Response and Function of Solanum lycopersicum L. SlSGR2 Gene under Cadmium Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials, Growth Conditions, and Stress Treatment
2.2. Ultrastructural Analysis
2.3. Subcellular Localization
2.4. Total RNA Extraction, cDNA Synthesis, and Isolation of SlSGR2 Gene
2.5. Bioinformatics Analysis
2.6. Expression Pattern Analysis of SlSGR2 Gene under Cadmium Treatment
2.7. Overexpression Vector Construction and A. thanliana Transformation
2.8. Mutant Identification
2.9. Determination of Plant Physiological Indicators
2.10. Statistical Analysis
3. Results
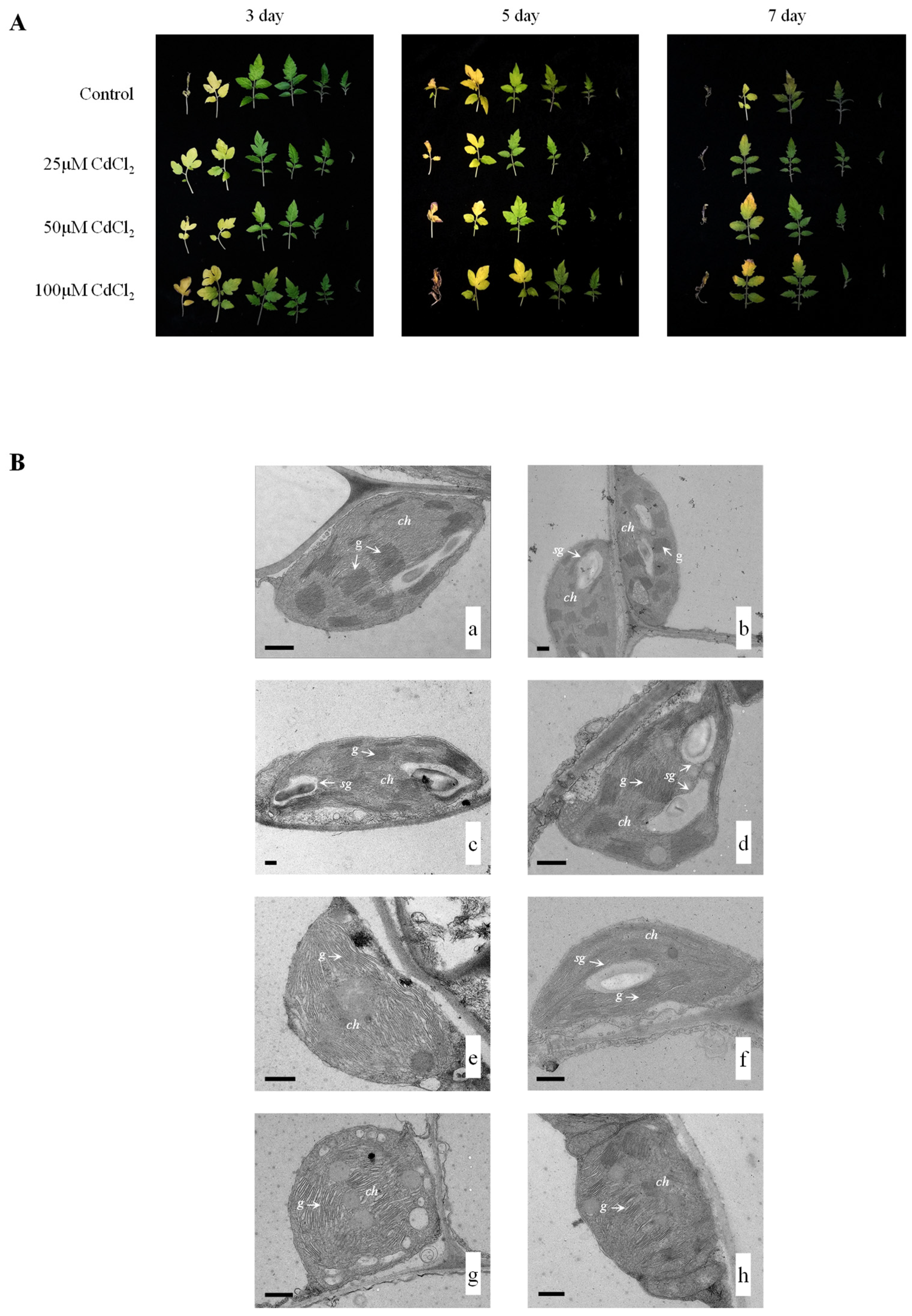
3.1. Phenotype of Tomato Leaves under Cadmium Stress
3.2. Ultrastructure of Tomato Leaves
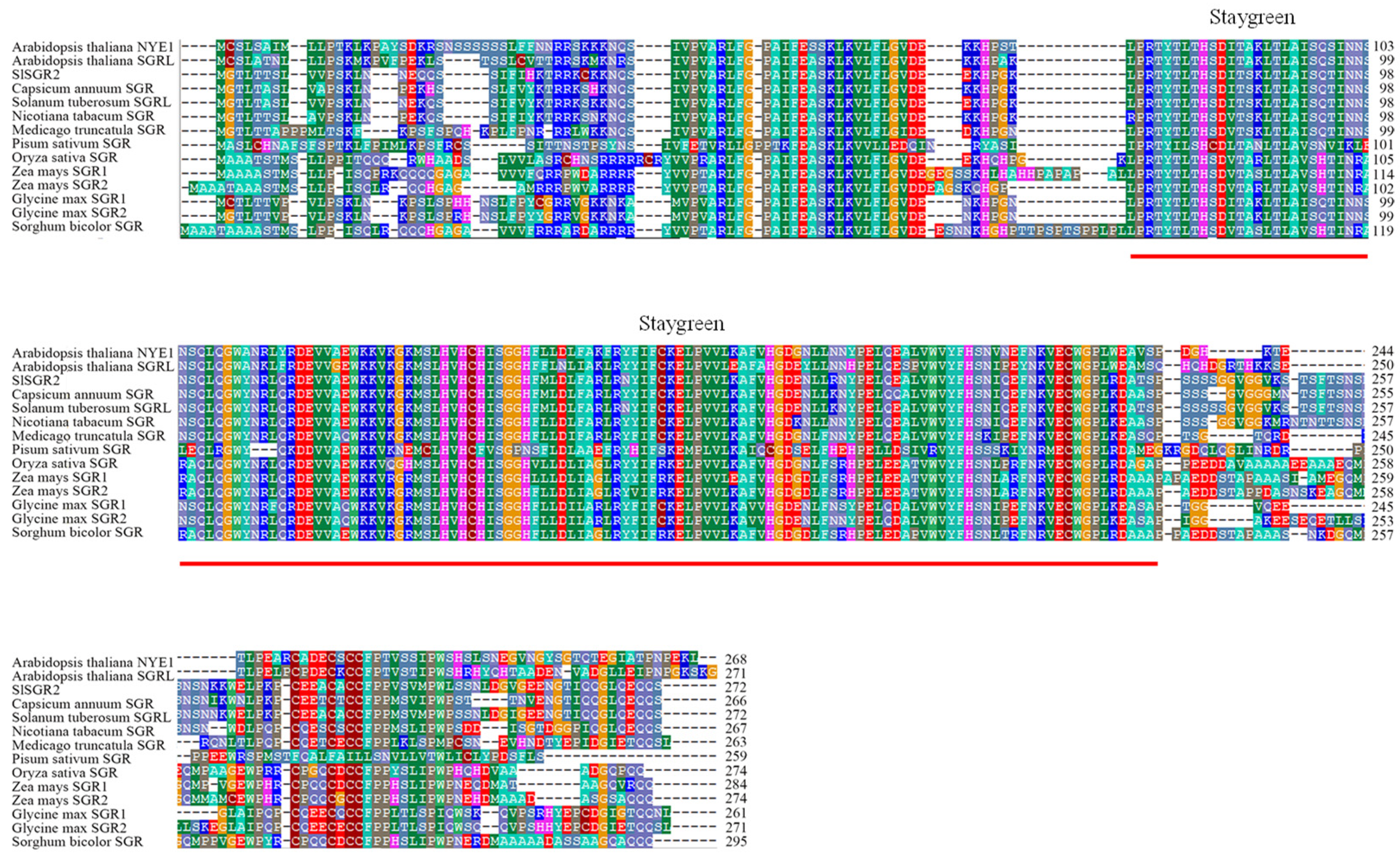
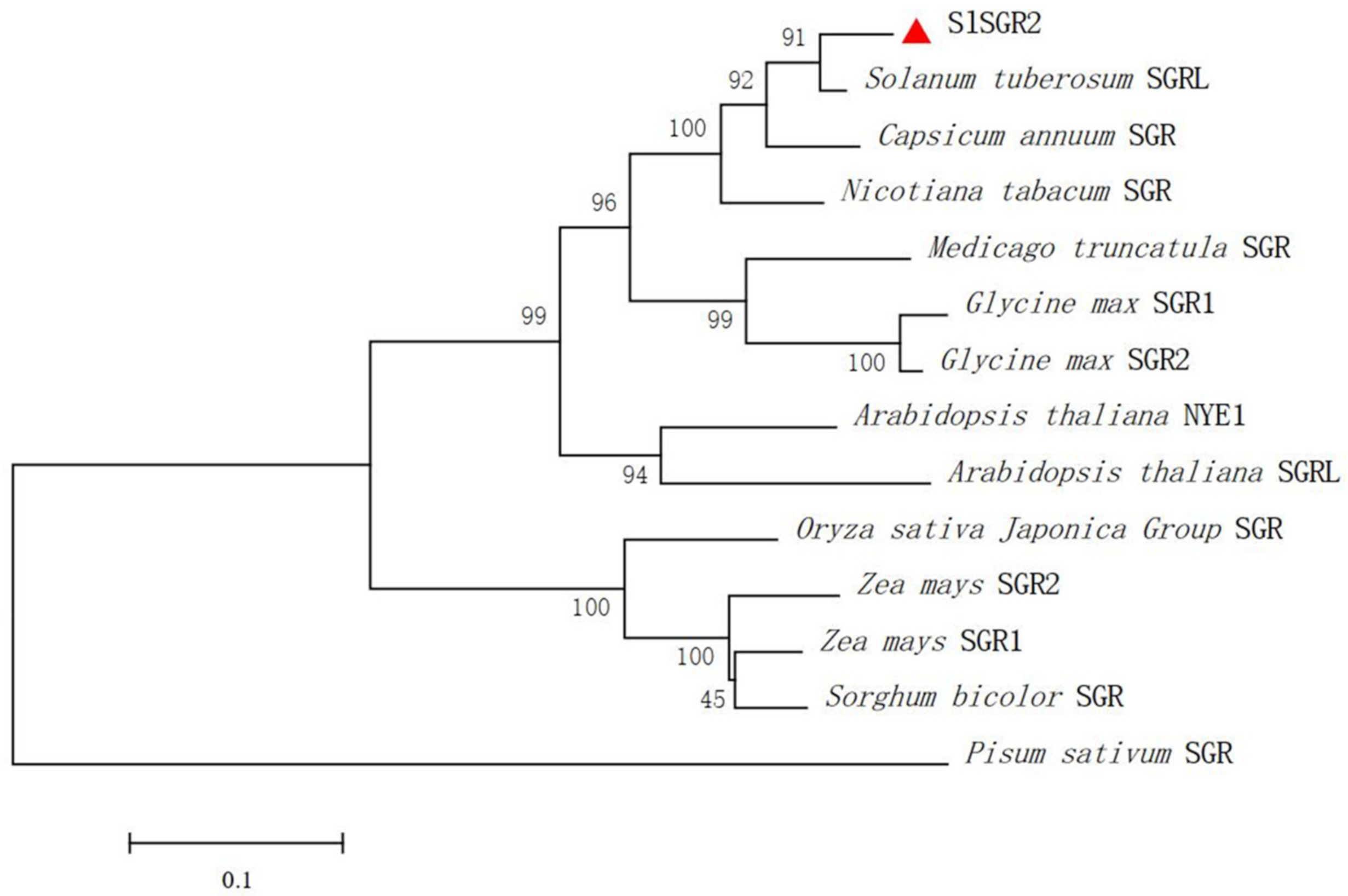
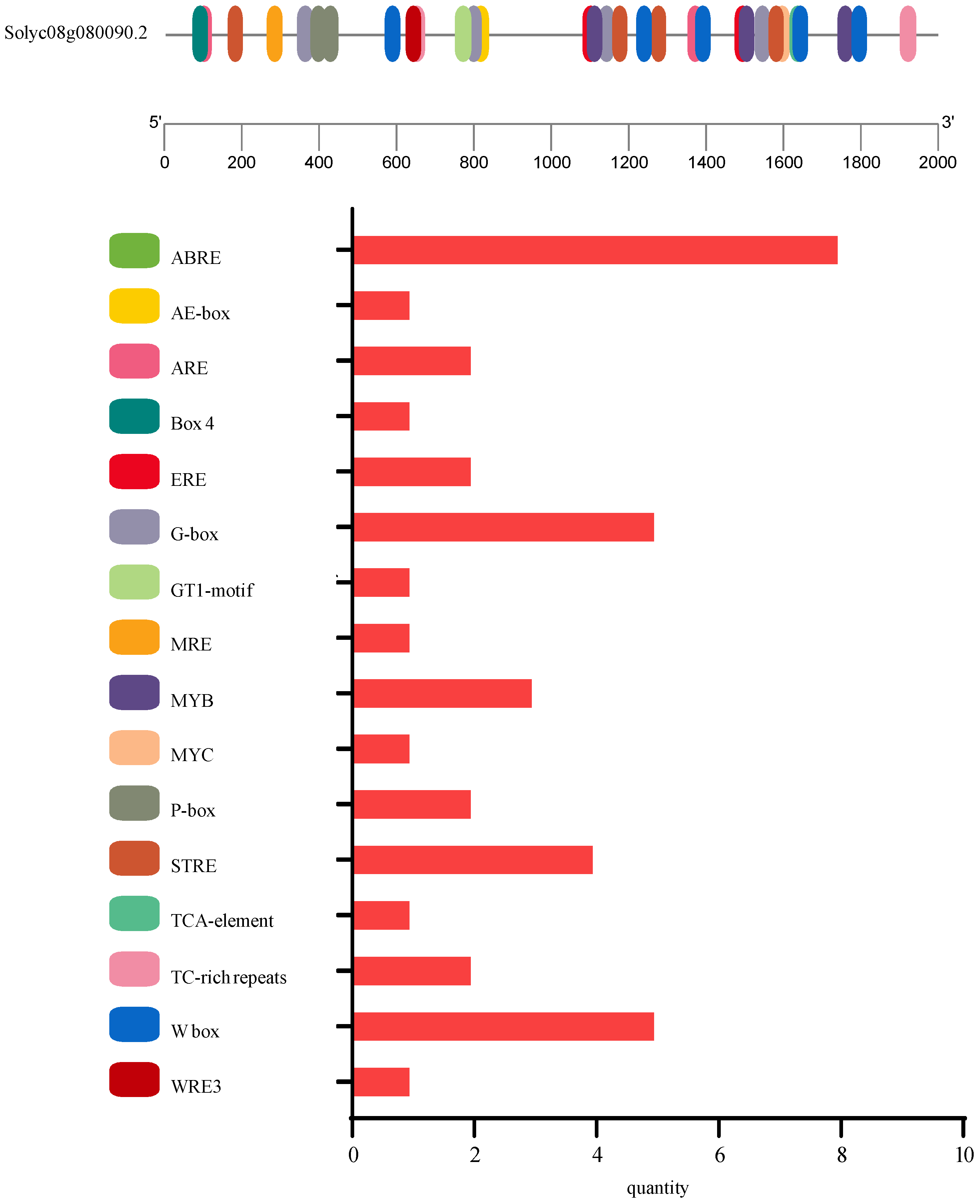
3.3. Analysis of SlSGR2 Sequence
3.4. Subcellular Localization Analysis of SlSGR2
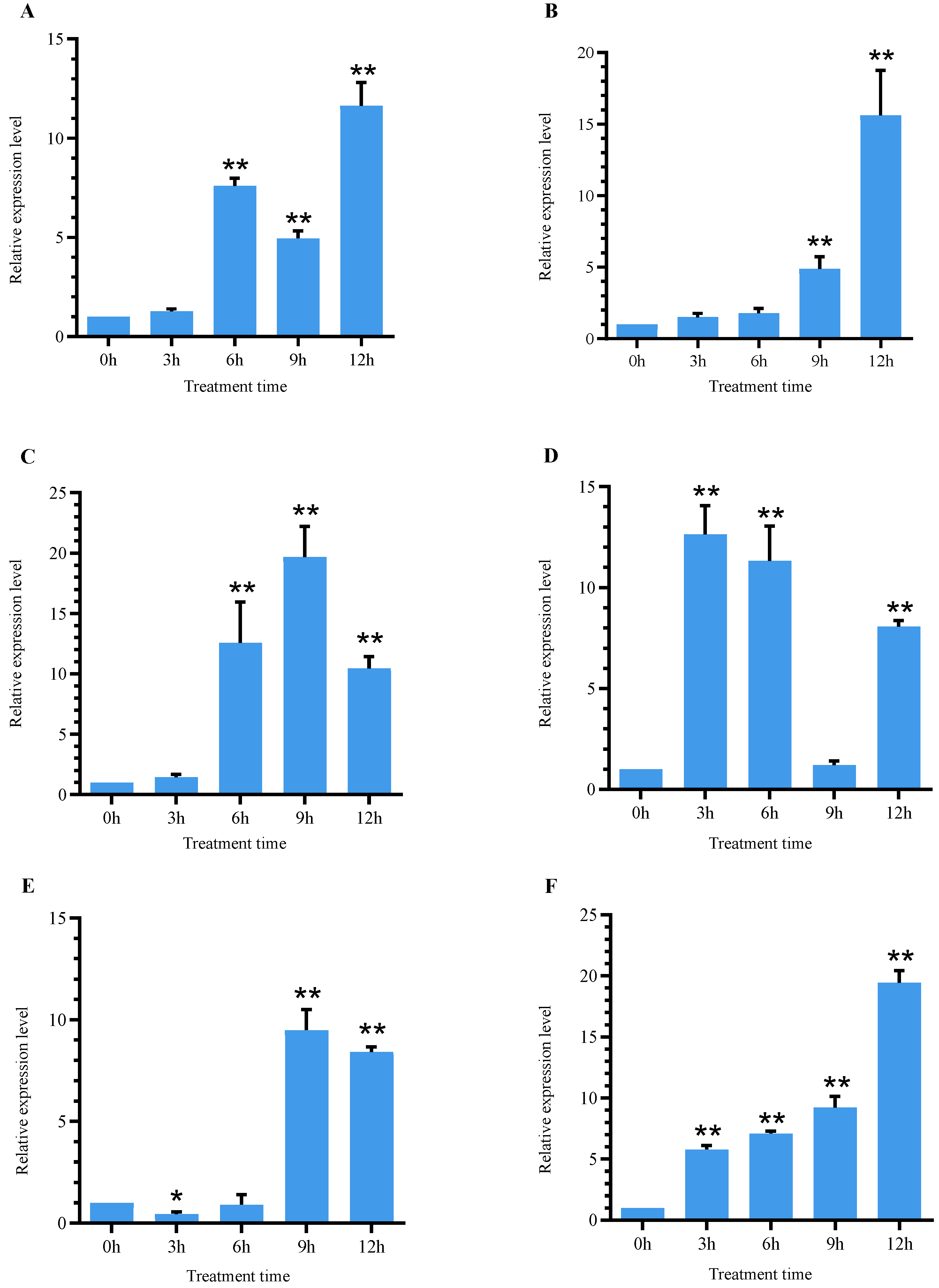
3.5. Expression Analysis of SlSGR2 under Cadmium Stress
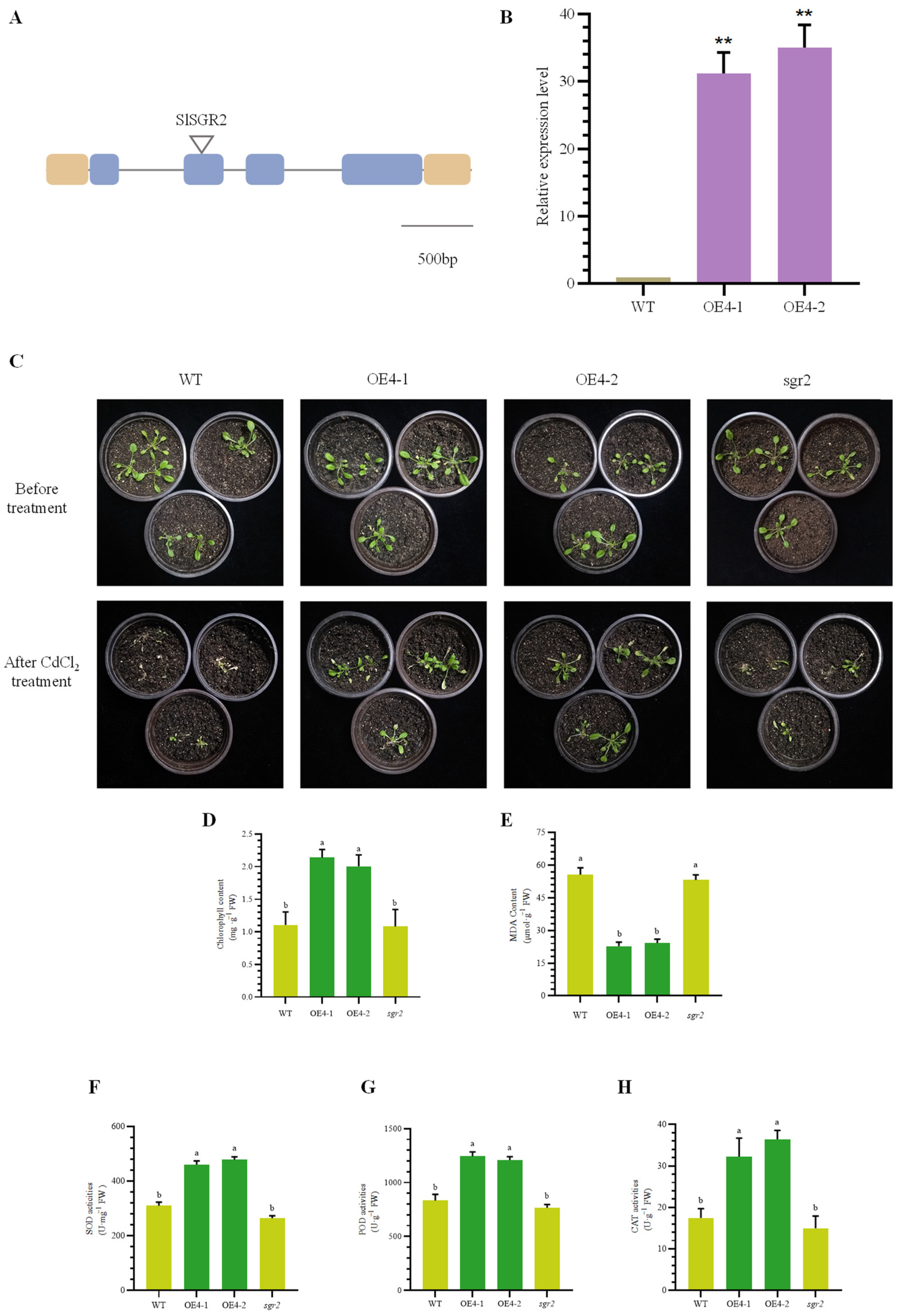
3.6. Identification of Transgenic and Mutant of A. thaliana
3.7. Overexpression of SlSGR2 Negatively Regulates Chlorophyll Degradation
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hechmi, N.; Aissa, N.B.; Abdenaceur, H.; Jedidi, N. Evaluating the phytoremediation potential of Phragmites australis grown in pentachlorophenol and cadmium co-contaminated soils. Environ. Sci. Pollut. Res. Int. 2014, 21, 1304–1313. [Google Scholar] [CrossRef]
- Huang, H.; Yu, N.; Wang, L.; Gupta, D.K.; He, Z.; Wang, K.; Zhu, Z.; Yan, X.; Li, T.; Yang, X.E. The phytoremediation potential of bioenergy crop Ricinus communis for DDTs and cadmium co-contaminated soil. Bioresour. Technol. 2011, 102, 11034–11038. [Google Scholar] [CrossRef] [PubMed]
- Wuana, R.A.; Okieimen, F.E. Heavy Metals in Contaminated Soils: A Review of Sources, Chemistry, Risks and Best Available Strategies for Remediation. ISRN Ecol. 2011, 2011, 402647. [Google Scholar] [CrossRef]
- Yamagata, N.; Shigematsu, I. Cadmium pollution in perspective. Bull. Inst. Public Health 1970, 19, 1–27. [Google Scholar]
- Wang, B.; Lin, C.; Xuan, Z.; Duan, X.; Wu, F. A soil ingestion pilot study for teenage children in China. Chemosphere 2018, 202, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.J.; Ma, Y.; Zhu, Y.G.; Zhong, T.; Mcgrath, S.P. Soil Contamination in China: Current Status and Mitigation Strategies. Environ. Sci. Technol. 2015, 49, 750. [Google Scholar] [CrossRef] [PubMed]
