New Insights into the Roles of Osmanthus Fragrans Heat-Shock Transcription Factors in Cold and Other Stress Responses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification of OfHSFs in O. fragrans
2.2. Integrated Analyses of Gene Structure, Motif Distribution and Phylogenetic Evolution of OfHSFs
2.3. Chromosomal Locations and Gene Duplications of OfHSF Genes
2.4. OfHSF Promoter Cis-Acting Elements Analysis
2.5. Expression Profiles of OfHSFs as Assessed by RNA-Seq
2.6. Plant Materials and Stress Treatments
2.7. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR) Analyses
2.8. Transient Overexpression of OfHSF11 and OfHSF43 in Tobacco for Subcellular Localization and Cold-Stress Biochemical Analysis
3. Results
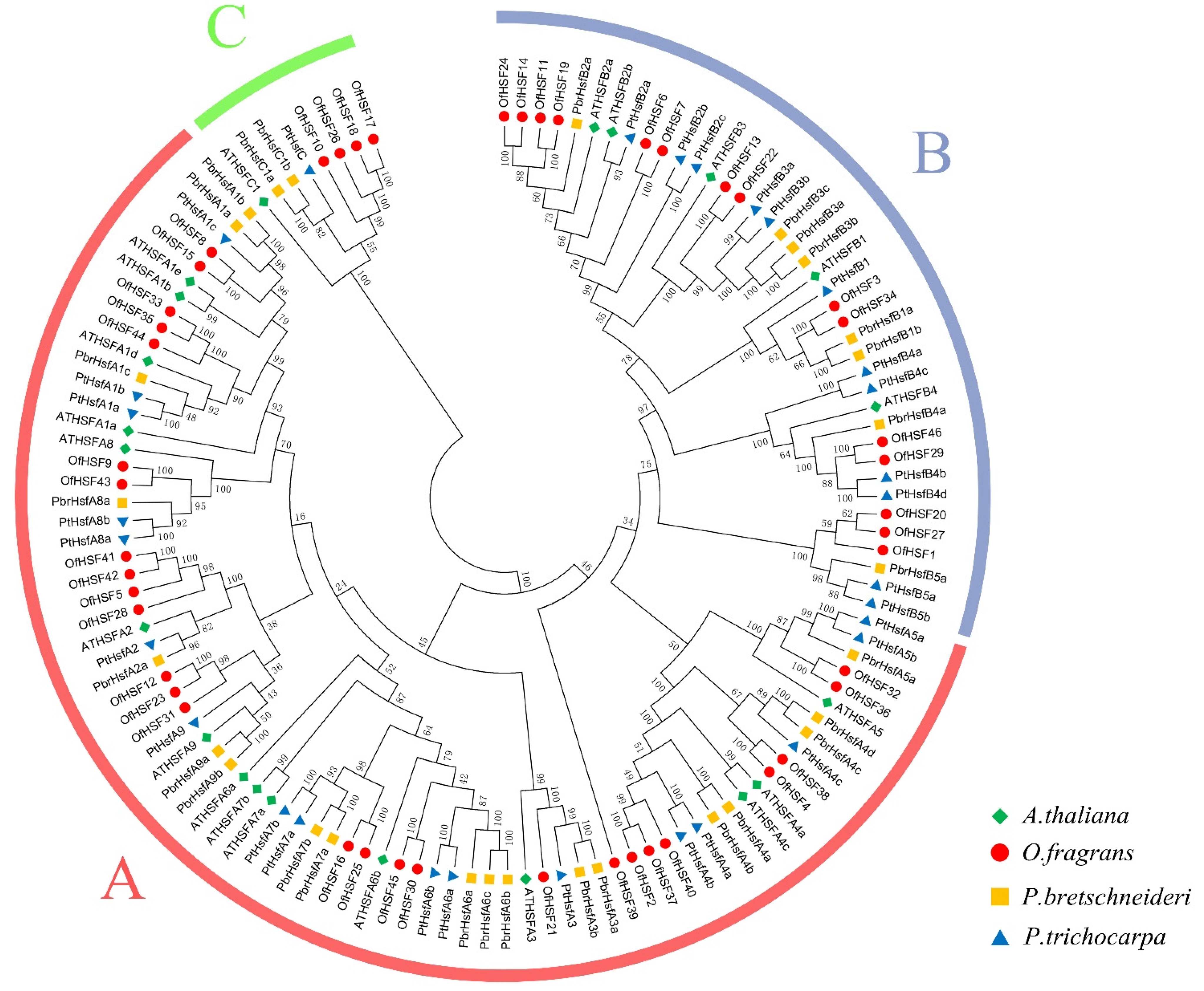
3.1. Genome-Wide Identification and Analysis of the HSF Gene Family in O. fragrans
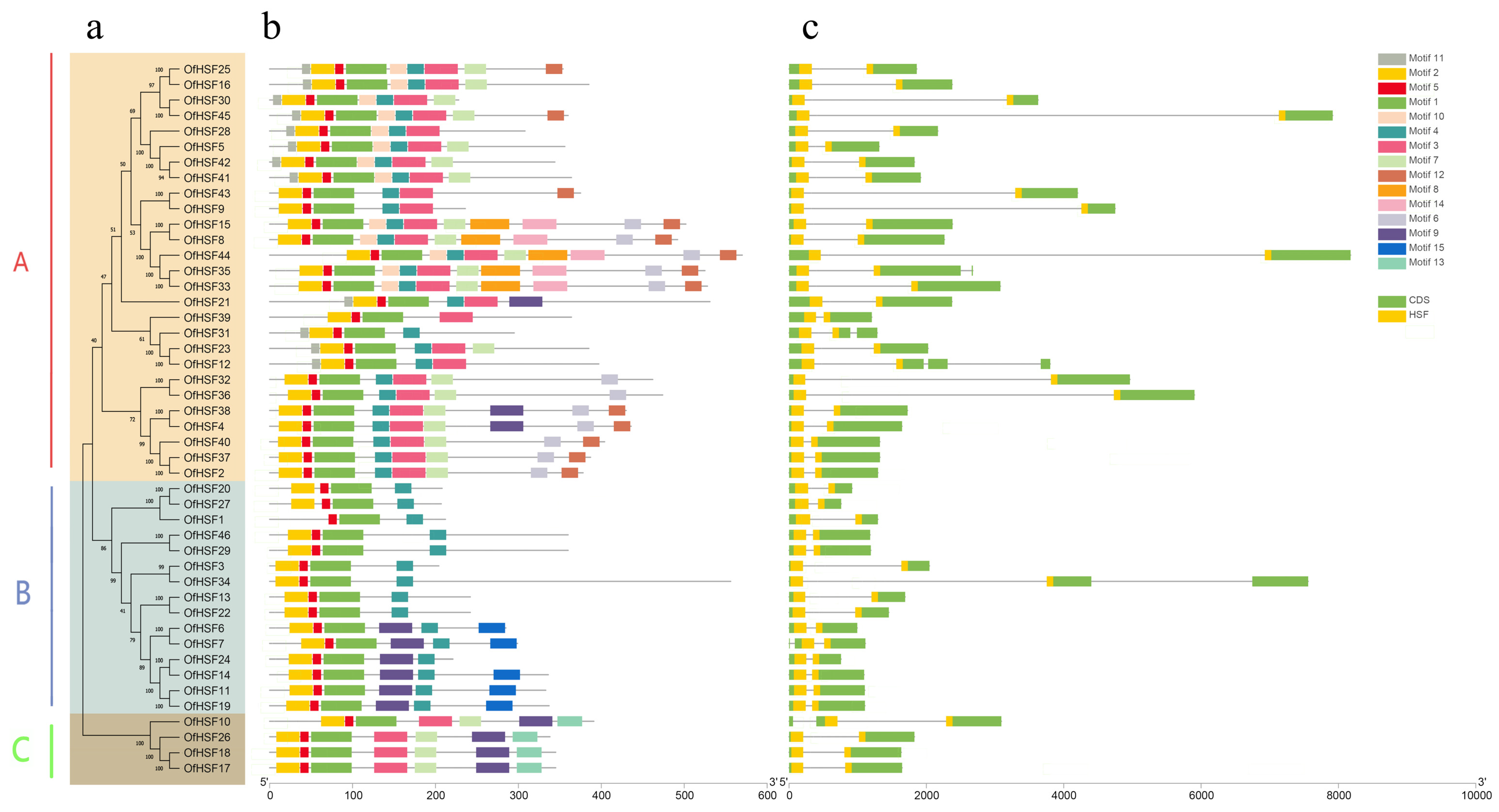
3.2. Phylogenetic Analysis, Intron–Exon Structures and Conserved Motifs of OfHSF Proteins
3.3. OfHSF Promoter Cis-Acting Element Analysis
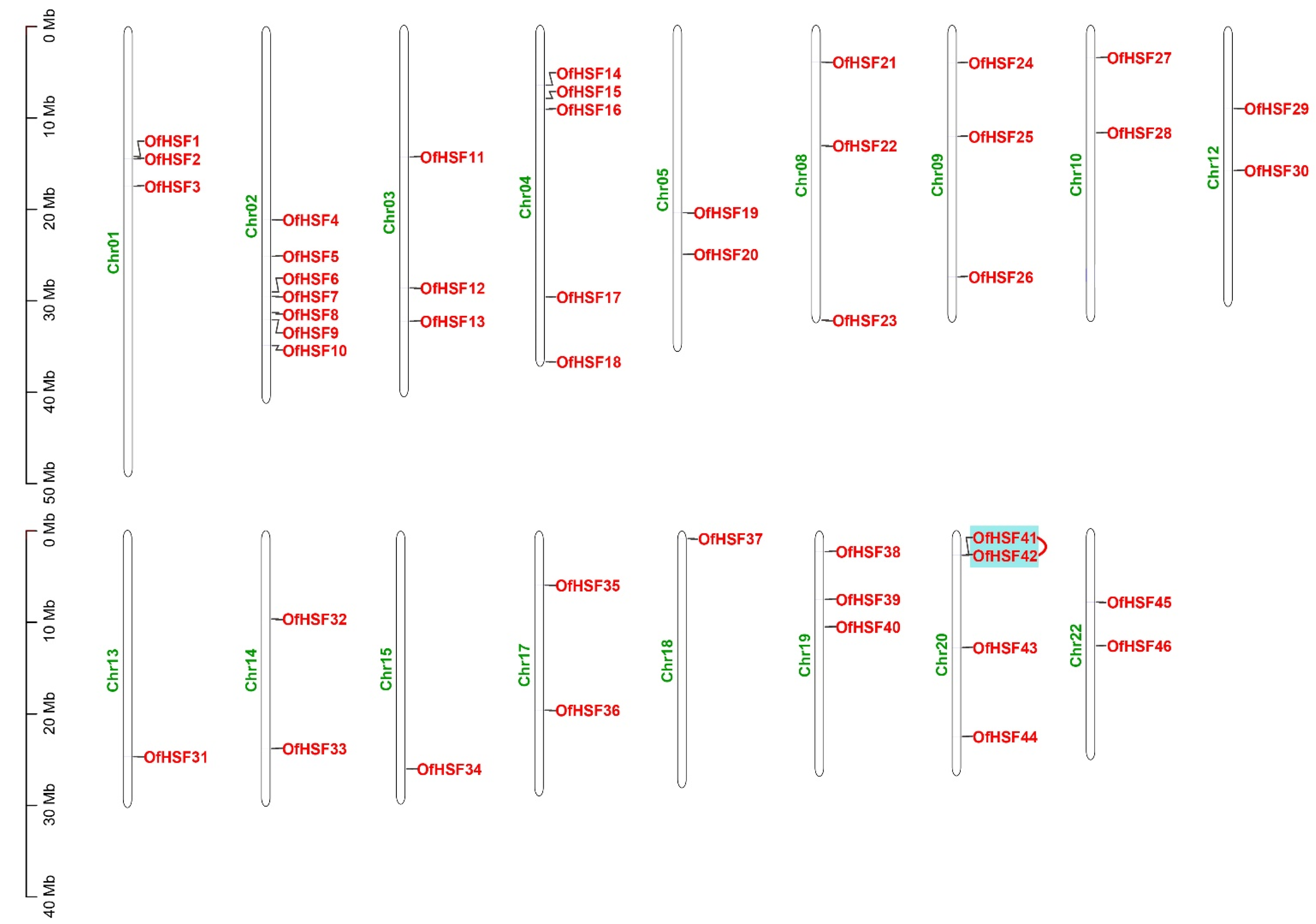
3.4. Chromosomal Location and Synteny Analysis of OfHSF Genes
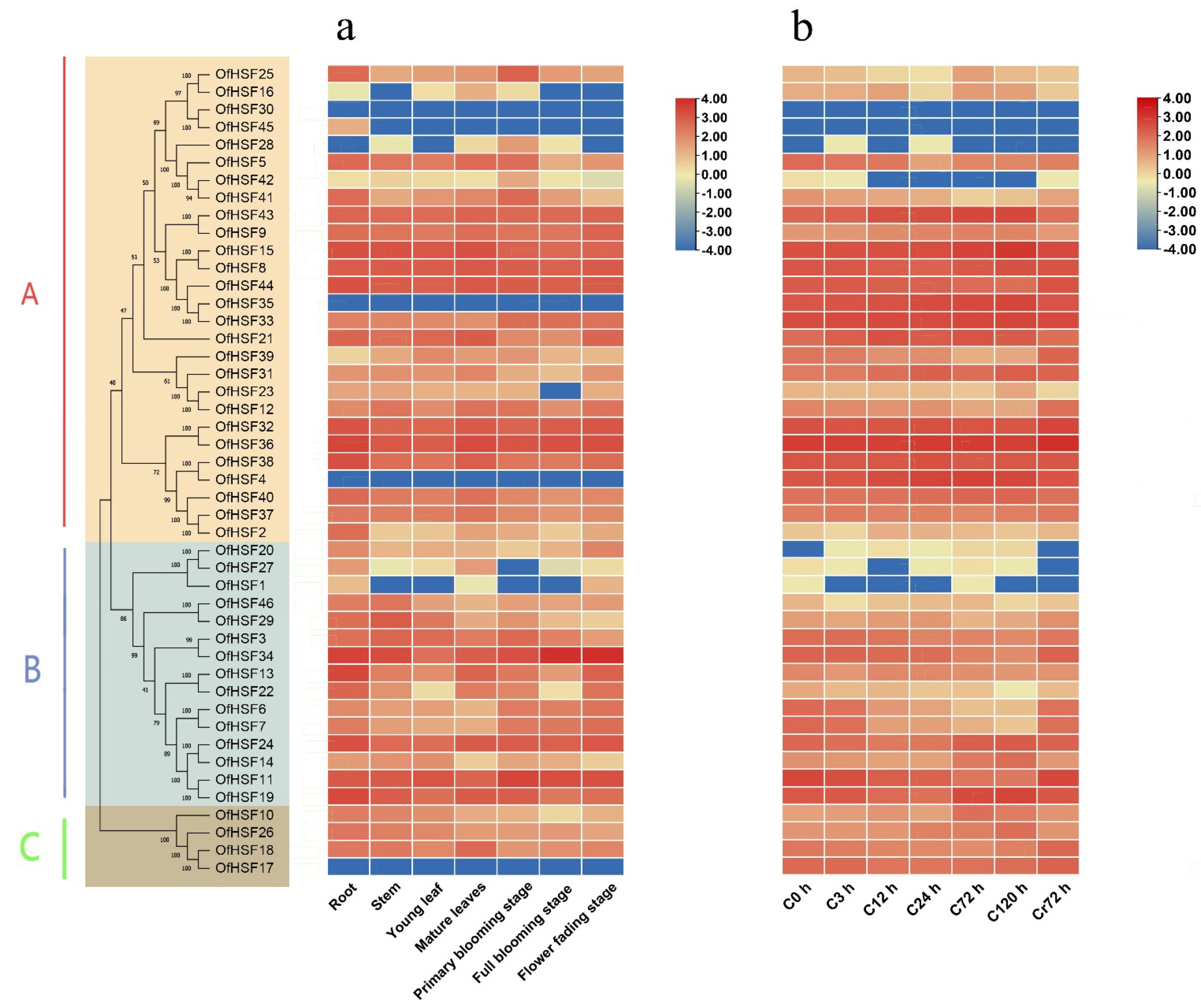
3.5. Expression Profiles of OfHSF Genes in Different Tissues
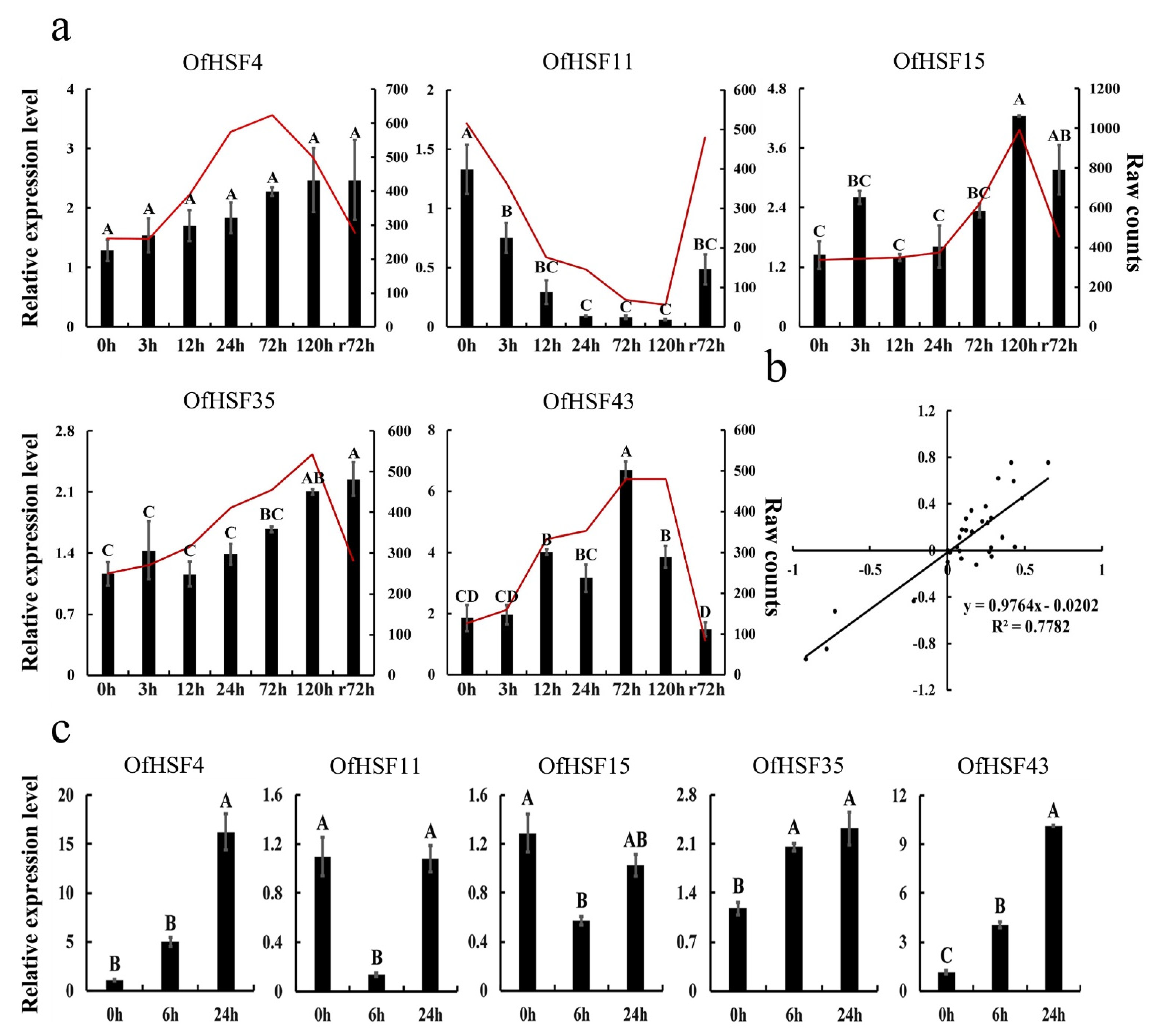
3.6. Expression Patterns of OfHSF Genes in Response to Cold Stress
3.7. The qRT-PCR Analysis of Five Candidate OfHSFs under Cold- and Heat-Stress Conditions
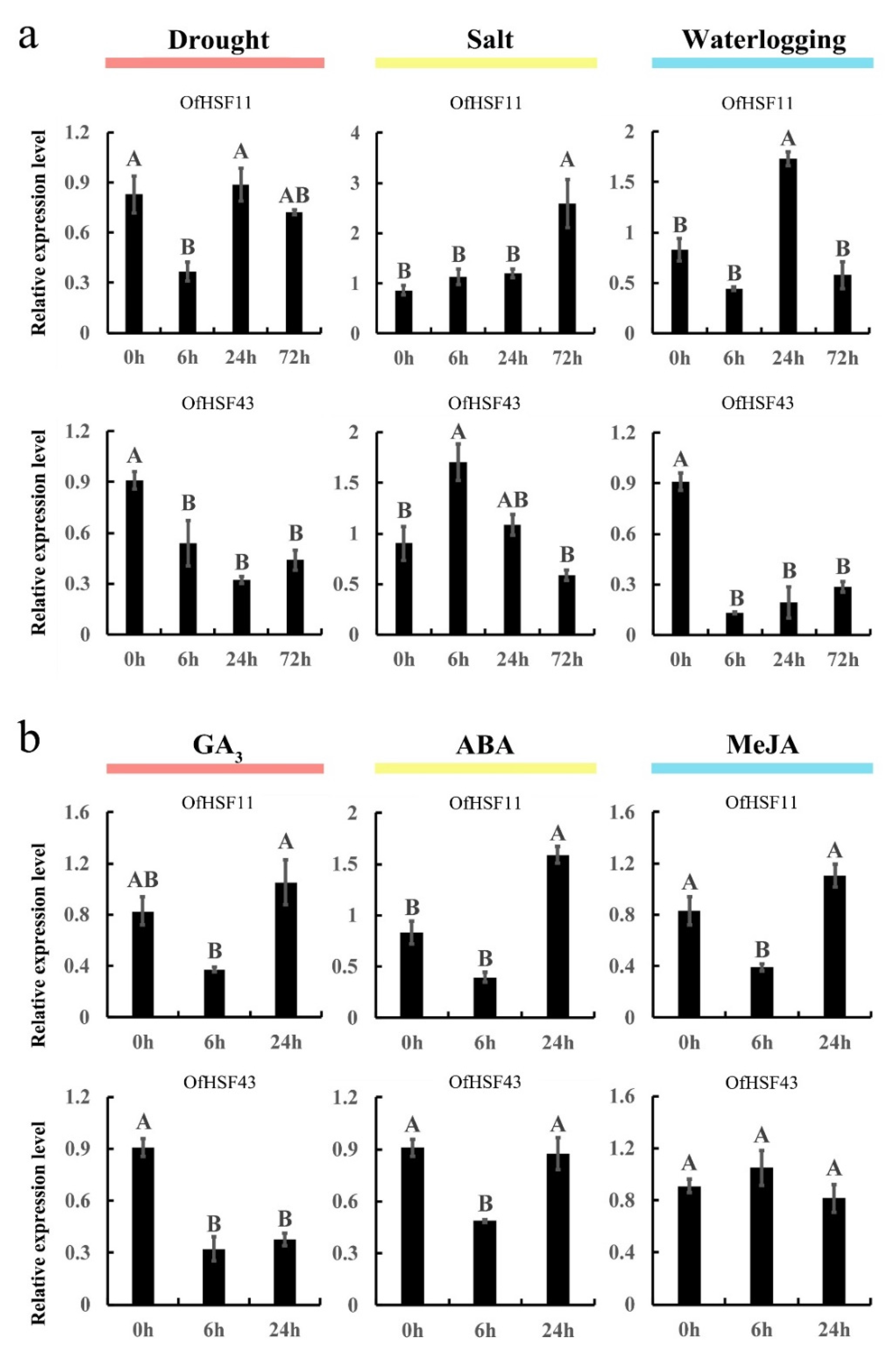
3.8. OfHSF11 and OfHSF43 Are Induced by Different Stresses
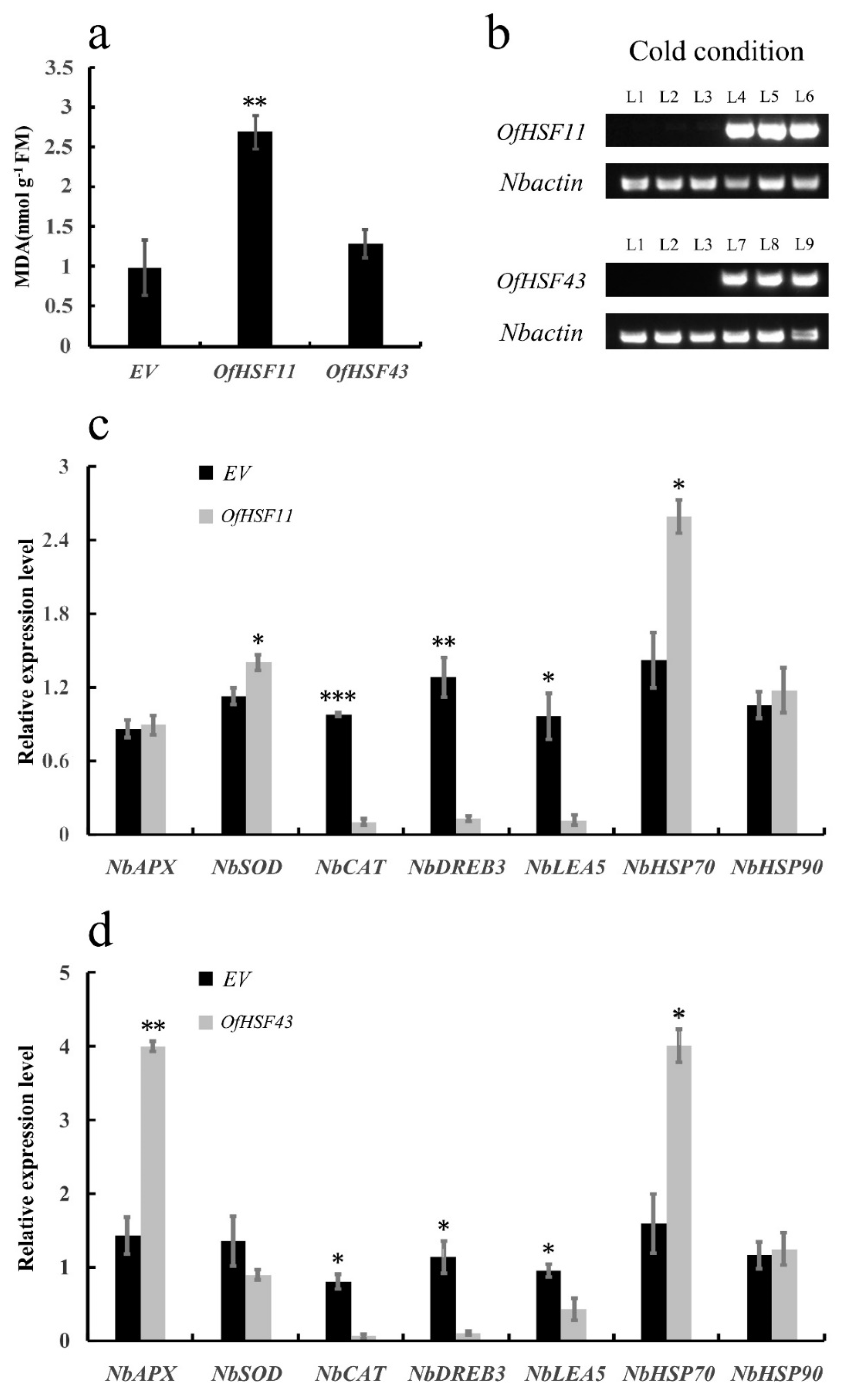
3.9. OfHSF11 and OfHSF43 Are Localized in the Nuclei
