Genome-Wide Identification, Evolution, and Expression Analysis of the TCP Gene Family in Rose (Rosa chinensis Jacq.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of RcTCPs Genes in R. chinensis
2.2. Chromosome Distribution and Synteny Analysis of RcTCP Genes
2.3. Physicochemical Properties and Subcellular Localization of RcTCP Genes
2.4. Phylogenetic Analysis
2.5. Motif, Domain and Gene Structure Analysis
2.6. RcTCP Cis-Regulatory Element (CRE) Analysis
2.7. Collinearity Analysis
2.8. Expression Patterns of RcTCP Genes
2.9. Plant Material and PEG Treatments
2.10. RNA Isolation and Real-Time Quantitative PCR Analysis
3. Results
3.1. Identification and Characteristics of RcTCP Genes
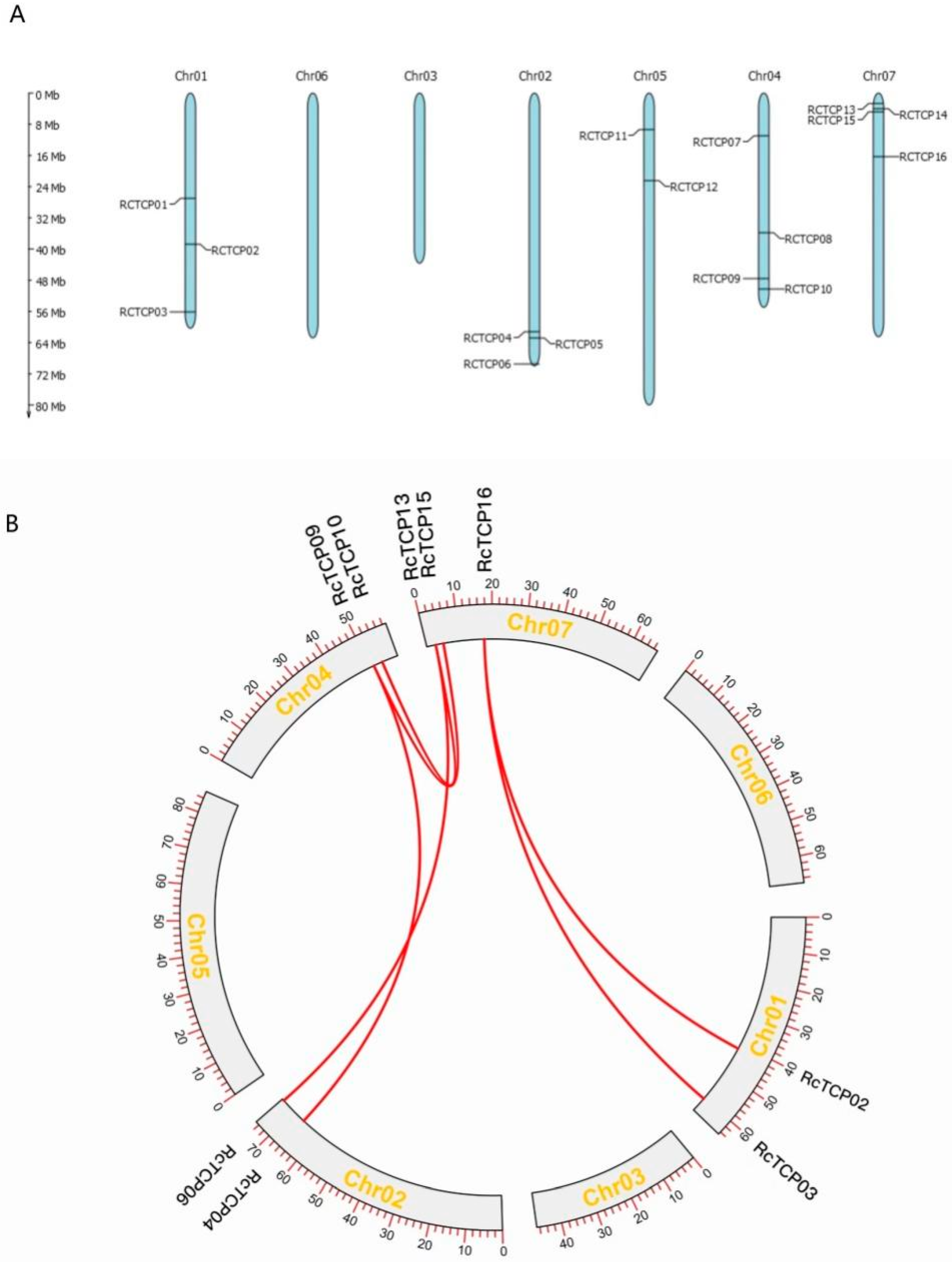
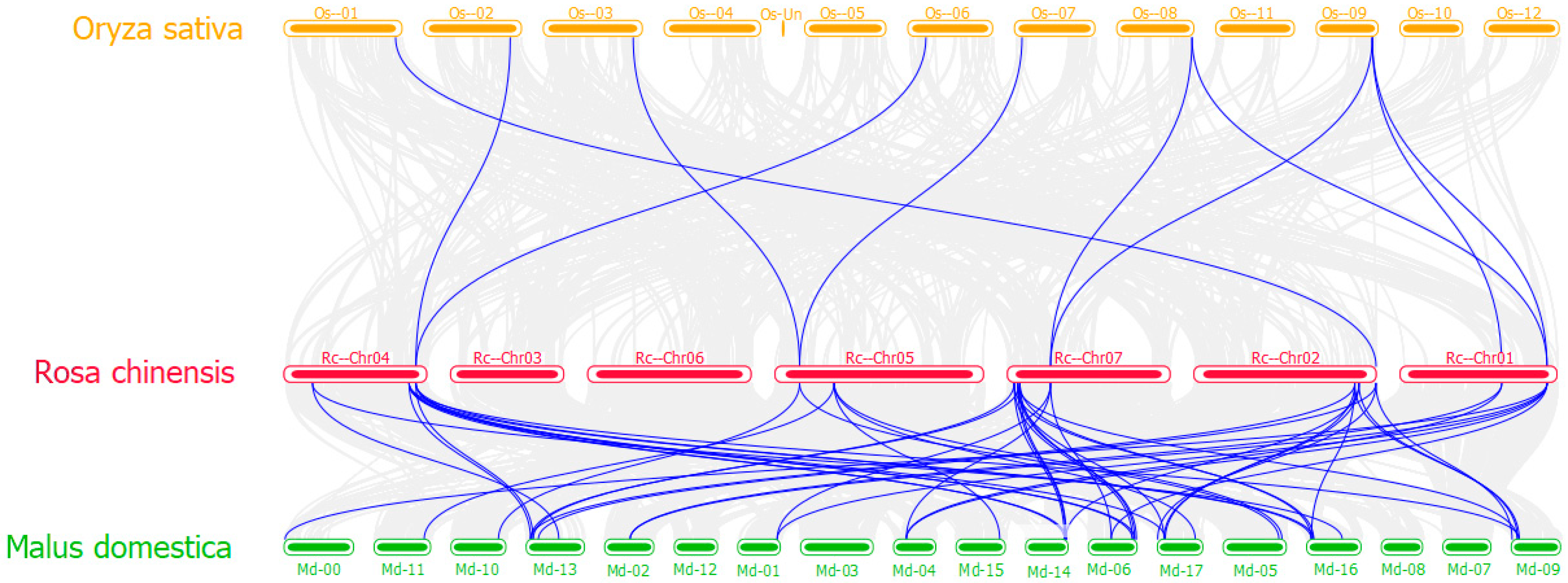
3.2. Chromosome Distribution and Synteny Analysis of RcTCP Genes
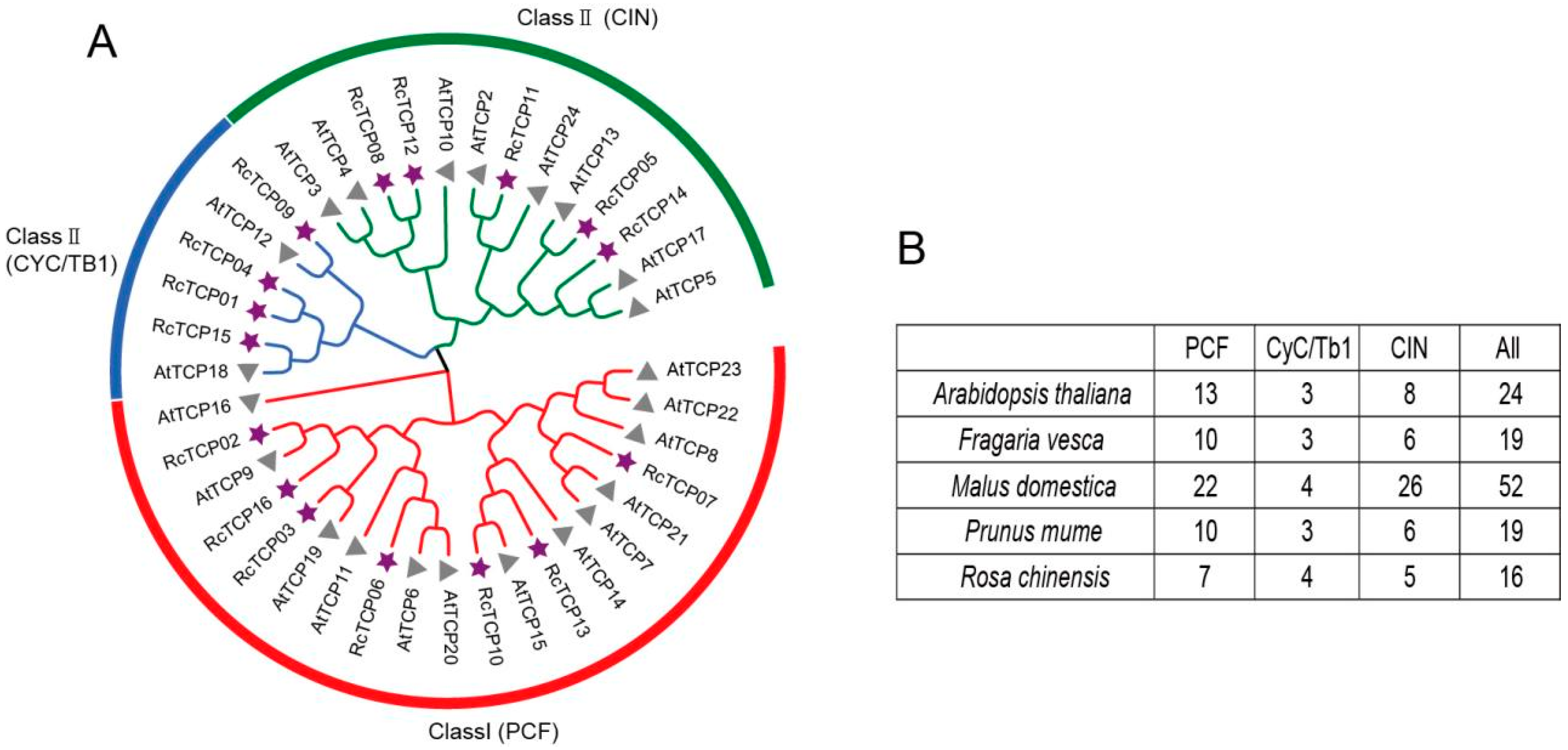
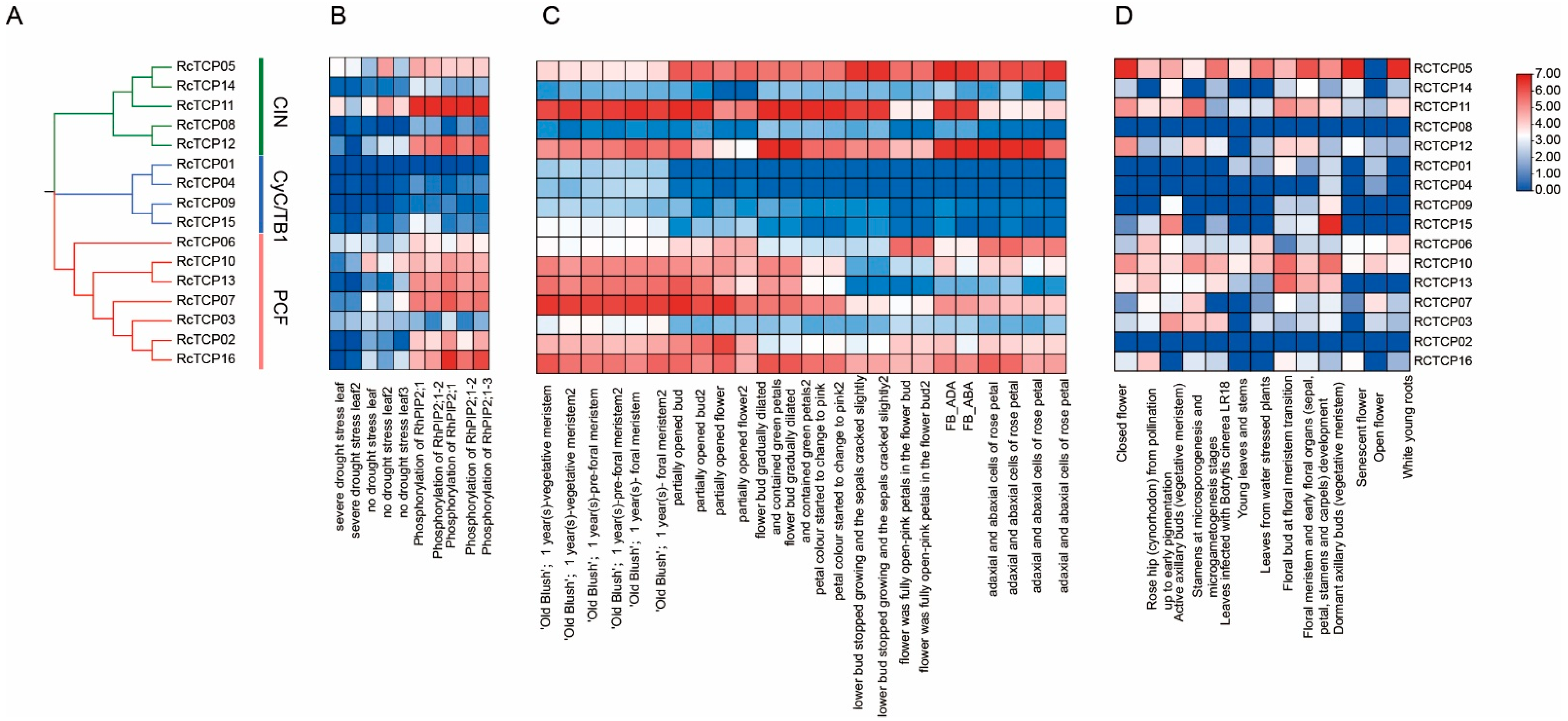
3.3. Phylogenetic Analysis of RcTCP Proteins
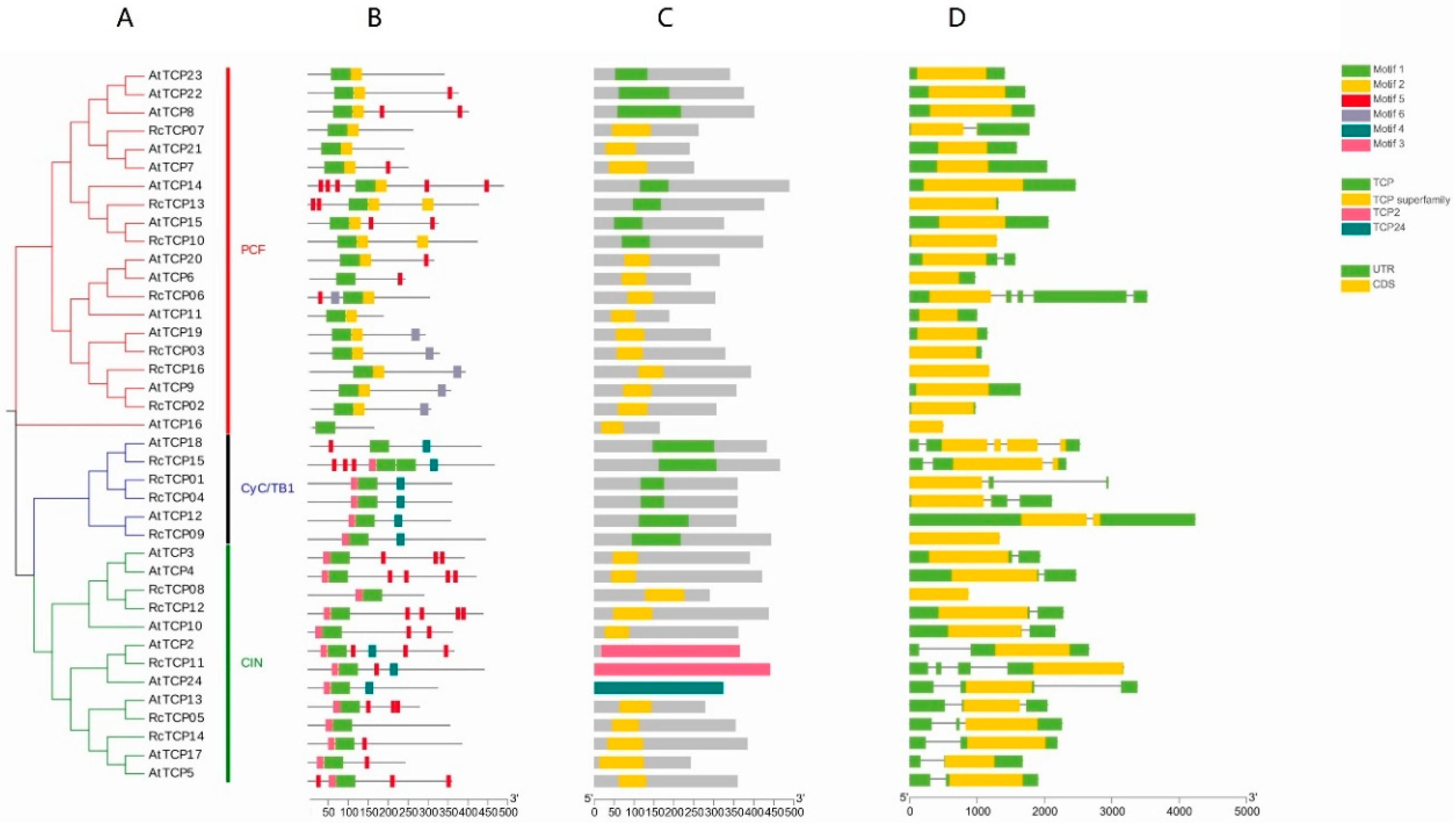
3.4. Conservative Motifs, Structural Domains, Gene Structure Analysis
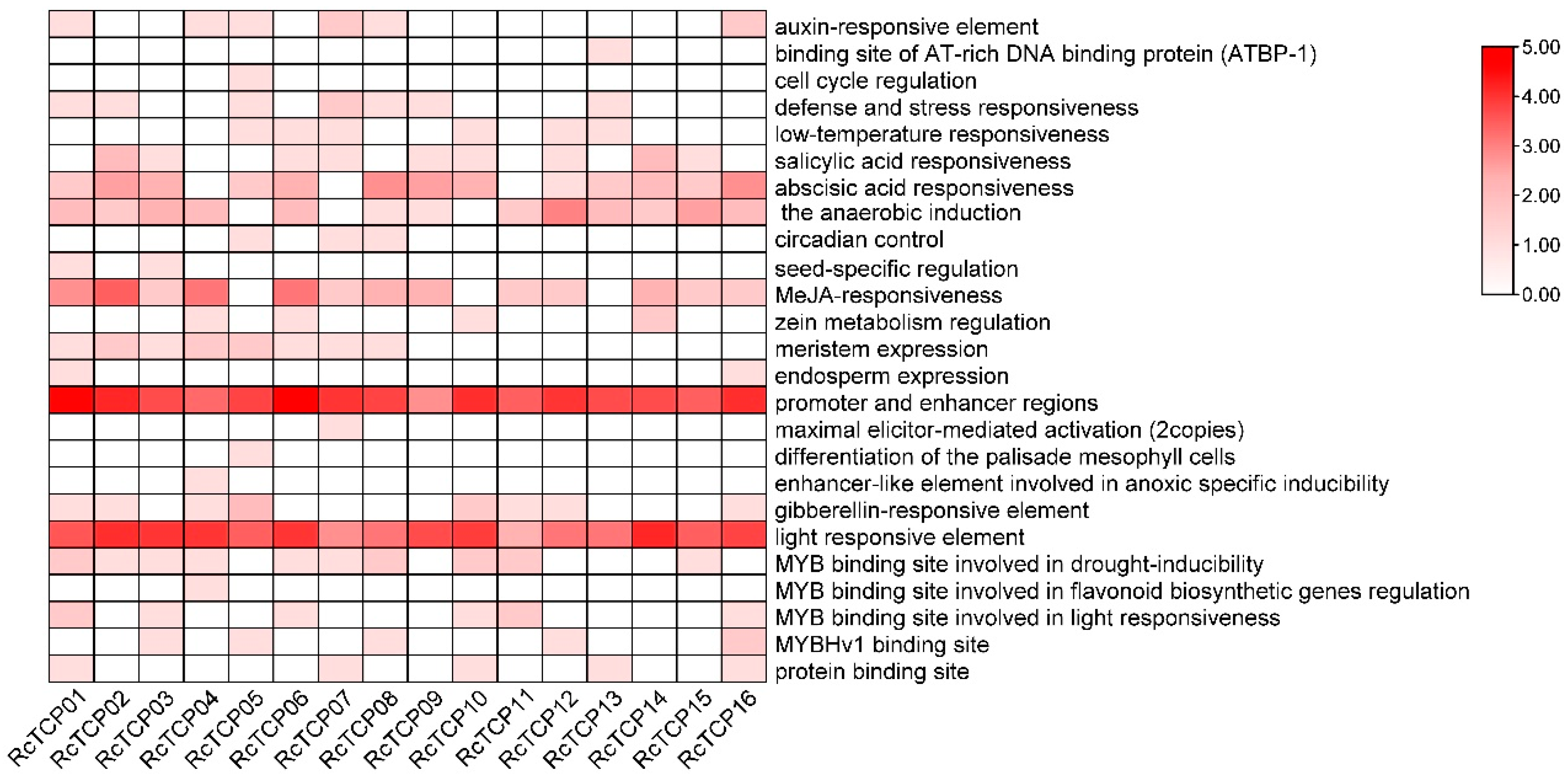
3.5. Promoter Cis-Regulatory Elements Analysis of RcTCP Genes
3.6. Collinearity Analysis of RcTCPs
3.7. Expression Patterns of RcTCP Genes
3.8. Expression Pattern of RcTCP Genes under PEG Treatments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dhaka, N.; Bhardwaj, V.; Sharma, M.K.; Sharma, R. Evolving Tale of TCPs: New Paradigms and Old Lacunae. Front. Plant Sci. 2017, 8, 479. [Google Scholar] [CrossRef]