- Huaiman, C. Heavy Metal Pollution in Soil-Plant Systems. Pedosphere 1998, 8, 72–75. [Google Scholar]
- Rizwan, M.; Ali, S.; Abbas, T.; Zia-Ur-Rehman, M.; Hannan, F.; Keller, C.; Al-Wabel, M.I.; Ok, Y.S. Cadmium minimization in wheat: A critical review. Ecotoxicol. Environ. Saf. 2016, 130, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, M.; Ali, S.; Adrees, M.; Ibrahim, M.; Tsang, D.C.W.; Zia-Ur-Rehman, M.; Zahir, Z.A.; Rinklebe, J.R.; Tack, F.M.G.; Ok, Y.S. A critical review on effects, tolerance mechanisms and management of cadmium in vegetables. Chemosphere 2017, 182, 90–105. [Google Scholar] [CrossRef]
- Hallenbeck, W. Human health effects of exposure to cadmium. Birkhäuser 1986, 50, 131–137. [Google Scholar] [CrossRef]
- Hongyan, L.; Weiqin, W.; Aibin, H.; Lixiao, N. Correlation of leaf and root senescence during ripening in dry seeded and transplanted rice. Rice Sci. 2018, 25, 279–285. [Google Scholar] [CrossRef]
- Kobayashi, E.; Suwazono, Y.; Dochi, M.; Honda, R.; Kido, T. Influence of consumption of cadmium-polluted rice or Jinzu River water on occurrence of renal tubular dysfunction and/or Itai-itai disease. Biol. Trace Elem. Res. 2009, 127, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Meharg, A.A.; Norton, G.J.; Deacon, C.; Williams, P.; Islam, M.R. Variation in rice cadmium related to human exposure. Environ. Sci. Technol. 2013, 47, 5613–5618. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Wang, Y.; Mao, W.; Sui, H.; Yong, L.; Yang, D.; Jiang, D.; Zhang, L.; Gong, Y. Dietary cadmium exposure assessment among the Chinese population. PLoS ONE 2017, 12, e0177978. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.A.; Hc, A.; Pmk, B.; Fjz, A. Cadmium contamination in agricultural soils of China and the impact on food safety. Environ. Pollut. 2019, 249, 1038–1048. [Google Scholar]
- EFSA Panel. Cadmium in food—Scientific opinion of the Panel on Contaminants in the Food Chain. EFSA J. 2009, 7, 1–139. [Google Scholar] [CrossRef]
- Clemens, S.; Aarts, M.G.M.; Thomine, S.; Verbruggen, N. Plant science: The key to preventing slow cadmium poisoning. Trends Plant Sci. 2013, 18, 92–99. [Google Scholar] [CrossRef]
- Hongping, C.; Xinping, Y.; Peng, W.; Zixuan, W.; Ming, L.; Fang-Jie, Z. Dietary cadmium intake from rice and vegetables and potential health risk: A case study in Xiangtan, southern China. Sci. Total Environ. 2018, 639, 271–277. [Google Scholar]
