose metabolism in the roots of Raphanus sativus L. Front. Plant Sci. 2021, 12, 716782. [Google Scholar] [CrossRef] [PubMed]
- Mitsui, Y.; Shimomura, M.; Komatsu, K.; Namiki, N.; Shibata-Hatta, M.; Imai, M.; Katayose, Y.; Mukai, Y.; Kanamori, H.; Kurita, K.; et al. The radish genome and comprehensive gene expression profile of tuberous root formation and development. Sci. Rep. 2015, 5, 10835. [Google Scholar] [CrossRef]
- Liu, Y.; Yin, X.; Yang, Y.; Wang, C.; Yang, Y. Molecular cloning and expression analysis of turnip (Brassica rapa var. rapa) sucrose transporter gene family. Plant Divers. 2017, 20, 123–129. [Google Scholar] [CrossRef]
- Chen, Q.; Hu, T.; Li, X.; Song, C.P.; Zhu, J.K.; Chen, L.; Zhao, Y. Phosphorylation of SWEET sucrose transporters regulates plant root:shoot ratio under drought. Nat. Plants 2022, 8, 68–77. [Google Scholar] [CrossRef]
- Ma, S.; Li, Y.X.; Li, X.; Sui, X.L.; Zhang, Z.X. Phloem unloading strategies and mechanisms in crop fruits. J. Plant Growth Regul. 2019, 38, 494–500. [Google Scholar] [CrossRef]
- Reuscher, S.; Akiyama, M.; Yasuda, T.; Makino, H.; Aoki, K.; Shibata, D.; Shiratake, K. The sugar transporter inventory of tomato: Genome-wide identification and expression analysis. Plant Cell Physiol. 2014, 55, 1123–1141. [Google Scholar] [CrossRef]
- Dinant, S.; Le, H.R. Delving deeper into the link between sugar transport, sugar signaling, and vascular system development. Physiol. Plant. 2022, 174, e13684. [Google Scholar] [CrossRef]
- Sauer, N. Molecular physiology of higher plant sucrose transporters. FEBS Lett. 2007, 581, 2309–2317. [Google Scholar] [CrossRef]
- Usha, B. Diverse expression of sucrose transporter gene family in Zea mays. J. Genet. 2015, 94, 151–154. [Google Scholar] [CrossRef]
- Riesmeier, J.W.; Willmitzer, L.; Frommer, W.B. Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 1992, 11, 4705–4713. [Google Scholar] [CrossRef]
- Milne, R.J.; Byrt, C.S.; Patrick, J.W.; Grof, C.P. Are sucrose transporter expression profiles linked with patterns of biomass partitioning in Sorghum phenotypes? Front. Plant Sci. 2013, 4, 223. [Google Scholar] [CrossRef]
- Jian, H.; Lu, K.; Yang, B.; Wang, T.; Zhang, L.; Zhang, A.; Wang, J.; Liu, L.; Qu, C.; Li, J. Genome-wide analysis and expression profiling of the SUC and SWEET gene families of sucrose transporters in oilseed rape (Brassica napus L.). Front. Plant Sci. 2016, 28, 1464. [Google Scholar] [CrossRef]
- Poudel, K.; Luo, X.; Chen, L.; Jing, D.; Xia, X.; Tang, L.; Li, H.; Cao, S. Identification of the SUT gene family in pomegranate (Punicagranatum L.) and functional analysis of PgL0145810.1. Int. J. Mol. Sci. 2020, 21, 6608. [Google Scholar] [CrossRef]
- Yan, Z.X.; Yang, H.Y.; Zhang, C.H.; Wu, W.L.; Li, W.L. Functional analysis of the blackberry sucrose transporter gene RuSUT2. Russ. J. Plant Physiol. 2021, 68, 246–253. [Google Scholar] [CrossRef]
- Kühn, C.; Grof, C.P. Sucrose transporters of higher plants. Curr. Opin. Plant Biol. 2010, 13, 288–298. [Google Scholar] [CrossRef]
- Stadler, R.; Truernit, E.; Gahrtz, M.; Sauer, N. The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in Arabidopsis. Plant J. 1999, 19, 269–278. [Google Scholar] [CrossRef]
- Sivitz, A.B.; Reinders, A.; Ward, J.M. Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiol. 2008, 147, 92–100. [Google Scholar] [CrossRef]
