Evolution of LysM-RLK Gene Family in Wild and Cultivated Peanut Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Identifying LysM-RLKs in Cultivated and Wild Peanut Genomes
2.2. Chromosomal Location of LysM-RLK Genes and the Synteny Analysis
2.3. Digital Expression Analysis of LysM-RLK Genes
2.4. Sequence Alignments and Phylogenetic Analyses
2.5. Ka/Ks Analysis
2.6. Plant Inoculation Assays
2.7. RNA Extraction and Expression Analysis
2.8. Statistical Analysis
3. Results
3.1. Identification of LysM-RLKs in Reference Genomes of A. hypogaea, A. duranensis, and A. ipaensis
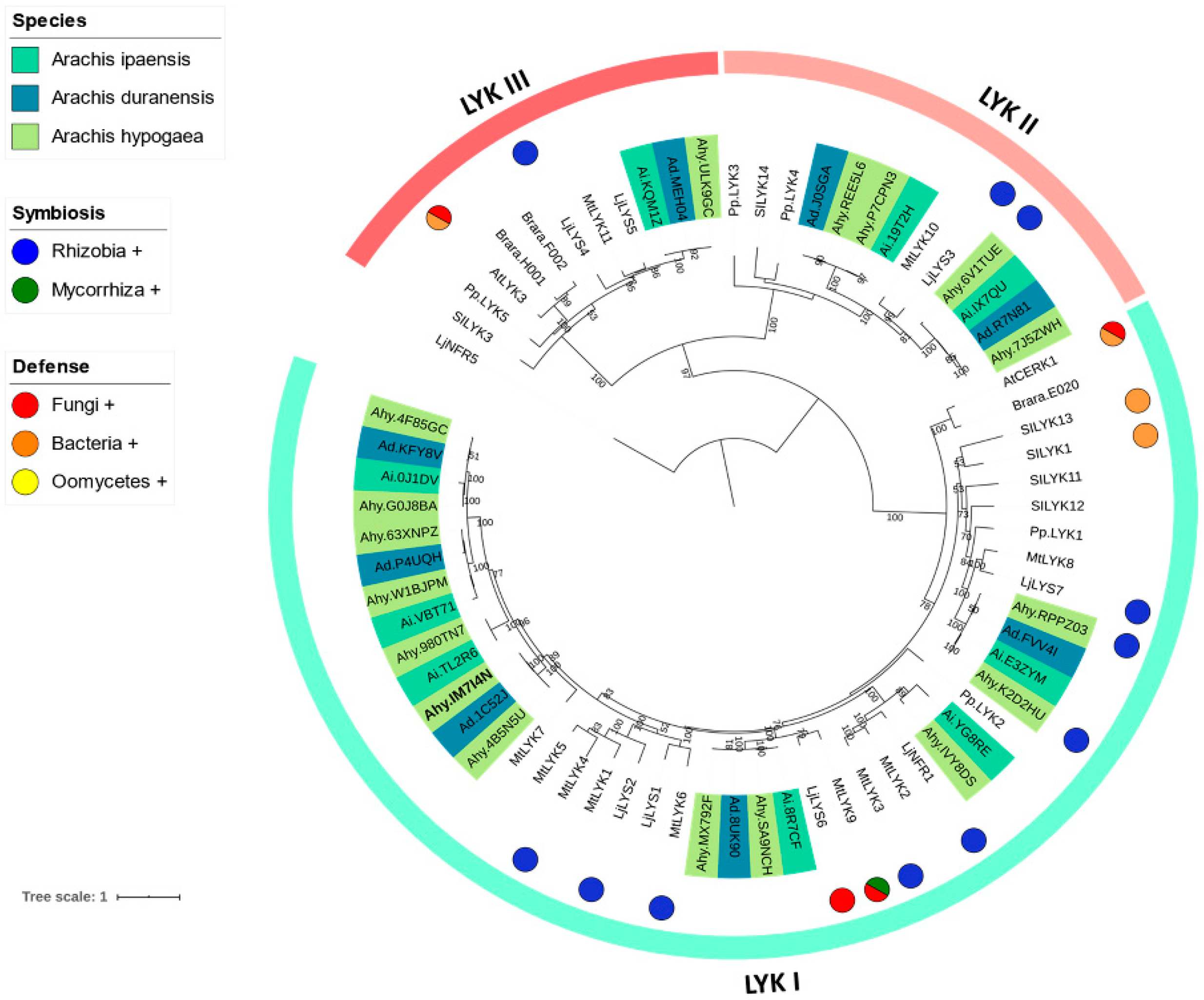
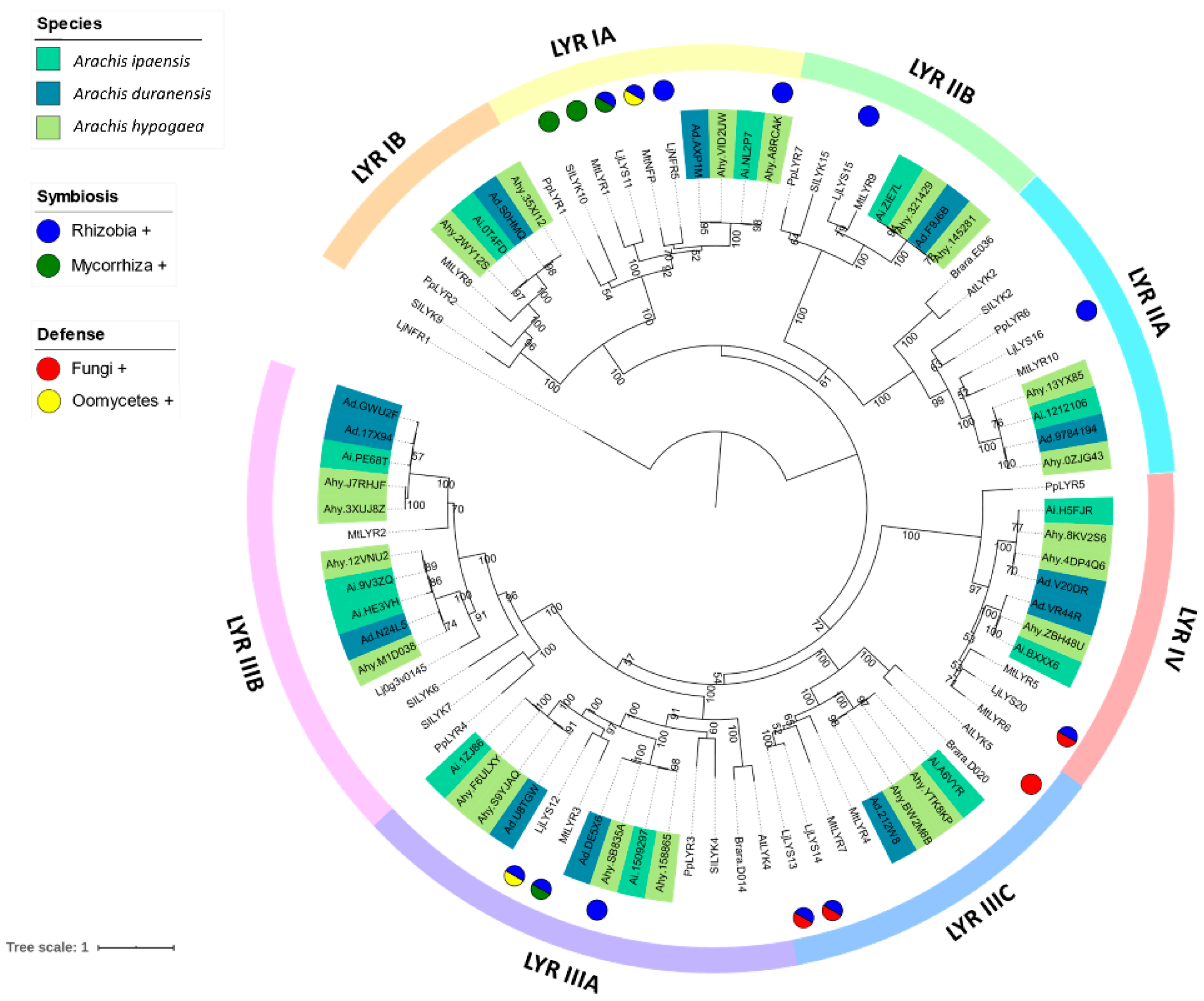
3.2. Phylogenetic Reconstruction of the LysM-RLK Family
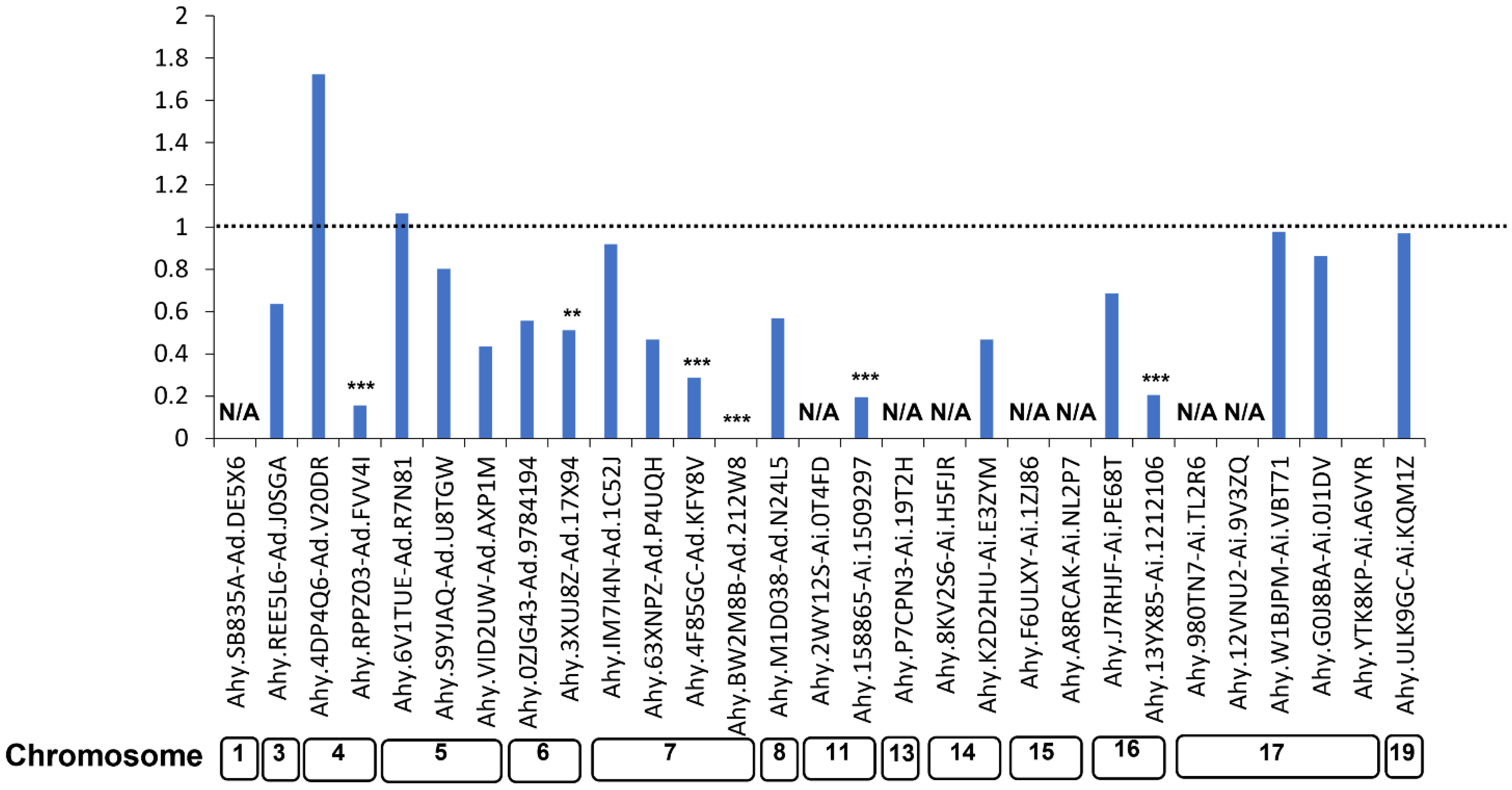
3.3. Ka/Ks Analysis of LysM-RLK Genes
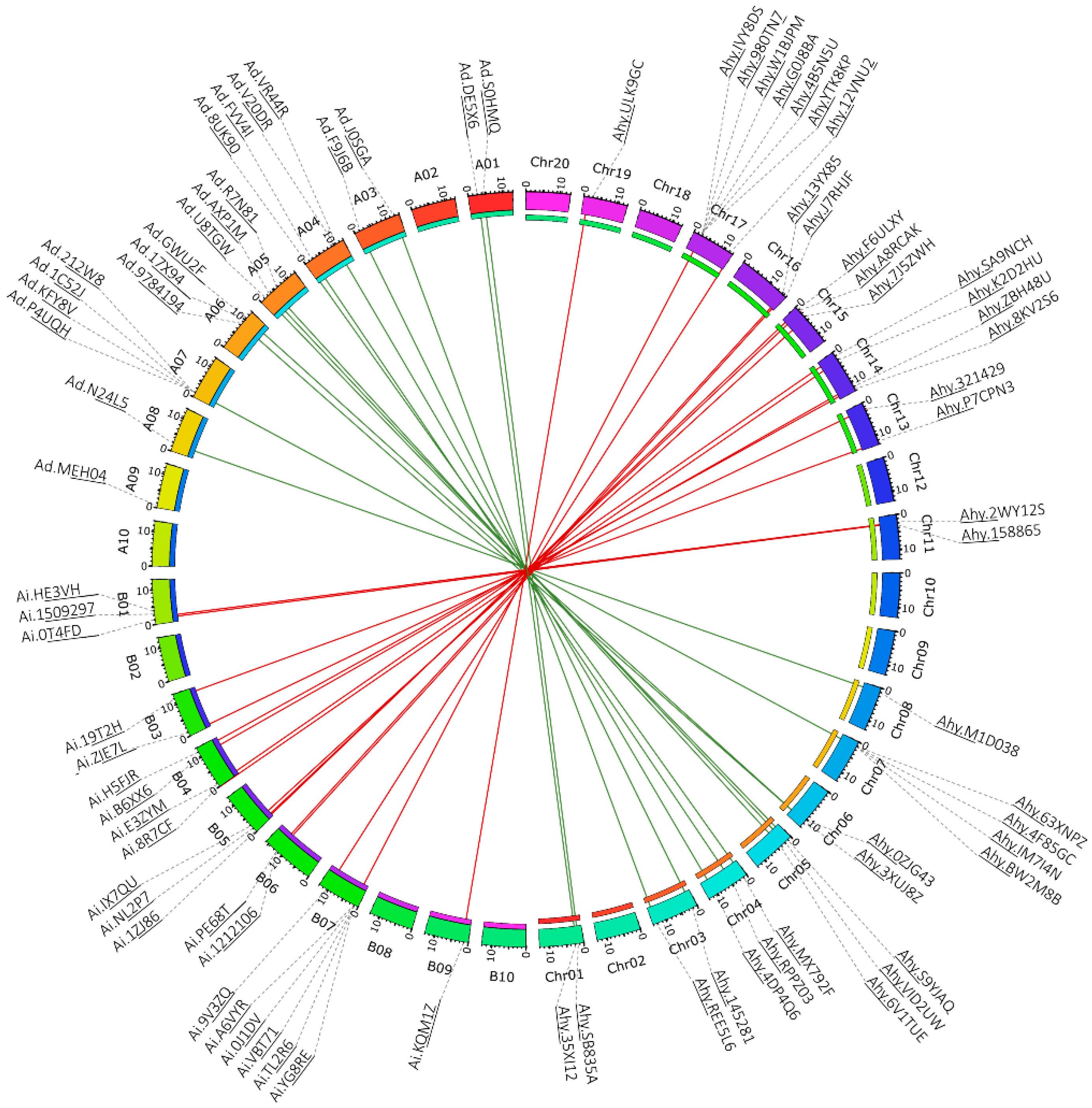
3.4. Chromosomal Location and Synteny Analysis of Peanut LysM-RLK Genes
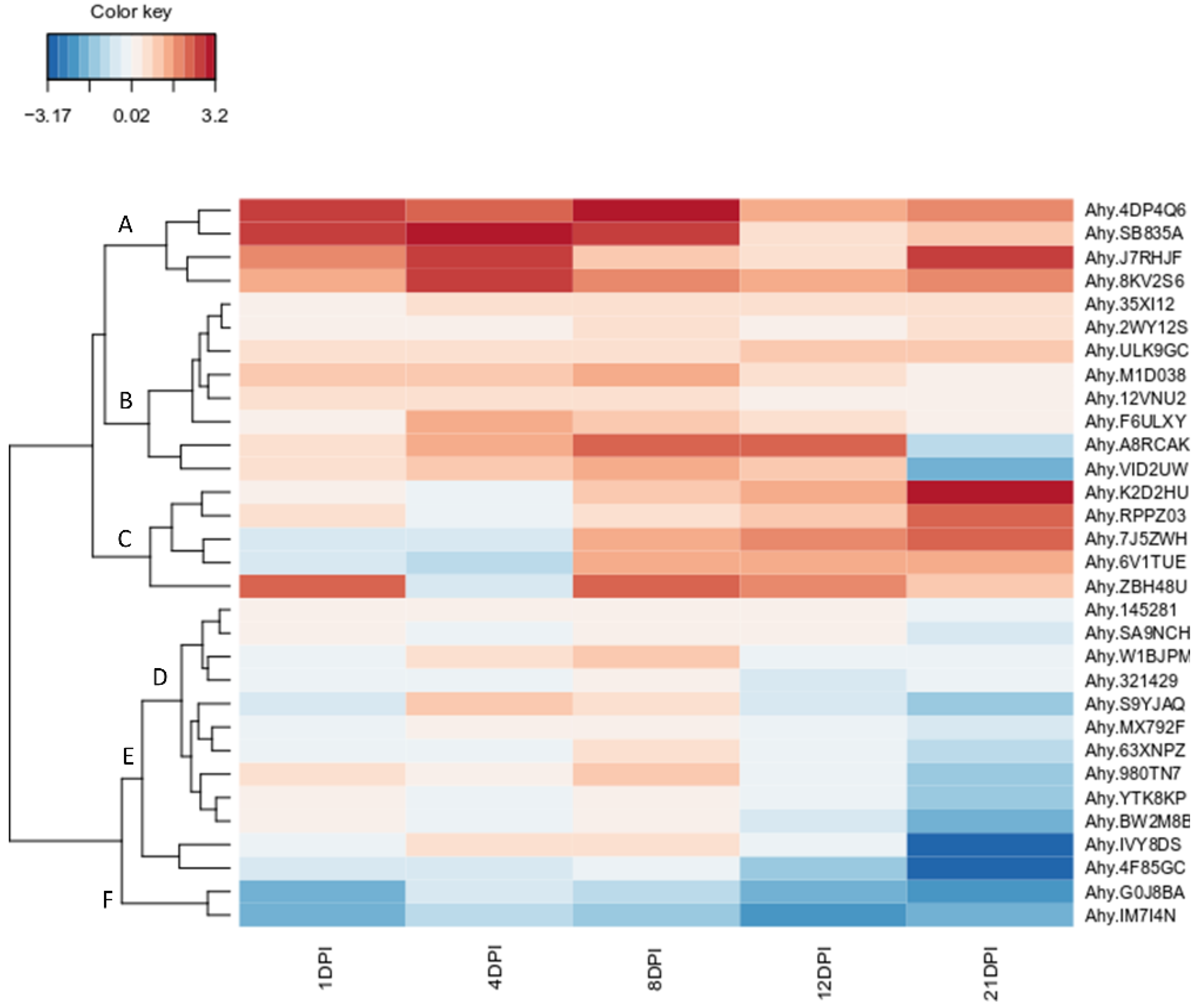
3.5. Temporal Expression Analysis of LysM-RLKs Genes in Peanut–Bradyrhizobia Interaction
3.6. Expression Analysis of Peanut Receptors
4. Discussion
4.1. Phylogenetic Reconstruction of the Peanut LysM-RLK Family and Digital Gene Expression Analysis
4.2. LysM-RLK LYK Group
4.3. LysM-RLK LYR Group
4.4. Temporal Expression Analysis of A. hypogaea LysM-RLK Receptors within the Progress of Symbiosis
4.5. Chromosomal Location of Peanut LysM-RLK and Synteny Analysis
4.6. Expression Analysis in NFs and Chitosan-Treated Plants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zipfel, C.; Oldroyd, G.E.D. Plant Signalling in Symbiosis and Immunity. Nature 2017, 543, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.H.; Paszkowski, U. Receptor-Like Kinases Sustain Symbiotic Scrutiny. Plant Physiol. 2020, 182, 1597–1612. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.; Tonelli, M.L.; Figueredo, M.S.; Ibáñez, F.; Fabra, A. The Lipopeptide Surfactin Triggers Induced Systemic Resistance and Priming State Responses in Arachis hypogaea L. Eur. J. Plant Pathol. 2018, 152, 845–851. [Google Scholar] [CrossRef]
- Antolín-Llovera, M.; Petutsching, E.K.; Ried, M.K.; Lipka, V.; Nürnberger, T.; Robatzek, S.; Parniske, M. Knowing Your Friends and Foes–Plant Receptor-like Kinases as Initiators of Symbiosis or Defence. New Phytol. 2014, 204, 791–802. [Google Scholar] [CrossRef]
- Kelly, S.; Radutoiu, S.; Stougaard, J. Legume LysM Receptors Mediate Symbiotic and Pathogenic Signalling. Curr. Opin. Plant Biol. 2017, 39, 152–158. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D. Speak, Friend, and Enter: Signalling Systems That Promote Beneficial Symbiotic Associations in Plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef]