- Shi, T.; Zhang, Y.; Gong, Y.; Haiying, J.; Xiao, W. Status of cadmium accumulation in agricultural soils across China (1975–2016): From temporal and spatial variations to risk assessment. Chemosphere 2019, 230, 136–143. [Google Scholar] [CrossRef]
- Ye, K.; Wu, R. Autumn snow cover variability over northern Eurasia and roles of atmospheric circulation. Adv. Atmos. Sci. 2017, 34, 847–858. [Google Scholar] [CrossRef]
- Andresen, E.; Küpper, H. Cadmium toxicity in plants. Met. Ions Life Sci. 2013, 11, 395–413. [Google Scholar] [CrossRef] [PubMed]
- Verbruggen, N.; Hermans, C.; Schat, H. Mechanisms to cope with arsenic or cadmium excess in plants. Curr. Opin. Plant Biol. 2009, 12, 364–372. [Google Scholar] [CrossRef]
- Sharma, P.; Dubey, R.S. Involvement of oxidative stress and role of antioxidative defense system in growing rice seedlings exposed to toxic concentrations of aluminum. Plant Cell Rep. 2007, 26, 2027–2038. [Google Scholar] [CrossRef] [PubMed]
- Lux, A.; Vaculík, M.; Martinka, M.; Lisková, D.; Staden, J.V. Cadmium induces hypodermal periderm formation in the roots of the monocotyledonous medicinal plant Merwilla plumbea. Ann. Bot. 2011, 107, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Wang, Z. Physiological mechanism of hypertolerance of cadmium in Kentucky bluegrass and tall fescue: Chemical forms and tissue distribution. Environ. Exp. Bot. 2013, 96, 35–42. [Google Scholar] [CrossRef]
- Baldantoni, D.; Morra, L.; Zaccardelli, M.; Alfani, A. Cadmium accumulation in leaves of leafy vegetables. Ecotoxicol. Environ. Saf. 2016, 123, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.S.; Dietz, K.J.; Mimura, T. Vacuolar compartmentalization as indispensable component of heavy metal detoxification in plants. Plant Cell Environ. 2016, 39, 1112–1126. [Google Scholar] [CrossRef]
- Carrier, P.; Baryla, A.; Havaux, M. Cadmium distribution and microlocalization in oilseed rape (Brassica napus) after long-term growth on cadmium-contaminated soil. Planta 2003, 216, 939–950. [Google Scholar] [CrossRef]
- Bačkor, M.; Váczi, P.; Barták, M.; Bud’ová, J.; Dzubaj, A. Uptake, photosynthetic characteristics and membrane lipid peroxidation levels in the lichen photobiont Trebouxia erici exposed to copper and cadmium. Bryologist 2007, 110, 100–107. [Google Scholar] [CrossRef]
- Yi, T.H.; Kao, C.H. Cadmium toxicity is reduced by nitric oxide in rice leaves. Plant Growth Regul. 2004, 42, 227–238. [Google Scholar]