- Leggewie, G.; Kolbe, A.; Lemoine, R.; Roessner, U.; Lytovchenko, A.; Zuther, E.; Kehr, J.; Frommer, W.B.; Riesmeier, J.W.; Willmitzer, L.; et al. Overexpression of the sucrose transporter SoSUT1 in potato results in alterations in leaf carbon partitioning and in tuber metabolism but has little impact on tuber morphology. Planta 2003, 217, 158–167. [Google Scholar] [CrossRef]
- Baker, R.F.; Leach, K.A.; Boyer, N.R.; Swyers, M.J.; Benitez-Alfonso, Y.; Skopelitis, T.; Luo, A.; Sylvester, A.; Jackson, D.; Braun, D.M. Sucrose transporter ZmSut1 expression and localization uncover new insights into sucrose phloem loading. Plant Physiol. 2016, 172, 1876–1898. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.M.; Kim, N.; Ahn, B.O.; Oh, M.; Chung, W.H.; Chung, H.; Jeong, S.; Lim, K.B.; Hwang, Y.J.; Kim, G.B.; et al. Elucidating the triplicated ancestral genome structure of radish based on chromosome-level comparison with the Brassica genomes. Theor. Appl. Genet. 2016, 129, 1357–1372. [Google Scholar] [CrossRef] [PubMed]
- Elisabeth, G.; Alexandre, G.; Christine, H.; Ivan, I.; Ron, D.A.; Amos, B. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar]
- Chen, Y.J.; Yu, P.; Luo, J.C.; Jiang, Y. Secreted protein prediction system combining CJ-SPHMM, TMHMM, and PSORT. Mamm. Genome 2003, 14, 859–865. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Machanick, P.; Bailey, T.L. MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 2011, 27, 1696–1697. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, X.; Gong, Y.; Xu, L.; Wang, Y.; Liu, L. Evaluation of reference genes for gene expression studies in radish (Raphanus sativus L.) using quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2012, 424, 398–403. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wu, Z.; Li, T.; Liu, X.Y.; Yuan, G.Z.; Hou, H.Z.; Teng, N.J. A novel R2R3-MYB transcription factor LlMYB305 from Lilium longiflorum plays a positive role in thermos tolerance via activating heat-protective genes. Environ. Exp. Bot. 2021, 184, 104399. [Google Scholar] [CrossRef]
- Zhang, W.T.; Li, J.X.; Dong, J.H.; Wang, Y.; Xu, L.; Li, K.X.; Yi, X.F.; Zhu, Y.L.; Liu, L.W. RsSOS1 responding to salt stress might be involved in regulating salt tolerance by maintaining Na+ homeostasis in radish (Raphanus sativus L.). Horticulturae 2021, 7, 458. [Google Scholar] [CrossRef]
- Yu, C.L.; Sun, C.D.; Shen, C.J.; Wang, S.K.; Liu, F.; Liu, Y.; Chen, Y.L.; Li, C.Y.; Qian, Q.; Aryal, B.; et al. The auxin transporter, OsAUX1, is involved in primary root and root hair elongation and in Cd stress responses in rice (Oryza sativa L.). Plant J. 2015, 83, 818–830. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 2010, 16, 735–743. [Google Scholar] [CrossRef]
- Li, W.; Sun, K.; Ren, Z.Y.; Song, C.X.; Pei, X.Y.; Liu, Y.A.; Wang, Z.Y.; He, K.L.; Zhang, F.; Zhou, X.J.; et al. Molecular evolution and stress and phytohormone responsiveness of SUT genes in Gossypium hirsutum. Front. Genet. 2018, 9, 494. [Google Scholar] [CrossRef]
- Vanbel, A.; Gamalei, Y.V. Ecophysiology of phloem loading in source leaves. Plant Cell Environ. 2010, 15, 265–270. [Google Scholar]
- Sun, L.; Deng, R.; Liu, J.; Lai, M.; Wu, J.; Liu, X.; Shahid, M.Q. An overview of sucrose transporter (SUT) genes family in rice. Mol. Biol. Rep. 2022, 49, 5685–5695. [Google Scholar] [CrossRef]