- Boller, T. Chemoperception of Microbial Signals in Plant Cells. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 189–214. [Google Scholar] [CrossRef]
- Fliegmann, J.; Bono, J.-J. Lipo-Chitooligosaccharidic Nodulation Factors and Their Perception by Plant Receptors. Glycoconj J. 2015, 32, 455–464. [Google Scholar] [CrossRef]
- Yin, H.; Du, Y.; Dong, Z. Chitin Oligosaccharide and Chitosan Oligosaccharide: Two Similar but Different Plant Elicitors. Front. Plant Sci. 2016, 7, 522. [Google Scholar] [CrossRef]
- Gust, A.A.; Willmann, R.; Desaki, Y.; Grabherr, H.M.; Nürnberger, T. Plant LysM Proteins: Modules Mediating Symbiosis and Immunity. Trends Plant Sci. 2012, 17, 495–502. [Google Scholar] [CrossRef]
- Kochert, G.; Stalker, H.T.; Gimenes, M.; Galgaro, L.; Lopes, C.R.; Moore, K. Rflp and cytogenetic evidence on the origin and evolution of allotetraploid domesticated Peanut, Arachis Hypogaea (Leguminosae). Am. J. Bot. 1996, 83, 1282–1291. [Google Scholar] [CrossRef]
- Seijo, J.G.; Lavia, G.I.; Fernandez, A.; Krapovickas, A.; Ducasse, D.; Moscone, E.A. Physical Mapping of the 5S and 18S-25S RRNA Genes by FISH as Evidence That Arachis Duranensis and A. Ipaensis Are the Wild Diploid Progenitors of A. Hypogaea (Leguminosae). Am. J. Bot. 2004, 91, 1294–1303. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, F.; Wall, L.; Fabra, A. Starting Points in Plant-Bacteria Nitrogen-Fixing Symbioses: Intercellular Invasion of the Roots. J. Exp. Bot. 2017, 68, 1905–1918. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The Genome Sequence of Segmental Allotetraploid Peanut Arachis Hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.-C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The Genome of Cultivated Peanut Provides Insight into Legume Karyotypes, Polyploid Evolution and Crop Domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The Genome Sequences of Arachis Duranensis and Arachis Ipaensis, the Diploid Ancestors of Cultivated Peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef]
- Peng, Z.; Liu, F.; Wang, L.; Zhou, H.; Paudel, D.; Tan, L.; Maku, J.; Gallo, M.; Wang, J. Transcriptome Profiles Reveal Gene Regulation of Peanut (Arachis Hypogaea L.) Nodulation. Sci. Rep. 2017, 7, 40066. [Google Scholar] [CrossRef]
- Karmakar, K.; Kundu, A.; Rizvi, A.Z.; Dubois, E.; Severac, D.; Czernic, P.; Cartieaux, F.; DasGupta, M. Transcriptomic Analysis with the Progress of Symbiosis in ‘Crack-Entry’ Legume Arachis Hypogaea Highlights Its Contrast with ‘Infection Thread’ Adapted Legumes. Mol. Plant Microbe Interact. 2019, 32, 271–285. [Google Scholar] [CrossRef]