3.10. Overexpression of OfHSF11 in Tobacco Resulted in Increased Cold Sensitivity and Changed Expression Level of Stress-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, S.A.; Li, M.Z.; Wang, S.M.; Yin, H.J. Revisiting the Role of Plant Transcription Factors in the Battle against Abiotic Stress. Int. J. Mol. Sci. 2018, 19, 1634. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Liu, J.H.; Ma, X.; Luo, D.X.; Gong, Z.H.; Lu, M.H. The Plant Heat Stress Transcription Factors (HSFs): Structure, Regulation, and Function in Response to Abiotic Stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotak, S.; Larkindale, J.; Lee, U.; von Koskull-Doring, P.; Vierling, E.; Scharf, K.D. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef]
- Scharf, K.D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta 2012, 1819, 104–119. [Google Scholar] [CrossRef]
- Nover, L.; Bharti, K.; Döring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 2001, 6, 177. [Google Scholar] [CrossRef]
- Peteranderl, R.; Rabenstein, M.; Shin, Y.K.; Liu, C.W.; Wemmer, D.E.; King, D.S.; Nelson, H.C. Biochemical and biophysical characterization of the trimerization domain from the heat shock transcription factor. Biochemistry 1999, 38, 3559–3569. [Google Scholar] [CrossRef] [PubMed]
- Boscheinen, O.; Lyck, R.; Queitsch, C.; Treuter, E.; Zimarino, V.; Scharf, K.D. Heat stress transcription factors from tomato can functionally replace HSF1 in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 1997, 255, 322–331. [Google Scholar] [CrossRef]
- Heerklotz, D.; Doring, P.; Bonzelius, F.; Winkelhaus, S.; Nover, L. The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Mol. Cell. Biol. 2001, 21, 1759–1768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotak, S.; Port, M.; Ganguli, A.; Bicker, F.; von Koskull-Doring, P. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 2004, 39, 98–112. [Google Scholar] [CrossRef]