- Cubas, P.; Lauter, N.; Doebley, J.; Coen, E. The TCP domain: A Motif Found in Proteins Regulating Plant Growth and Development. Plant J. 1999, 18, 215–222. [Google Scholar] [CrossRef]
- Shutian, L. The Arabidopsis thaliana TCP Transcription Factors: A Broadening Horizon Beyond Development. Plant Signal. Behav. 2015, 10, e1044192. [Google Scholar]
- Manassero, N.G.; Viola, I.L.; Welchen, E.; Gonzalez, D.H. TCP Transcription Factors: Architectures of Plant Form. Biomol. Concepts. 2013, 4, 111–127. [Google Scholar] [CrossRef]
- Zhao, Y.; Su, X.; Wang, X.; Wang, M.; Chi, X.; Aamir Manzoor, M.; Li, G.; Cai, Y. Comparative Genomic Analysis of TCP Genes in Six Rosaceae Species and Expression Pattern Analysis in Pyrus bretschneideri. Front. Genet. 2021, 12, 669959. [Google Scholar] [CrossRef]
- Liu, M.M.; Wang, M.M.; Yang, J.; Wen, J.; Guo, P.C.; Wu, Y.W.; Ke, Y.Z.; Li, P.F.; Li, J.N.; Du, H. Evolutionary and Comparative Expression Analyses of TCP Transcription Factor Gene Family in Land Plants. Int. J. Mol. Sci. 2019, 20, 3591. [Google Scholar] [CrossRef]
- Citerne, H.L.; Luo, D.; Pennington, R.T.; Coen, E.; Cronk, Q.C. A phylogenomic investigation of CYCLOIDEA-like TCP genes in the Leguminosae. Plant Physiol. 2003, 131, 1042–1053. [Google Scholar] [CrossRef] [PubMed]
- Han, L. Genome-wide identification and characterization of TCP transcription factors and study on the role of EIN3 in upland cotton (Gossypium hirsutum). Sci. Rep. 2017, 7, 10118. [Google Scholar]