- Sharma, V.; Bhattacharyya, S.; Kumar, R.; Kumar, A.; Ibañez, F.; Wang, J.; Guo, B.; Sudini, H.K.; Gopalakrishnan, S.; DasGupta, M.; et al. Molecular Basis of Root Nodule Symbiosis between Bradyrhizobium and “Crack-Entry” Legume Groundnut (Arachis hypogaea L.). Plants 2020, 9, 276. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-Scale Protein Function Classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam Protein Families Database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Rose, L.E. The Evolutionary History of LysM-RLKs (LYKs/LYRs) in Wild Tomatoes. BMC Evol. Biol. 2019, 19, 141. [Google Scholar] [CrossRef] [PubMed]
- Buendia, L.; Girardin, A.; Wang, T.; Cottret, L.; Lefebvre, B. LysM Receptor-Like Kinase and LysM Receptor-Like Protein Families: An Update on Phylogeny and Functional Characterization. Front. Plant Sci. 2018, 9, 1531. [Google Scholar] [CrossRef] [PubMed]
- Sela, I.; Ashkenazy, H.; Katoh, K.; Pupko, T. GUIDANCE2: Accurate Detection of Unreliable Alignment Regions Accounting for the Uncertainty of Multiple Parameters. Nucleic Acids Res. 2015, 43, W7–W14. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v3: An Online Tool for the Display and Annotation of Phylogenetic and Other Trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiao, J.; Wu, J.; Zhang, H.; Liu, G.; Wang, X.; Dai, L. ParaAT: A Parallel Tool for Constructing Multiple Protein-Coding DNA Alignments. Biochem. Biophys. Res. Commun. 2012, 419, 779–781. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Zhao, X.-Q.; Wang, J.; Wong, G.K.-S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through Model Selection and Model Averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Spaink, H.P.; Sheeley, D.M.; van Brussel, A.A.N.; Glushka, J.; York, W.S.; Tak, T.; Geiger, O.; Kennedy, E.P.; Reinhold, V.N.; Lugtenberg, B.J.J. A Novel Highly Unsaturated Fatty Acid Moiety of Lipo-Oligosaccharide Signals Determines Host Specificity of Rhizobium. Nature 1991, 354, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Limpens, E.; Franken, C.; Smit, P.; Willemse, J.; Bisseling, T.; Geurts, R. LysM Domain Receptor Kinases Regulating Rhizobial Nod Factor-Induced Infection. Science (1979) 2003, 302, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Petutschnig, E.K.; Jones, A.M.E.; Serazetdinova, L.; Lipka, U.; Lipka, V. The Lysin Motif Receptor-like Kinase (LysM-RLK) CERK1 Is a Major Chitin-Binding Protein in Arabidopsis Thaliana and Subject to Chitin-Induced Phosphorylation. J. Biol. Chem. 2010, 285, 28902–28911. [Google Scholar] [CrossRef]