- Qiao, X.; Li, M.; Li, L.; Yin, H.; Wu, J.; Zhang, S. Genome-wide identification and comparative analysis of the heat shock transcription factor family in Chinese white pear (Pyrus bretschneideri) and five other Rosaceae species. BMC Plant Biol. 2015, 15, 12. [Google Scholar] [CrossRef] [Green Version]
- Giorno, F.; Guerriero, G.; Baric, S.; Mariani, C. Heat shock transcriptional factors in Malus domestica: Identification, classification and expression analysis. BMC Genom. 2012, 13, 639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.X.; Yan, H.W.; Wang, Y.Y.; Feng, L.; Chen, Z.; Xiang, Y. Genome Duplication and Evolution of Heat Shock Transcription Factor (HSF) Gene Family in Four Model Angiosperms. J. Plant Growth Regul. 2016, 35, 903–920. [Google Scholar] [CrossRef]
- Jin, G.H.; Gho, H.J.; Jung, K.H. A systematic view of rice heat shock transcription factor family using phylogenomic analysis. J. Plant Physiol. 2013, 170, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Mittler, R. Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann. Bot. 2006, 98, 279–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.X.; Xu, Z.S.; Li, P.S.; Yang, L.; Wei, Y.Q.; Chen, M.; Li, L.C.; Zhang, G.S.; Ma, Y.Z. Overexpression of TaHSF3 in Transgenic Arabidopsis Enhances Tolerance to Extreme Temperatures. Plant Mol. Biol. Rep. 2013, 31, 688–697. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- John, R.; Anjum, N.A.; Sopory, S.K.; Akram, N.A.; Ashraf, M. Some key physiological and molecular processes of cold acclimation. Biol. Plantarum. 2016, 60, 603–618. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, W.; Liu, J.; Song, S.; Hou, X.; Jia, C.; Li, J.; Miao, H.; Wang, Z.; Tie, W.; et al. An aquaporin gene MaPIP2-7 is involved in tolerance to drought, cold and salt stresses in transgenic banana (Musa acuminata L.). Plant Physiol. Biochem. 2020, 147, 66–76. [Google Scholar] [CrossRef]