- Toppi, L.; Gabbrielli, R. Response to cadmium in higher plants. Environ. Exp. Bot. 1999, 41, 105–130. [Google Scholar] [CrossRef]
- Küpper, H.; Küpper, F.; Spiller, M. Environmental relevance of heavy metal-substituted chlorophylls using the example of water plants. J. Exp. Bot. 1996, 47, 259–266. [Google Scholar] [CrossRef]
- Faller, P.; Kienzler, K.; Krieger-Liszkay, A. Mechanism of Cd2+ toxicity: Cd2+ inhibits photoactivation of Photosystem II by competitive binding to the essential Ca2+ site. Biochim. Biophys. Acta 2005, 1706, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Sigfridsson, K.G.; Bernát, G.; Mamedov, F.; Styring, S. Molecular interference of Cd(2+) with Photosystem II. Biochim. Biophys. Acta 2004, 1659, 19–31. [Google Scholar] [CrossRef]
- Horie, Y.; Ito, H.; Kusaba, M.; Tanaka, R.; Tanaka, A. Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J. Biol. Chem. 2009, 284, 17449–17456. [Google Scholar] [CrossRef]
- Kusaba, M.; Ito, H.; Morita, R.; Iida, S.; Sato, Y.; Fujimoto, M.; Kawasaki, S.; Tanaka, R.; Hirochika, H.; Nishimura, M.; et al. Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 2007, 19, 1362–1375. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Morita, R.; Katsuma, S.; Nishimura, M.; Tanaka, A.; Kusaba, M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2009, 57, 120–131. [Google Scholar] [CrossRef]
- Meguro, M.; Ito, H.; Takabayashi, A.; Tanaka, R.; Tanaka, A. Identification of the 7-hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis. Plant Cell 2011, 23, 3442–3453. [Google Scholar] [CrossRef]
- Morita, R.; Sato, Y.; Masuda, Y.; Nishimura, M.; Kusaba, M. Defect in non-yellow coloring 3, an alpha/beta hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 2009, 59, 940–952. [Google Scholar] [CrossRef]
- Schelbert, S.; Aubry, S.; Burla, B.; Agne, B.; Kessler, F.; Krupinska, K.; Hörtensteiner, S. Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 2009, 21, 767–785. [Google Scholar] [CrossRef]
- Pruzinská, A.; Anders, I.; Aubry, S.; Schenk, N.; Tapernoux-Lüthi, E.; Müller, T.; Kräutler, B.; Hörtensteiner, S. In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 2007, 19, 369–387. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, R.; Hirashima, M.; Satoh, S.; Tanaka, A. The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: Inhibition of the pheophorbide a oxygenase activity does not lead to the “stay-green” phenotype in Arabidopsis. Plant Cell Physiol. 