- Klaus-Heisen, D.; Nurisso, A.; Pietraszewska-Bogiel, A.; Mbengue, M.; Camut, S.; Timmers, T.; Pichereaux, C.; Rossignol, M.; Gadella, T.W.J.; Imberty, A.; et al. Structure-Function Similarities between a Plant Receptor-like Kinase and the Human Interleukin-1 Receptor-Associated Kinase-4. J. Biol. Chem. 2011, 286, 11202–11210. [Google Scholar] [CrossRef]
- Madsen, L.H.; Tirichine, L.; Jurkiewicz, A.; Sullivan, J.T.; Heckmann, A.B.; Bek, A.S.; Ronson, C.W.; James, E.K.; Stougaard, J. The Molecular Network Governing Nodule Organogenesis and Infection in the Model Legume Lotus Japonicus. Nat. Commun. 2010, 1, 10. [Google Scholar] [CrossRef]
- Zeng, L.; Velásquez, A.C.; Munkvold, K.R.; Zhang, J.; Martin, G.B. A Tomato LysM Receptor-like Kinase Promotes Immunity and Its Kinase Activity Is Inhibited by AvrPtoB. Plant J. 2012, 69, 92–103. [Google Scholar] [CrossRef]
- Arrighi, J.-F.; Barre, A.; ben Amor, B.; Bersoult, A.; Soriano, L.C.; Mirabella, R.; de Carvalho-Niebel, F.; Journet, E.-P.; Ghérardi, M.; Huguet, T.; et al. The Medicago Truncatula Lysin [Corrected] Motif-Receptor-like Kinase Gene Family Includes NFP and New Nodule-Expressed Genes. Plant Physiol. 2006, 142, 265–279. [Google Scholar] [CrossRef]
- Cao, Y.; Liang, Y.; Tanaka, K.; Nguyen, C.T.; Jedrzejczak, R.P.; Joachimiak, A.; Stacey, G. The Kinase LYK5 Is a Major Chitin Receptor in Arabidopsis and Forms a Chitin-Induced Complex with Related Kinase CERK1. Elife 2014, 3, e03766. [Google Scholar] [CrossRef]
- Malkov, N.; Fliegmann, J.; Rosenberg, C.; Gasciolli, V.; Timmers, A.C.J.; Nurisso, A.; Cullimore, J.; Bono, J.-J. Molecular Basis of Lipo-Chitooligosaccharide Recognition by the Lysin Motif Receptor-like Kinase LYR3 in Legumes. Biochem. J. 2016, 473, 1369–1378. [Google Scholar] [CrossRef]
- Lohmann, G.V.; Shimoda, Y.; Nielsen, M.W.; Jørgensen, F.G.; Grossmann, C.; Sandal, N.; Sørensen, K.; Thirup, S.; Madsen, L.H.; Tabata, S.; et al. Evolution and Regulation of the Lotus Japonicus LysM Receptor Gene Family. Mol. Plant Microbe Interact. 2010, 23, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Bozsoki, Z.; Cheng, J.; Feng, F.; Gysel, K.; Vinther, M.; Andersen, K.R.; Oldroyd, G.; Blaise, M.; Radutoiu, S.; Stougaard, J. Receptor-Mediated Chitin Perception in Legume Roots Is Functionally Separable from Nod Factor Perception. Proc. Natl. Acad. Sci. USA 2017, 114, E8118–E8127. [Google Scholar] [CrossRef] [PubMed]