- Dong, C.; Zhang, Z.; Qin, Y.; Ren, J.P.; Huang, J.F.; Wang, B.L.; Lu, H.L.; Cai, B.H.; Tao, J.M. VaCBF1 from Vitis amurensis associated with cold acclimation and cold tolerance. Acta Physiol. Plant 2013, 35, 2975–2984. [Google Scholar] [CrossRef]
- Swindell, W.R.; Huebner, M.; Weber, A.P. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genom. 2007, 8, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Hirochika, H.; Iwabuchi, M.; Oda, K. Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta 2008, 227, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Niu, C.Y.; Yang, C.R.; Jinn, T.L. The Heat Stress Factor HSFA6b Connects ABA Signaling and ABA-Mediated Heat Responses. Plant Physiol. 2016, 172, 1182–1199. [Google Scholar] [CrossRef]
- Bechtold, U.; Albihlal, W.S.; Lawson, T.; Fryer, M.J.; Sparrow, P.A.; Richard, F.; Persad, R.; Bowden, L.; Hickman, R.; Martin, C.; et al. Arabidopsis HEAT SHOCK TRANSCRIPTION FACTORA1b overexpression enhances water productivity, resistance to drought, and infection. J. Exp. Bot. 2013, 64, 3467–3481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Yang, X.; Xie, J.; Ding, W.; Li, Y.; Yue, Y.; Wang, L. Biochemical and Comparative Transcriptome Analyses Reveal Key Genes Involved in Major Metabolic Regulation Related to Colored Leaf Formation in Osmanthus fragrans ’Yinbi Shuanghui’ during Development. Biomolecules 2020, 10, 549. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.; Mai, R.Z.; Zou, J.J.; Zhang, H.Y.; Zeng, X.L.; Zheng, R.R.; Wang, C.Y. Analysis of aroma-active compounds in three sweet osmanthus (Osmanthus fragrans) cultivars by GC-olfactometry and GC-MS. J. Zhejiang Univ. Sci. B 2014, 15, 638–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Wang, S.; Xia, H.X.; Guo, P.; Wang, Y.H.; Shang, F.D.; Li, Y. Comparative analysis of candidate genes for multi-season flowering in two varieties of sweet osmanthus. Sci. Hortic. 2021, 285, 110175. [Google Scholar] [CrossRef]