2003, 44, 1266–1274. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Park, S.Y.; Paek, N.C. The Divergent Roles of STAYGREEN (SGR) Homologs in Chlorophyll Degradation. Mol. Cells 2015, 38, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Armstead, I.; Donnison, I.; Aubry, S.; Harper, J.; Hörtensteiner, S.; James, C.; Mani, J.; Moffet, M.; Ougham, H.; Roberts, L.; et al. Cross-species identification of Mendel’s I locus. Science 2007, 315, 73. [Google Scholar] [CrossRef] [PubMed]
- Cha, K.W.; Lee, Y.J.; Koh, H.J.; Lee, B.M.; Nam, Y.W.; Paek, N.C. Isolation, characterization, and mapping of the stay green mutant in rice. Theor. Appl. Genet. 2002, 104, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; An, K.; Liao, Y.; Zhou, X.; Cao, Y.; Zhao, H.; Ge, X.; Kuai, B. Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol. 2007, 144, 1429–1441. [Google Scholar] [CrossRef]
- Jiang, H.; Li, M.; Liang, N.; Yan, H.; Wei, Y.; Xu, X.; Liu, J.; Xu, Z.; Chen, F.; Wu, G. Molecular cloning and function analysis of the stay green gene in rice. Plant J. 2007, 52, 197–209. [Google Scholar] [CrossRef]
- Barry, C.S.; McQuinn, R.P.; Chung, M.Y.; Besuden, A.; Giovannoni, J.J. Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 2008, 147, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Guo, Y.; Kuai, B. Isolation and characterization of a chlorophyll degradation regulatory gene from tall fescue. Plant Cell Rep. 2011, 30, 1201–1207. [Google Scholar] [CrossRef]
- Zhou, C.; Han, L.; Pislariu, C.; Nakashima, J.; Fu, C.; Jiang, Q.; Quan, L.; Blancaflor, E.B.; Tang, Y.; Bouton, J.H.; et al. From model to crop: Functional analysis of a STAY-GREEN gene in the model legume Medicago truncatula and effective use of the gene for alfalfa improvement. Plant Physiol. 2011, 157, 1483–1496. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Li, C.; Li, W.; Wang, Z.; Zhou, Z.; Shen, Y.; Wu, M.; Wu, Y.; Li, G.; Kong, L.A.; et al. Concerted evolution of D1 and D2 to regulate chlorophyll degradation in soybean. Plant J. 2014, 77, 700–712. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Park, S.Y.; Kim, Y.S.; Wang, S.H.; Yoo, S.C.; Hörtensteiner, S.; Paek, N.C. Arabidopsis STAY-GREEN2 is a negative regulator of chlorophyll degradation during leaf senescence. Mol. Plant 2014, 7, 1288–1302. [Google Scholar] [CrossRef] [PubMed]