- Parapunova, V.; Busscher, M.; Busscher-Lange, J.; Lammers, M.; Karlova, R.; Bovy, A.G.; Angenent, G.C.; de Maagd, R.A. Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 2014, 14, 157. [Google Scholar] [CrossRef] [PubMed]
- Jiu, S.; Xu, Y.; Wang, J.; Wang, L.; Wang, S.; Ma, C.; Guan, L.; Abdullah, M.; Zhao, M.; Xu, W. Genome-Wide Identification, Characterization, and Transcript Analysis of the TCP Transcription Factors in Vitis vinifera. Front. Genet. 2019, 10, 1276. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, Z.; Zhao, K.; Yang, W.; Cheng, T.; Wang, J.; Zhang, Q. Genome-Wide Identification, Characterization and Expression Analysis of the TCP Gene Family in Prunus mume. Front. Plant Sci. 2016, 7, 1301. [Google Scholar] [CrossRef]
- Bendahmane, M.; Dubois, A.; Raymond, O.; Bris, M.L. Genetics and Genomics of Flower Initiation and Development in Roses. J. Exp. Bot. 2013, 64, 847–857. [Google Scholar] [CrossRef]
- Smulders, M.J.M.; Arens, P.; Bourke, P.M.; Debener, T.; Linde, M.; Riek, J.; Leus, L.; Ruttink, T.; Baudino, S.; Hibrant Saint-Oyant, L.; et al. In the Name of the Rose: A Roadmap for Rose Research in the Genome Era. Hortic. Res. 2019, 6, 65. [Google Scholar] [CrossRef]
- Liu, J.; Wu, S.; Sun, J.J.; Sun, J.R.; Wang, H.; Cao, X.; Lu, J.; Jalal, A.; Wang, C. Genome-Wide Analysis Reveals Widespread Roles for Rcrem Genes in Floral Organ Development in Rosa Chinensis. Genomics 2021, 113, 3881–3894. [Google Scholar] [CrossRef]