- Gibelin-Viala, C.; Amblard, E.; Puech-Pages, V.; Bonhomme, M.; Garcia, M.; Bascaules-Bedin, A.; Fliegmann, J.; Wen, J.; Mysore, K.S.; Signor, C.; et al. The Medicago Truncatula LysM Receptor-like Kinase LYK9 Plays a Dual Role in Immunity and the Arbuscular Mycorrhizal Symbiosis. New Phytol. 2019, 223, 1516–1529. [Google Scholar] [CrossRef] [PubMed]
- Shu, H.; Luo, Z.; Peng, Z.; Wang, J. The Application of CRISPR/Cas9 in Hairy Roots to Explore the Functions of AhNFR1 and AhNFR5 Genes during Peanut Nodulation. BMC Plant Biol. 2020, 20, 417. [Google Scholar] [CrossRef] [PubMed]
- Ben, C.; Debellé, F.; Berges, H.; Bellec, A.; Jardinaud, M.-F.; Anson, P.; Huguet, T.; Gentzbittel, L.; Vailleau, F. MtQRRS1, AnR-Locus Required ForMedicago Truncatulaquantitative Resistance ToRalstonia Solanacearum. New Phytol. 2013, 199, 758–772. [Google Scholar] [CrossRef]
- Liu, Y.; Hassan, S.; Kidd, B.N.; Garg, G.; Mathesius, U.; Singh, K.B.; Anderson, J.P. Ethylene Signaling Is Important for Isoflavonoid-Mediated Resistance to Rhizoctonia Solani in Roots of Medicago Truncatula. Mol. Plant Microbe Interact. 2017, 30, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Kaku, H.; Nishizawa, Y.; Ishii-Minami, N.; Akimoto-Tomiyama, C.; Dohmae, N.; Takio, K.; Minami, E.; Shibuya, N. Plant Cells Recognize Chitin Fragments for Defense Signaling through a Plasma Membrane Receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 11086–11091. [Google Scholar] [CrossRef]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM Receptor Kinase, Is Essential for Chitin Elicitor Signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef] [PubMed]
- Fliegmann, J.; Canova, S.; Lachaud, C.; Uhlenbroich, S.; Gasciolli, V.; Pichereaux, C.; Rossignol, M.; Rosenberg, C.; Cumener, M.; Pitorre, D.; et al. Lipo-Chitooligosaccharidic Symbiotic Signals Are Recognized by LysM Receptor-Like Kinase LYR3 in the Legume Medicago Truncatula. ACS Chem. Biol. 2013, 8, 1900–1906. [Google Scholar] [CrossRef] [PubMed]
- Bono, J.J.; Fliegmann, J.; Gough, C.; Cullimore, J. Expression and Function of the Medicago Truncatula Lysin Motif Receptor-like Kinase (LysM-RLK) Gene Family in the Legume–Rhizobia Symbiosis. Model Legume Med. Truncatula 2019, 439–447. [Google Scholar] [CrossRef]
- de Mita, S.; Streng, A.; Bisseling, T.; Geurts, R. Evolution of a Symbiotic Receptor through Gene Duplications in the Legume–Rhizobium Mutualism. New Phytol. 2013, 201, 961–972. [Google Scholar] [CrossRef] [PubMed]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Felle, H.H.; Umehara, Y.; Grønlund, M.; Sato, S.; Nakamura, Y.; Tabata, S.; Sandal, N.; et al. Plant Recognition of Symbiotic Bacteria Requires Two LysM Receptor-like Kinases. Nature 2003, 425, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Price, P.A.; Tanner, H.R.; Dillon, B.A.; Shabab, M.; Walker, G.C.; Griffitts, J.S. Rhizobial Peptidase HrrP Cleaves Host-Encoded Signaling Peptides and Mediates Symbiotic Compatibility. Proc. Natl. Acad. Sci. USA 2015, 112, 15244–15249. [Google Scholar] [CrossRef] [PubMed]
