Abstract
Chlorophyll is vital for photosynthesis to produce sugars and other useful biochemical products in green plants. However, the molecular effects of chlorophyll deficiency in Chrysanthemum are largely unknown. In this study, we identified a bud sport mutant chrysanthemum belonging to the variety ‘Nannong Binyun’, which has yellow branches. Plant physiological studies have shown that the yellow color is revealed due to chlorophyll loss. RNA extracts of yellow and green tissues were analyzed using high-throughput RNA-sequencing, and a total of 11,649 tissue enriched unigenes that respond to chlorophyll deficiency were identified, including 4803 unigenes upregulated in yellow tissues and 6846 unigenes in green tissues. GO analysis revealed that these tissue-enriched genes may involve in the physiological processes of chlorophyll accumulation and photosynthesis. In addition, many DEGs from the families of AP2-EREBP, bHLH, MYB, and FAR1 that are associated with plant development and stress response were detected. Our study found that most of the genes from the GRAS family were downregulated in yellow leaves, indicating their putative roles in stem cell maintenance and possible contribution to leaf size determination.
1. Introduction
Green plants contain chlorophyll that can absorb and convert light through photosynthesis into chemical energy that plants use to produce sugars and other biochemical products for growth and development. When a green plant encounters stress conditions or at certain developmental stages, the color of the plant may change from green to yellow due to the lack/loss of chlorophyll. Severe stress conditions may cause the death of the yellow leaves, or even the death of the entire plant.
Various types of mutations that contribute to defects in chlorophyll synthesis or chloroplast morphogenesis have been identified in several plant species [1]. For example, some mutants were reported to exhibit chlorophyll deficiency phenotypes in response to certain environmental factors such as light and nutrients as well as phytohormones [2,3,4,5,6]. Additionally, permanent chlorophyll-deficient mutations are common in a wide variety of cultivated and natural plant species, as well as in artificially induced mutant cultivars [1,7,8]. These mutants would contribute to the discovery of genes controlling chloroplast development and facilitate our understanding of the mechanisms underlying chlorophyll deficiency. To date, numerous genes have been identified to control chlorophyll synthesis or chloroplast morphogenesis in an age-triggered or stress-induced manner, sequentially leading to senescence/death. For example, members of NAC (NAM, ATAF1/2, and CUC2) transcription factor (TF) family, WRKY, bHLH, and MYB families are essential factors that modulate transcriptional changes of genes involved in chlorophyll synthesis or degradation [9,10,11].
Chrysanthemum (Chrysanthemum morifolium Ramat.) is a major ornamental plant species that has been widely used in cut flowers, potted plants, and landscaping [12,13]. The leaves of cut chrysanthemum tend to turn yellow through chlorophyll degradation [14,15], which has negative impacts on the ornamental and economic value of this plant. Many investigations have been focused on the mechanisms that contribute to the chlorophyll degradation; however, to date, little attempt has been made to investigate the gene expression regulation in response to chlorophyll deficiency in chrysanthemum. Our study focused on a bud sport mutant in chrysanthemum. A bud sport mutant of the variety ‘Nannong Binyun’ with yellow branches was identified in our cultivation field. We confirmed that the distinct yellow color was due to lack of chlorophyll. We performed RNA-seq analysis using samples of yellow and green part of leaves from the same plant to identify genes that respond to chlorophyll deficiency. We then explored the functional features of the unigenes upregulated in green and yellow tissues to understand the underlying molecular mechanism in response to chlorophyll deficiency in chrysanthemum. In addition, the expression of some key genes was validated by using qRT-PCR. Therefore, this study aimed to identify genes and biological pathways that are affected by chlorophyll deficiency.
2. Materials and Methods
2.1. Plant Materials and Cultivation Conditions
The green and yellow tissues of bud sport mutant of Chrysanthemum morifolium cv. ‘Nannong Binyun’ were obtained from the Chrysanthemum Germplasm Resource Preserving Center of Nanjing Agricultural University (Nanjing, China). Rooted seedlings were transferred into a soil mixture containing peat and perlite and cultivated for two weeks under a 16 h/8 h (day (sunlight)/night) photoperiod at a constant temperature of 23 °C.