- Annick, D.; Sebastien, C.; Olivier, R.; Benjamin, P.; Ludovic, C.; Aymeric, R.; Jean-Paul, O.; Soulaiman, S.; Rossitza, A.; Sylvie, B. Transcriptome Database Resource and Gene Expression Atlas for the Rose. BMC Genom. 2012, 13, 638. [Google Scholar]
- Zhang, S.; Feng, M.; Chen, W.; Zhou, X.; Lu, J.; Wang, Y.; Li, Y.; Jiang, C.Z.; Gan, S.S.; Ma, N.; et al. In rose, transcription factor PTM balances growth and drought survival via PIP2;1 aquaporin. Nat. Plants 2019, 5, 290–299. [Google Scholar] [CrossRef]
- Li, W.; Fu, L.; Geng, Z.; Zhao, X.; Liu, Q.; Jiang, X. Physiological Characteristic Changes and Full-Length Transcriptome of Rose (Rosa chinensis) Roots and Leaves in Response to Drought Stress. Plant Cell Physiol. 2021, 61, 2153–2166. [Google Scholar] [CrossRef]
- Han, Y.; Yong, X.; Yu, J.; Cheng, T.; Wang, J.; Yang, W.; Pan, H.; Zhang, Q. Identification of Candidate Adaxial-Abaxial-Related Genes Regulating Petal Expansion During Flower Opening in Rosa chinensis “Old Blush”. Front. Plant Sci. 2019, 10, 1098. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, C.; Li, X.; Xu, H.; Liang, Y.; Ma, N.; Fei, Z.; Gao, J.; Jiang, C.Z.; Ma, C. Transcriptome Profiling of Petal Abscission Zone and Functional Analysis of an Aux/IAA Family Gene RhIAA16 Involved in Petal Shedding in Rose. Front. Plant Sci. 2016, 7, 1375. [Google Scholar] [CrossRef]
- Han, Y.; Wan, H.; Cheng, T.; Wang, J.; Yang, W.; Pan, H.; Zhang, Q. Comparative RNA-seq Analysis of Transcriptome Dynamics During Petal Development in Rosa chinensis. Sci. Rep. 2017, 7, 43382. [Google Scholar] [CrossRef]