- Chao, W.S.; Liu, V.; Thomson, W.W.; Platt, K.; Walling, L.L. The Impact of Chlorophyll-Retention Mutations, d1d2 and cyt-G1, during Embryogeny in Soybean. Plant Physiol. 1995, 107, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Guiamét, J.J.; Schwartz, E.; Pichersky, E.; Noodén, L.D. Characterization of Cytoplasmic and Nuclear Mutations Affecting Chlorophyll and Chlorophyll-Binding Proteins during Senescence in Soybean. Plant Physiol. 1991, 96, 227–231. [Google Scholar] [CrossRef]
- Clerkx, E.J.; Vries, H.B.; Ruys, G.J.; Groot, S.P.; Koornneef, M. Characterization of green seed, an enhancer of abi3-1 in Arabidopsis that affects seed longevity. Plant Physiol. 2003, 132, 1077–1084. [Google Scholar] [CrossRef] [PubMed]
- Johnson-Flanagan, A.M.; Spencer, M.S. Ethylene Production during Development of Mustard (Brassica juncea) and Canola (Brassica napus) Seed. Plant Physiol. 1994, 106, 601–606. [Google Scholar] [CrossRef]
- Delmas, F.; Sankaranarayanan, S.; Deb, S.; Widdup, E.; Bournonville, C.; Bollier, N.; Northey, J.G.; McCourt, P.; Samuel, M.A. ABI3 controls embryo degreening through Mendel’s I locus. Proc. Natl. Acad. Sci. USA 2013, 110, E3888–E3894. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, T.; Ren, G.; Hörtensteiner, S.; Zhou, Y.; Cahoon, E.B.; Zhang, C. Chlorophyll degradation: The tocopherol biosynthesis-related phytol hydrolase in Arabidopsis seeds is still missing. Plant Physiol. 2014, 166, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Zhang, J.; Li, J.; Yang, C.; Wang, T.; Ouyang, B.; Li, H.; Giovannoni, J.; Ye, Z. A STAY-GREEN protein SlSGR1 regulates lycopene and β-carotene accumulation by interacting directly with SlPSY1 during ripening processes in tomato. New Phytol. 2013, 198, 442–452. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Lee, S.H.; Kim, Y.S.; Park, O.K.; Hörtensteiner, S.; Paek, N.C. Delayed degradation of chlorophylls and photosynthetic proteins in Arabidopsis autophagy mutants during stress-induced leaf yellowing. J. Exp. Bot. 2014, 65, 3915–3925. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Kim, D.; Kim, Y.S.; Hörtensteiner, S.; Paek, N.C. Arabidopsis STAYGREEN-LIKE (SGRL) promotes abiotic stress-induced leaf yellowing during vegetative growth. FEBS Lett. 2014, 588, 3830–3837. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, H.; Huang, X.; Xing, J.; Yao, J.; Yin, T.; Jiang, J.; Wang, P.; Xu, B. STAYGREEN-mediated chlorophyll a catabolism is critical for photosystem stability during heat-induced leaf senescence in perennial ryegrass. Plant Cell Environ. 2022, 45, 1412–1427. [Google Scholar] [CrossRef] [PubMed]