2.2. Total Chlorophyll Content Measurement
Leaves were harvested and snap-frozen in liquid nitrogen. To measure the total chlorophyll content, 300 mg of each frozen leaves sample were extracted in 5 mL 95% ethanol (v/v) solution and incubated at room temperature for 48 h in the dark. After centrifugation at 4000 rpm for 10 min, the total chlorophyll content (mg/g) was determined by measuring the absorbance at 663 and 645 nm and calculated as previously described [16]. Total chlorophyll content (mg/g) = (8.04A663 + 20.29A645) × V × N/(1000 × m), where A663 indicates the absorbance value at 663 nm, A645 indicates the absorbance value at 645 nm, V is the total volume of the extraction solution (mL), N is the times of dilution, and m is the weight of sample (g).
2.3. Fv/Fm Measurement
Chlorophyll fluorescence efficiency was examined after a 30 min dark acclimation period using an imaging pulse amplitude modulated (PAM) fluorimeter (Imaging-PAM; WALZ, Effeltrich, Germany), according to a previously described protocol [17]. The Fv/Fm was calculated using the reported formula [18]. Images of Fv/Fm were obtained and the representative leaves are shown in Figure 1.
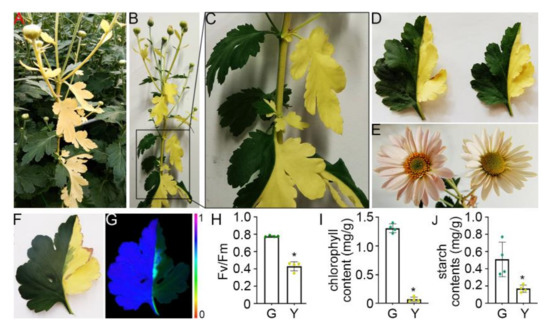
Figure 1.
Phenotypic characterization of chrysanthemum bud sport mutant. (A) The bud sport mutant plant with yellow branches in the field-grown chrysanthemum variety ‘Nannong Binyun’; (B) The bud sport mutant plant shows an asymmetrical half-green and half-yellow appearance in the main stem and branches; (C) an enlarged view of the boxed region in (B); (D) the phenotype of leaves with half-green and half-yellow appearance; (E) the phenotype of flowers generated from green (left) and yellow (right) branches in the same plant; (F) a bud sport mutant leaf used for Fv/Fm measurement; (G) images of chlorophyll fluorescence parameters Fv/Fm of the bud sport mutant leaf. The color code depicted at the bottom of the image ranges from 0 (red) to (1) purple; (H) the photochemical activity of Fv/Fm of four biological replicates; (I) chlorophyll content in the green and yellow tissues based on three biological replicates, and (J) starch content of the green and yellow tissues based on three biological replicates. * p < 0.05 when compared to the control by one-tailed unpaired t-test. Error bars represent SD. G indicates green tissues, Y indicates yellow tissues.
2.4. Starch Content Analysis
Samples of frozen leaves (0.1 g) were used for starch extraction and the absorbance of starch was measured at 620 nm, as described in the manual of Starch Content Assay Kit (BC0700-50T, Solarbio, China). Starch content was calculated as follows: starch content (mg/g fresh leaves) = 0.289 × (A620 + 0.0295)/W, where A620 indicates the absorbance value at 620 nm and W indicates the weight (g) of fresh leaves.
2.5. RNA Extraction and Real-Time PCR
The green and yellow tissues from three leaves in a single plant were bulked and total RNA was extracted using an RNeasy plant kit (Qiagen, Germantown, MD, USA). First-strand cDNA was synthesized using oligo-(dT)18 primers and M-MLV reverse transcriptase (Takara, Beijing, China). Real-time PCR was performed for each sample in three technical replicates, according to the manufacturer’s instructions. The expression levels of gene candidates were normalized to that of CmEF1α and CmUbiquitin [19,20]. We confirmed that the expression of these two control genes is stable between green and yellow tissues in RNA-seq data. Primers used for qPCR are listed in Supplementary Table S1, the detailed sequence information of unigenes is listed in Supplementary Table S2.