- Guo, X.; Yu, C.; Luo, L.; Wan, H.; Zhen, N.; Xu, T.; Tan, J.; Pan, H.; Zhang, Q. Transcriptome of the floral transition in Rosa chinensis ‘Old Blush’. BMC Genom. 2017, 18, 199. [Google Scholar] [CrossRef]
- Liu, J.; Fu, X.; Dong, Y.; Lu, J.; Ren, M.; Zhou, N.; Wang, C. MIKC-Type Mads-Box Genes in Rosa chinensis: The Remarkable Expansion of ABCDE Model Genes and Their Roles in Floral Organogenesis. Hortic. Res. 2018, 5, 25. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Z.; Ma, N.; Gao, J. Regulation of the Rose Rh-PIP2;1 Promoter by Hormones and Abiotic Stresses in Arabidopsis. Plant Cell Rep. 2009, 28, 185–196. [Google Scholar] [CrossRef]
- Ma, N.; Xue, J.; Li, Y.; Liu, X.; Dai, F.; Jia, W.; Luo, Y.; Gao, J. Rh-PIP2;1, a Rose Aquaporin Gene, Is Involved in Ethylene-Regulated Petal Expansion. Plant Physiol. 2008, 148, 894–907. [Google Scholar] [CrossRef]
- Lehti-Shiu, M.D.; Panchy, N.; Wang, P.; Uygun, S.; Shiu, S.-H. Diversity, Expansion, and Evolutionary Novelty of Plant DNA-binding Transcription Factor Families. BBA Gene Regul. Mech. 2017, 1860, 3–20. [Google Scholar] [CrossRef]
- Danisman, S.; van der Wal, F.; Dhondt, S.; Waites, R.; de Folter, S.; Bimbo, A.; van Dijk, A.D.; Muino, J.M.; Cutri, L.; Dornelas, M.C. Arabidopsis Class I and Class II TCP Transcription Factors Regulate Jasmonic Acid Metabolism and Leaf Development Antagonistically. Plant Physiol. 2012, 159, 1511–1523. [Google Scholar] [CrossRef]
- Yao, X.; Ma, H.; Wang, J.; Zhang, D. Genome-Wide Comparative Analysis and Expression Pattern of TCP Gene Families in Arabidopsis thaliana and Oryza sativa. J. Integr. Plant Biol. 2007, 49, 885–897. [Google Scholar] [CrossRef]