- Czernic, P.; Gully, D.; Cartieaux, F.; Moulin, L.; Guefrachi, I.; Patrel, D.; Pierre, O.; Fardoux, J.; Chaintreuil, C.; Nguyen, P.; et al. Convergent Evolution of Endosymbiont Differentiation in Dalbergioid and Inverted Repeat-Lacking Clade Legumes Mediated by Nodule-Specific Cysteine-Rich Peptides. Plant Physiol. 2015, 169, 1254–1265. [Google Scholar] [CrossRef]
- Pan, H.; Wang, D. Nodule Cysteine-Rich Peptides Maintain a Working Balance during Nitrogen-Fixing Symbiosis. Nat. Plants 2017, 3, 17048. [Google Scholar] [CrossRef]
- Kawaharada, Y.; Nielsen, M.W.; Kelly, S.; James, E.K.; Andersen, K.R.; Rasmussen, S.R.; Füchtbauer, W.; Madsen, L.H.; Heckmann, A.B.; Radutoiu, S.; et al. Differential Regulation of the Epr3 Receptor Coordinates Membrane-Restricted Rhizobial Colonization of Root Nodule Primordia. Nat. Commun. 2017, 8, 14534. [Google Scholar] [CrossRef]
- Kawaharada, Y.; Kelly, S.; Nielsen, M.W.; Hjuler, C.T.; Gysel, K.; Muszyński, A.; Carlson, R.W.; Thygesen, M.B.; Sandal, N.; Asmussen, M.H.; et al. Receptor-Mediated Exopolysaccharide Perception Controls Bacterial Infection. Nature 2015, 523, 308–312. [Google Scholar] [CrossRef]
- Liang, Y.; Cao, Y.; Tanaka, K.; Thibivilliers, S.; Wan, J.; Choi, J.; Kang, C.H.; Qiu, J.; Stacey, G. Nonlegumes Respond to Rhizobial Nod Factors by Suppressing the Innate Immune Response. Science (1979) 2013, 341, 1384–1387. [Google Scholar] [CrossRef]
- Paparella, C.; Savatin, D.V.; Marti, L.; de Lorenzo, G.; Ferrari, S. The Arabidopsis LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE3 Regulates the Cross Talk between Immunity and Abscisic Acid Responses. Plant Physiol. 2014, 165, 262–276. [Google Scholar] [CrossRef]
- Amor, B.B.; Shaw, S.L.; Oldroyd, G.E.D.; Maillet, F.; Penmetsa, R.V.; Cook, D.; Long, S.R.; Denarie, J.; Gough, C. The NFP Locus of Medicago Truncatula Controls an Early Step of Nod Factor Signal Transduction Upstream of a Rapid Calcium Flux and Root Hair Deformation. Plant J. 2003, 34, 495–506. [Google Scholar] [CrossRef]