- Van Eck, J.; Kirk, D.D.; Walmsley, A.M. Tomato (Lycopersicum esculentum). Methods Mol. Biol. 2006, 343, 459–473. [Google Scholar] [CrossRef] [PubMed]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper Enzymes in Isolated Chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Dongfang, L.; Xiaole, C.; Naixing, W. Determination of Malondialdehyde in Biological Samples by 2.5th Order Differential Voltammetry. Prog. Biochem. Biophys. 1997, 24, 361–363. [Google Scholar]
- Yazdi, M.; Kolahi, M.; Mohajel Kazemi, E.; Goldson Barnaby, A. Study of the contamination rate and change in growth features of lettuce (Lactuca sativa Linn.) in response to cadmium and a survey of its phytochelatin synthase gene. Ecotoxicol. Environ. Saf. 2019, 180, 295–308. [Google Scholar] [CrossRef]
- Tran, T.A.; Popova, L.P. Functions and toxicity of cadmium in plants: Recent advances and future prospects. Doga Turk. J. Bot. 2013, 37, 1–13. [Google Scholar] [CrossRef]
- Lee, H.Y.; Back, K. Cadmium Disrupts Subcellular Organelles, Including Chloroplasts, Resulting in Melatonin Induction in Plants. Molecules 2017, 22, 1791. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Schelbert, S.; Park, S.Y.; Han, S.H.; Lee, B.D.; Andrès, C.B.; Kessler, F.; Hörtensteiner, S.; Paek, N.C. STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell 2012, 24, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Yu, G.; Li, H.; Xie, Z.; Wen, W.; Zhang, J.; Huang, B. Knockdown of STAYGREEN in Perennial Ryegrass (Lolium perenne L.) Leads to Transcriptomic Alterations Related to Suppressed Leaf Senescence and Improved Forage Quality. Plant Cell Physiol. 2019, 60, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.E.; Kadonaga, J.T. The RNA polymerase II core promoter: A key component in the regulation of gene expression. Genes Dev. 2002, 16, 2583–2592. [Google Scholar] [CrossRef]
- Peoples, M.B.; Dalling, M.J. The interplay between proteolysis and amino acid metabolism during senescence and nitrogen reallocation. In Senescence and Aging in Plants; Academic Press: San Diego, CA, USA, 1988; pp. 181–217. [Google Scholar]
- Mur, L.A.; Aubry, S.; Mondhe, M.; Kingston-Smith, A.; Gallagher, J.; Timms-Taravella, E.; James, C.; Papp, I.; Hörtensteiner, S.; Thomas, H.; et al. Accumulation of chlorophyll catabolites photosensitizes the hypersensitive response elicited by Pseudomonas syringae in Arabidopsis. New Phytol. 2010, 188, 161–174. [Google Scholar] [CrossRef]