- Xu, R.; Sun, P.; Jia, F.; Lu, L.; Li, Y.; Zhang, S.; Huang, J. Genomewide Analysis of TCP Transcription Factor Gene Family in Malus domestica. J. Genet. 2014, 93, 733–746. [Google Scholar] [CrossRef]
- Leng, X.; Wei, H.; Xu, X.; Ghuge, S.A.; Jia, D.; Liu, G.; Wang, Y.; Yuan, Y. Genome-wide Identification and Transcript Analysis of TCP Transcription Factors in Grapevine. BMC Genom. 2019, 20, 786. [Google Scholar] [CrossRef]
- Ma, X.; Ma, J.; Fan, D.; Li, C.; Jiang, Y.; Luo, K. Genome-wide Identification of TCP Family Transcription Factors from Populus euphratica and Their Involvement in Leaf Shape Regulation. Sci. Rep. 2016, 6, 32795. [Google Scholar] [CrossRef]
- Zhou, J.; Li, J.; Li, X.; Xiao, D.; Li, M. Genome-Wide Identification, Evolution and Expression Analysis of TCP Gene Family in Celery (Apium graveolens L.). J. Sichuan Agric. Univ. 2022, 40, 145–155. [Google Scholar]
- Rath, M.; Challa, K.R.; Sarvepalli, K.; Nath, U. CINCINNATA-Like TCP Transcription Factors in Cell Growth—An Expanding Portfolio. Front. Plant Sci. 2022, 13, 825341. [Google Scholar] [CrossRef]
- Huang, T.; Irish, V.F. Temporal Control of Plant Organ Growth by TCP Transcription Factors. Curr. Biol. 2015, 25, 1765–1770. [Google Scholar] [CrossRef]
- Zhang, Z.; Lu, S.; Ma, Z.; Zhou, Q.; Hw, H.; Chen, B.; Mao, J. Bioinformatics Identifcation and Expression Analysis of TCP Transcription Factor Family in Strawberry. Acta Bot. Boreal. 2020, 40, 2031–2043. [Google Scholar]