- Madsen, E.B.; Madsen, L.H.; Radutoiu, S.; Olbryt, M.; Rakwalska, M.; Szczyglowski, K.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; et al. A Receptor Kinase Gene of the LysM Type Is Involved in Legumeperception of Rhizobial Signals. Nature 2003, 425, 637–640. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, S.R.; Füchtbauer, W.; Novero, M.; Volpe, V.; Malkov, N.; Genre, A.; Bonfante, P.; Stougaard, J.; Radutoiu, S. Intraradical Colonization by Arbuscular Mycorrhizal Fungi Triggers Induction of a Lipochitooligosaccharide Receptor. Sci. Rep. 2016, 6, 29733. [Google Scholar] [CrossRef]
- Fuechtbauer, W.; Yunusov, T.; Bozsóki, Z.; Gavrin, A.; James, E.K.; Stougaard, J.; Schornack, S.; Radutoiu, S. LYS12 LysM Receptor DeceleratesPhytophthora Palmivoradisease Progression InLotus Japonicus. Plant J. 2017, 93, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Rose, C.M.; Venkateshwaran, M.; Volkening, J.D.; Grimsrud, P.A.; Maeda, J.; Bailey, D.J.; Park, K.; Howes-Podoll, M.; den Os, D.; Yeun, L.H.; et al. Rapid Phosphoproteomic and Transcriptomic Changes in the Rhizobia-Legume Symbiosis. Mol. Cell. Proteom. 2012, 11, 724–744. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Kaku, H.; Shimoda, Y.; Sugiyama, A.; Shimamura, M.; Takanashi, K.; Yazaki, K.; Aoki, T.; Shibuya, N.; Kouchi, H. From Defense to Symbiosis: Limited Alterations in the Kinase Domain of LysM Receptor-like Kinases Are Crucial for Evolution of Legume-Rhizobium Symbiosis. Plant J. 2010, 65, 169–180. [Google Scholar] [CrossRef]
- Fávero, A.P.; dos Santos, R.F.; Simpson, C.E.; Valls, J.F.M.; Vello, N.A. Successful Crosses between Fungal-Resistant Wild Species of Arachis (Section Arachis) and Arachis Hypogaea. Genet. Mol. Biol. 2015, 38, 353–365. [Google Scholar] [CrossRef]
- Chen, T.; Duan, L.; Zhou, B.; Yu, H.; Zhu, H.; Cao, Y.; Zhang, Z. Interplay of Pathogen-Induced Defense Responses and Symbiotic Establishment in Medicago Truncatula. Front. Microbiol. 2017, 8, 973. [Google Scholar] [CrossRef]
- Op den Camp, R.; Streng, A.; de Mita, S.; Cao, Q.; Polone, E.; Liu, W.; Ammiraju, J.S.S.; Kudrna, D.; Wing, R.; Untergasser, A.; et al. LysM-Type Mycorrhizal Receptor Recruited for Rhizobium Symbiosis in Nonlegume Parasponia. Science (1979) 2011, 331, 909–912. [Google Scholar] [CrossRef]
- Young, N.D.; Debellé, F.; Oldroyd, G.E.D.; Geurts, R.; Cannon, S.B.; Udvardi, M.K.; Benedito, V.A.; Mayer, K.F.X.; Gouzy, J.; Schoof, H.; et al. The Medicago Genome Provides Insight into the Evolution of Rhizobial Symbioses. Nature 2011, 480, 520–524. [Google Scholar] [CrossRef]
- Ivanov, S.; Fedorova, E.E.; Limpens, E.; de Mita, S.; Genre, A.; Bonfante, P.; Bisseling, T. Rhizobium-Legume Symbiosis Shares an Exocytotic Pathway Required for Arbuscule Formation. Proc. Natl. Acad. Sci. USA 2012, 109, 8316–8321. [Google Scholar] [CrossRef]