- Zou, J.J.; Zhou, Y.; Cai, X.; Wang, C.Y. Increase in DNA fragmentation and the role of ethylene and reactive oxygen species in petal senescence of Osmanthus fragrans. Postharvest. Biol. Technol. 2014, 93, 97–105. [Google Scholar] [CrossRef]
- Geng, X.M.; Zhang, Y.M.; Wang, L.G.; Yang, X.L. Pretreatment with High-Dose Gamma Irradiation on Seeds Enhances the Tolerance of Sweet Osmanthus Seedlings to Salinity Stress. Forests 2019, 10, 406. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.L.; Li, L.; Ding, W.J.; Li, H.Y.; Shi, T.T.; Yang, X.L.; Wang, L.G.; Yue, Y.Z. Genome-wide identification of Osmanthus fragrans bHLH transcription factors and their expression analysis in response to abiotic stress. Environ. Exp. Bot. 2020, 172, 103990. [Google Scholar] [CrossRef]
- Yang, X.; Yue, Y.; Li, H.; Ding, W.; Chen, G.; Shi, T.; Chen, J.; Park, M.S.; Chen, F.; Wang, L. The chromosome-level quality genome provides insights into the evolution of the biosynthesis genes for aroma compounds of Osmanthus fragrans. Hortic. Res. 2018, 5, 72. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Yue, Y.Z.; Li, L.; Li, Y.L.; Li, H.Y.; Ding, W.J.; Shi, T.T.; Chen, G.W.; Yang, X.L.; Wang, L.G. Genome-Wide Analysis of NAC Transcription Factors and Characterization of the Cold Stress Response in Sweet Osmanthus. Plant Mol. Biol. Rep. 2020, 38, 314–330. [Google Scholar] [CrossRef]
- Singh, V.K.; Mangalam, A.K.; Dwivedi, S.; Naik, S. Primer premier: Program for design of degenerate primers from a protein sequence. Biotechniques 1998, 24, 318–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, Y.; Liu, J.; Shi, T.; Chen, M.; Li, Y.; Du, J.; Jiang, H.; Yang, X.; Hu, H.; Wang, L. Integrating Transcriptomic and GC-MS Metabolomic Analysis to Characterize Color and Aroma Formation during Tepal Development in Lycoris longituba. Plants 2019, 8, 53. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Fu, J.; Wang, Y.; Bao, Z.; Zhao, H. Identification of Suitable Reference Genes for Gene Expression Normalization in the Quantitative Real-Time PCR Analysis of Sweet Osmanthus (Osmanthus fragrans Lour.). PLoS ONE 2015, 10, e0136355. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Jin, C.; Huang, X.S.; Li, K.Q.; Yin, H.; Li, L.T.; Yao, Z.H.; Zhang, S.L. Overexpression of a bHLH1 Transcription Factor of Pyrus ussuriensis Confers Enhanced Cold Tolerance and Increases Expression of Stress-Responsive Genes. Front. Plant Sci. 2016, 7, 441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Q.; Xiang, X.; Liu, D.; Yang, A.; Wang, Y. Tobacco Transcription Factor NtbHLH123 Confers Tolerance to Cold Stress by Regulating the NtCBF Pathway and Reactive Oxygen Species Homeostasis. Front. Plant Sci. 2018, 9, 381. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Wall, P.K.; Leebens-Mack, J.H.; Lindsay, B.G.; Soltis, D.E.; Doyle, J.J.; Soltis, P.S.; Carlson, J.E.; Arumuganathan, K.; Barakat, A.; et al. Widespread genome duplications throughout the history of flowering plants. Genome Res. 2006, 16, 738–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Xu, W.; Ni, D.; Wang, M.; Guo, G. Genome-wide characterization of tea plant (Camellia sinensis) Hsf transcription factor family and role of CsHsfA2 in heat tolerance. BMC Plant Biol 2020, 20, 244. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wan, X.L.; Yu, J.Y.; Wang, K.L.; Zhang, J. Genome-Wide Identification, Classification, and Expression Analysis of the Hsf Gene Family in Carnation (Dianthus caryophyllus). Int. J. Mol. Sci. 2019, 20, 5233. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, D.; Yamaguchi, K.; Nishiuchi, T. High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J. Exp. Bot. 2007, 58, 3373–3383. [Google Scholar] [CrossRef] [Green Version]