- Han, J.; Cao, X.; Liu, H.; Fan, G. Analysis Genes in the Paulownia fortunei TCP Family and Their Response to Witches’ Broom and Drought Stress. J. For. Environ. 2022, 42, 337–345. [Google Scholar]

| Protein Name | Gene ID | Molecular Mass (Da) | Isoelectric Point | Amino Acid (aa) | Average Hydrophilic Coefficient | Aliphatic Index | Instability Index | Subcellular Localization Prediction |
|---|---|---|---|---|---|---|---|---|
| RcTCP01 | RC1G0213700 | 39,668.36 | 8.40 | 360 | −0.510 | 70.19 | 50.85 | Nucleus. |
| RcTCP02 | RC1G0312100 | 32,831.23 | 8.57 | 307 | −0.391 | 66.81 | 60.89 | Nucleus. |
| RcTCP03 | RC1G0531500 | 34,056.20 | 6.60 | 328 | −0.226 | 76.13 | 47.93 | Nucleus. |
| RcTCP04 | RC2G0577900 | 39,859.44 | 9.17 | 360 | −0.632 | 64.78 | 53.90 | Nucleus. |
| RcTCP05 | RC2G0599700 | 39,158.12 | 6.62 | 355 | −0.800 | 63.46 | 55.30 | Nucleus. |
| RcTCP06 | RC2G0685400 | 32,176.69 | 9.27 | 304 | −0.648 | 65.89 | 57.49 | Nucleus. |
| RcTCP07 | RC4G0096800 | 27,591.84 | 8.82 | 262 | −0.445 | 63.82 | 41.51 | Nucleus. |
| RcTCP08 | RC4G0266300 | 32,732.35 | 9.08 | 290 | −0.064 | 80.41 | 44.72 | Nucleus. |
| RcTCP09 | RC4G0400400 | 49,609.69 | 9.45 | 444 | −0.837 | 58.90 | 48.24 | Nucleus. |
| RcTCP10 | RC4G0435200 | 44,618.17 | 6.57 | 423 | −0.598 | 60.28 | 56.15 | Nucleus. |
| RcTCP11 | RC5G0134300 | 47,923.63 | 8.42 | 441 | −0.826 | 57.82 | 49.47 | Nucleus. |
| RcTCP12 | RC5G0279600 | 48,250.47 | 6.68 | 438 | −0.807 | 57.56 | 54.65 | Nucleus. |