- Deol, K.K.; Mukherjee, S.; Gao, F.; Brûlé-Babel, A.; Stasolla, C.; Ayele, B.T. Identification and characterization of the three homeologues of a new sucrose transporter in hexaploid wheat (Triticum aestivum L.). BMC Plant Biol. 2013, 13, 181. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Wei, Q.; Wan, H.; Sun, C. Phylogenetic relationships of sucrose transporters (SUTs) in plants and genome-wide characterization of SUT genes in Orchidaceae reveal roles in floral organ development. Peer. J. 2021, 9, e11961. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Julius, B.T.; Leach, K.A.; Tran, T.M.; Mertz, R.A.; Braun, D.M. Sugar transporters in plants: New insights and discoveries. Plant Cell Physiol. 2017, 58, 1442–1460. [Google Scholar] [CrossRef]
- Chincinska, I.A.; Liesche, J.; Krügel, U.; Michalska, J.; Geigenberger, P.; Grimm, B.; Kühn, C. Sucrose transporter StSUT4 from potato affects flowering, tuberization, and shade avoidance response. Plant Physiol. 2008, 146, 515–528. [Google Scholar] [CrossRef]
- Kühn, C.; Hajirezaei, M.R.; Fernie, A.R.; Roessner-Tunali, U.; Czechowski, T.; Hirner, B.; Frommer, W.B. The sucrose transporter StSUT1 localizes to sieve elements in potato tuber phloem and influences tuber physiology and development. Plant Physiol. 2003, 131, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Hoang, N.V.; Park, C.; Kamran, M.; Lee, J.Y. Gene regulatory network guided investigations and engineering of storage root development in root crops. Front. Plant Sci. 2020, 17, 762. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.F.; Qi, X.X.; Huang, X.S.; Xu, L.L.; Jin, C.; Wu, J.; Zhang, S.L. Overexpression of sucrose transporter gene PbSUT2 from Pyrus bretschneideri, enhances sucrose content in Solanum lycopersicum fruit. Plant Physiol. Biochem. 2016, 105, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.J.; Sun, M.H.; Liu, Y.J.; Hu, D.G.; Hao, Y.J. Molecular cloning and functional characterization of the apple sucrose transporter gene MdSUT2. Plant Physiol. Bioch. 2016, 109, 442–451. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; Zhang, X.; Cao, Y.; Xin, R.; Ma, Y.; Wang, L.; Xu, L.; Wang, Y.; Liu, R.; Liu, L. Genome-Wide Identification of Sucrose Transporter Genes and Functional Analysis of RsSUC1b in Radish (Raphanus sativus L.). Horticulturae 2022, 8, 1058. https://doi.org/10.3390/horticulturae8111058
Zhu X, Zhang X, Cao Y, Xin R, Ma Y, Wang L, Xu L, Wang Y, Liu R, Liu L. Genome-Wide Identification of Sucrose Transporter Genes and Functional Analysis of RsSUC1b in Radish (Raphanus sativus L.). Horticulturae. 2022; 8(11):1058. https://doi.org/10.3390/horticulturae8111058
Chicago/Turabian StyleZhu, Xiaofeng, Xiaoli Zhang, Yang Cao, Ruixian Xin, Yinbo Ma, Lun Wang, Liang Xu, Yan Wang, Rui Liu, and Liwang Liu. 2022. "Genome-Wide Identification of Sucrose Transporter Genes and Functional Analysis of RsSUC1b in Radish (Raphanus sativus L.)" Horticulturae 8, no. 11: 1058. https://doi.org/10.3390/horticulturae8111058
APA StyleZhu, X., Zhang, X., Cao, Y., Xin, R., Ma, Y., Wang, L., Xu, L., Wang, Y., Liu, R., & Liu, L. (2022). Genome-Wide Identification of Sucrose Transporter Genes and Functional Analysis of RsSUC1b in Radish (Raphanus sativus L.). Horticulturae, 8(11), 1058. https://doi.org/10.3390/horticulturae8111058