- Pirkkala, L.; Nykanen, P.; Sistonen, L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001, 15, 1118–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Li, M.Y.; Wang, F.; Xu, Z.S.; Huang, W.; Wang, G.L.; Ma, J.; Xiong, A.S. Heat shock factors in carrot: Genome-wide identification, classification, and expression profiles response to abiotic stress. Mol. Biol. Rep. 2015, 42, 893–905. [Google Scholar] [CrossRef]
- Liu, A.L.; Zou, J.; Zhang, X.W.; Zhou, X.Y.; Wang, W.F.; Xiong, X.Y.; Chen, L.Y.; Chen, X.B. Expression Profiles of Class A Rice Heat Shock Transcription Factor Genes Under Abiotic Stresses. J. Plant Biol. 2010, 53, 142–149. [Google Scholar] [CrossRef]
- Mittal, D.; Chakrabarti, S.; Sarkar, A.; Singh, A.; Grover, A. Heat shock factor gene family in rice: Genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol. Biochem. 2009, 47, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Han, Y.T.; Wei, W.; Li, Y.J.; Zhang, K.; Gao, Y.R.; Zhao, F.L.; Feng, J.Y. Identification, isolation, and expression analysis of heat shock transcription factors in the diploid woodland strawberry Fragaria vesca. Front. Plant Sci. 2015, 6, 736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, X.N.; Xiao, Y.Y.; Fan, Z.Q.; Chen, J.Y.; Lu, W.J.; Kuang, J.F. A banana fruit transcriptional repressor MaERF10 interacts with MaJAZ3 to strengthen the repression of JA biosynthetic genes involved in MeJA-mediated cold tolerance. Postharvest Biol. Technol. 2016, 120, 222–231. [Google Scholar] [CrossRef]
- Hu, Z.; Weijian, L.; Yali, F.; Huiquan, L. Gibberellic acid enhances postharvest toon sprout tolerance to chilling stress by increasing the antioxidant capacity during the short-term cold storage. Sci. Hortic. 2018, 237, 184–191. [Google Scholar] [CrossRef]