| RcTCP13 | RC7G0042700 | 45,529.80 | 6.81 | 426 | −0.734 | 54.39 | 64.76 | Nucleus. |
| RcTCP14 | RC7G0061200 | 42,654.47 | 8.93 | 384 | −0.685 | 65.76 | 51.74 | Nucleus. |
| RcTCP15 | RC7G0073800 | 52,440.83 | 7.04 | 466 | −0.869 | 59.27 | 52.95 | Nucleus. |
| RcTCP16 | RC7G0209000 | 40,894.44 | 6.40 | 393 | −0.474 | 60.38 | 62.10 | Nucleus. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, Y.; Fan, C.; Sun, J.; Chang, Y.; Lu, J.; Sun, J.; Wang, C.; Liu, J. Genome-Wide Identification, Evolution, and Expression Analysis of the TCP Gene Family in Rose (Rosa chinensis Jacq.). Horticulturae 2022, 8, 961. https://doi.org/10.3390/horticulturae8100961
Hou Y, Fan C, Sun J, Chang Y, Lu J, Sun J, Wang C, Liu J. Genome-Wide Identification, Evolution, and Expression Analysis of the TCP Gene Family in Rose (Rosa chinensis Jacq.). Horticulturae. 2022; 8(10):961. https://doi.org/10.3390/horticulturae8100961
Chicago/Turabian StyleHou, Yi, Chunguo Fan, Jingrui Sun, Yufei Chang, Jun Lu, Jingjing Sun, Changquan Wang, and Jinyi Liu. 2022. "Genome-Wide Identification, Evolution, and Expression Analysis of the TCP Gene Family in Rose (Rosa chinensis Jacq.)" Horticulturae 8, no. 10: 961. https://doi.org/10.3390/horticulturae8100961
APA StyleHou, Y., Fan, C., Sun, J., Chang, Y., Lu, J., Sun, J., Wang, C., & Liu, J. (2022). Genome-Wide Identification, Evolution, and Expression Analysis of the TCP Gene Family in Rose (Rosa chinensis Jacq.). Horticulturae, 8(10), 961. https://doi.org/10.3390/horticulturae8100961