- Munné-Bosch, S.; Alegre, L. Die and let live: Leaf senescence contributes to plant survival under drought stress. Funct. Plant Biol. 2004, 31, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Mecey, C.; Hauck, P.; Trapp, M.; Pumplin, N.; Plovanich, A.; Yao, J.; He, S.Y. A critical role of STAYGREEN/Mendel’s I locus in controlling disease symptom development during Pseudomonas syringae pv tomato infection of Arabidopsis. Plant Physiol. 2011, 157, 1965–1974. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.L.; Deng, L.; Yan, B.; Pan, Y.; Luo, M.; Chen, X.Q.; Hu, T.Z.; Chen, G.P. Silencing of the LeSGR1 gene in tomato inhibits chlorophyll degradation and exhibits a stay-green phenotype. Biol. Plant. 2011, 55, 27–34. [Google Scholar] [CrossRef]
- Pic, E.; de La Serve, B.T.; Tardieu, F.; Turc, O. Leaf senescence induced by mild water deficit follows the same sequence of macroscopic, biochemical, and molecular events as monocarpic senescence in pea. Plant Physiol. 2002, 128, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, S.Q.; Liu, Y.F.; Liu, P.; Lei, G.; He, S.J.; Ma, B.; Zhang, W.K.; Zhang, J.S.; Chen, S.Y. Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 2010, 62, 316–329. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef] [PubMed]
- Xing, B.; Gu, C.; Zhang, T.; Zhang, Q.; Yu, Q.; Jiang, J.; Liu, G. Functional Study of BpPP2C1 Revealed Its Role in Salt Stress in Betula platyphylla. Front. Plant Sci. 2021, 11, 617635. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Dong, X.; Yu, L.; Zhang, Y. Response and Function of Solanum lycopersicum L. SlSGR2 Gene under Cadmium Stress. Horticulturae 2022, 8, 1002. https://doi.org/10.3390/horticulturae8111002
Ma J, Dong X, Yu L, Zhang Y. Response and Function of Solanum lycopersicum L. SlSGR2 Gene under Cadmium Stress. Horticulturae. 2022; 8(11):1002. https://doi.org/10.3390/horticulturae8111002
Chicago/Turabian StyleMa, Jianyu, Xuanming Dong, Lijie Yu, and Yuhong Zhang. 2022. "Response and Function of Solanum lycopersicum L. SlSGR2 Gene under Cadmium Stress" Horticulturae 8, no. 11: 1002. https://doi.org/10.3390/horticulturae8111002
APA StyleMa, J., Dong, X., Yu, L., & Zhang, Y. (2022). Response and Function of Solanum lycopersicum L. SlSGR2 Gene under Cadmium Stress. Horticulturae, 8(11), 1002. https://doi.org/10.3390/horticulturae8111002