- Girardin, A.; Wang, T.; Ding, Y.; Keller, J.; Buendia, L.; Gaston, M.; Ribeyre, C.; Gasciolli, V.; Auriac, M.-C.; Vernié, T.; et al. LCO Receptors Involved in Arbuscular Mycorrhiza Are Functional for Rhizobia Perception in Legumes. Curr. Biol. 2019, 29, 4249–4259.e5. [Google Scholar] [CrossRef] [PubMed]
- Hadwiger, L.A. Pea–Fusarium Solani Interactions Contributions of a System Toward Understanding Disease Resistance. Phytopathology 2008, 98, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Hadwiger, L.A. Multiple Effects of Chitosan on Plant Systems: Solid Science or Hype. Plant Sci. 2013, 208, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Bozsoki, Z.; Gysel, K.; Hansen, S.B.; Lironi, D.; Krönauer, C.; Feng, F.; de Jong, N.; Vinther, M.; Kamble, M.; Thygesen, M.B.; et al. Ligand-Recognizing Motifs in Plant LysM Receptors Are Major Determinants of Specificity. Science (1979) 2020, 369, 663–670. [Google Scholar] [CrossRef]
- Murakami, E.; Cheng, J.; Gysel, K.; Bozsoki, Z.; Kawaharada, Y.; Hjuler, C.T.; Sørensen, K.K.; Tao, K.; Kelly, S.; Venice, F.; et al. Epidermal LysM Receptor Ensures Robust Symbiotic Signalling in Lotus Japonicus. Elife 2018, 7, e33506. [Google Scholar] [CrossRef]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Jurkiewicz, A.; Fukai, E.; Quistgaard, E.M.H.; Albrektsen, A.S.; James, E.K.; Thirup, S.; Stougaard, J. LysM Domains Mediate Lipochitin-Oligosaccharide Recognition and Nfr Genes Extend the Symbiotic Host Range. EMBO J. 2007, 26, 3923–3935. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez Melo, J.; Tonelli, M.L.; Barbosa, M.C.; Ariel, F.; Zhao, Z.; Wang, J.; Fabra, A.; Ibañez, F. Evolution of LysM-RLK Gene Family in Wild and Cultivated Peanut Species. Horticulturae 2022, 8, 1000. https://doi.org/10.3390/horticulturae8111000
Rodríguez Melo J, Tonelli ML, Barbosa MC, Ariel F, Zhao Z, Wang J, Fabra A, Ibañez F. Evolution of LysM-RLK Gene Family in Wild and Cultivated Peanut Species. Horticulturae. 2022; 8(11):1000. https://doi.org/10.3390/horticulturae8111000
Chicago/Turabian StyleRodríguez Melo, Johan, María Laura Tonelli, María Carolina Barbosa, Federico Ariel, Zifan Zhao, Jianping Wang, Adriana Fabra, and Fernando Ibañez. 2022. "Evolution of LysM-RLK Gene Family in Wild and Cultivated Peanut Species" Horticulturae 8, no. 11: 1000. https://doi.org/10.3390/horticulturae8111000
APA StyleRodríguez Melo, J., Tonelli, M. L., Barbosa, M. C., Ariel, F., Zhao, Z., Wang, J., Fabra, A., & Ibañez, F. (2022). Evolution of LysM-RLK Gene Family in Wild and Cultivated Peanut Species. Horticulturae, 8(11), 1000. https://doi.org/10.3390/horticulturae8111000