- Gusta, L.V.; Trischuk, R.; Weiser, C.J. Plant cold acclimation: The role of abscisic acid. J. Plant Growth Regul. 2005, 24, 308–318. [Google Scholar] [CrossRef]
- Mellacheruvu, S.; Tamirisa, S.; Vudem, D.R.; Khareedu, V.R. Pigeonpea Hybrid-Proline-Rich Protein (CcHyPRP) Confers Biotic and Abiotic Stress Tolerance in Transgenic Rice. Front. Plant Sci. 2015, 6, 1167. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Cao, H.; Qian, W.; Yao, L.; Hao, X.; Li, N.; Yang, Y.; Wang, X. Identification of a novel bZIP transcription factor in Camellia sinensis as a negative regulator of freezing tolerance in transgenic arabidopsis. Ann. Bot. 2017, 119, 1195–1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bin, J.; Zhu, M.; Ding, H.; Zai, Z.; Shi, T.; Wang, L.; Yang, X.; Yue, Y. New Insights into the Roles of Osmanthus Fragrans Heat-Shock Transcription Factors in Cold and Other Stress Responses. Horticulturae 2022, 8, 80. https://doi.org/10.3390/horticulturae8010080
Bin J, Zhu M, Ding H, Zai Z, Shi T, Wang L, Yang X, Yue Y. New Insights into the Roles of Osmanthus Fragrans Heat-Shock Transcription Factors in Cold and Other Stress Responses. Horticulturae. 2022; 8(1):80. https://doi.org/10.3390/horticulturae8010080
Chicago/Turabian StyleBin, Jing, Meilin Zhu, Huifen Ding, Zhouying Zai, Tingting Shi, Lianggui Wang, Xiulian Yang, and Yuanzheng Yue. 2022. "New Insights into the Roles of Osmanthus Fragrans Heat-Shock Transcription Factors in Cold and Other Stress Responses" Horticulturae 8, no. 1: 80. https://doi.org/10.3390/horticulturae8010080
APA StyleBin, J., Zhu, M., Ding, H., Zai, Z., Shi, T., Wang, L., Yang, X., & Yue, Y. (2022). New Insights into the Roles of Osmanthus Fragrans Heat-Shock Transcription Factors in Cold and Other Stress Responses. Horticulturae, 8(1), 80. https://doi.org/10.3390/horticulturae8010080