2.6. RNA-Seq and Analysis
The green and yellow tissues from a single leaf were collected. Three biological replicates were collected from three leaves in a single plant. Total RNA (4 µg) was used to prepare RNA library and multiplexed libraries were sequenced using the DNBSEQ platform (MGI). RNA-seq clean reads were assembled de novo using Trinity [21,22]. The unigenes were assembled, and their expression levels were determined using reads per kilobase of transcript per million mapped reads (RPKM) value. The unigenes that are upregulated in yellow or green leaves were determined using the following criteria: fold change (FC, RPKM in yellow tissue/RPKM in green tissue) ≥ 2 or FC ≤ 0.5, and with a Q value ≤ 0.05. Heat maps, dot plots, and Venn diagrams were generated using the R software version 4.0.4.
2.7. Statistical Analyses
Data are shown as means ± SD (standard deviation). Statistical analysis was performed using SPSS 16.0 software. Differences between samples were analyzed using p < 0.05 level as significant according to t-test.
3. Results
3.1. Phenotypic Characterization of Chrysanthemum Bud Sport Mutant with Yellow Branches
The cultivation of chrysanthemum for commercial purposes has produced many cultivars of bud sport mutants [23]. Recently, we identified a bud sport mutant of the variety ‘Nannong Binyun’ that had yellow branches (Figure 1A). The main stem of this mutant exhibited an asymmetrical half-green and half-yellow appearance, with the regular green branches from the green zone, yellow branches from the yellow zone, as well as half-green and half-yellow branches from the green-yellow transition zone (Figure 1B,C). Under normal conditions, yellow leaves senesced significantly faster and had smaller average leaf size compared to that of green leaves (Figure 1D). Both the green and yellow branches could produce flower, with lighter flower color in the yellow branches (Figure 1E). Although the Fv/Fm parameter could not be used to determine the quantum efficiency of PSII photochemistry [24], it usually correlates with the yield of PSII [25]. To examine the efficiency of photosynthesis, we compared the Fv/Fm ratio of green and yellow parts in the same leaf. The Fv/Fm ratio of the yellow part was markedly lower than that in the green part (Figure 1F–H, Supplementary Table S3). The yellow branches can grow in the Murashige and Skoog (MS) medium, and produce new yellow branches; however, it could not independently grow in the soil, probably due to the lack of chlorophyll that impairs the efficiency of photosynthesis (Figure 1F–I). The analysis of starch content revealed a distinct decrease in the accumulation of photosynthetic products in yellow leaves (Figure 1J). Collectively, our data indicated that the chrysanthemum mutant sectors have a deficiency in chlorophyll synthesis or maintenance, resulting in the decline in photosynthetic efficiency and the lack of energy supply that collectively attenuates its growth.
3.2. Paired-End Sequencing
We performed an RNA-seq assay to identify upregulated genes in green or yellow part of the leaf and to examine the function of these genes in response to chlorophyll deficiency. To identify the transcripts of interest, six cDNA libraries were constructed using RNA samples of leaves with half-green and half-yellow feature, including three yellow samples (Y1, Y2, and Y3) and three green samples (G1, G2, and G3), respectively, and were subjected to RNA-seq analysis using the DNBSEQ platform (MGI). A total of 287.48 million short reads, each of approximately 150 bp, were generated from the six samples (Figure S1). After stringent quality assessment and data filtering, such as adaptor sequence removal, reads with N > 5% clearing and low quality reads discarding, resulted in an overall clean reads of 255.07 million with a base quality score of 20% which were selected for subsequent analysis (Figure S1). Principal component analysis (PCA) is used to reduce the dimensionality of large datasets and to increase interpretability, which has been widely employed to intuitively evaluate RNA-seq data [26,27]. We performed PCA to explore the data structures of these RNA-seq gene expression profiles and results showed that the Y and G samples could be clustered into two sub-groups according to the gene expression profile of each sample (Figure 2A), indicating that the chlorophyll deficiency was the key driving factor to separate the datasets, and also indicating a good reproducibility of RNA-seq data among the biological replicates of this mutant. A total of 153,251 unigenes were assembled yielding total length of 173,229,779 bp and average transcript length of 1130 bp (Figure 2B), with average GC content of 39.21%. Based on the blast results of all 153,251 unigenes using different databases yielded a total of 100,400 (NR, ftp://ftp.ncbi.nlm.nih.gov/blast/db, accessed on 22 March 2021, 65.51%), 70,732 (NT, ftp://ftp.ncbi.nlm.nih.gov/blast/db, accessed on 22 March 2021, 46.15%), 68,724 (SwissProt, https://www.uniprot.org/, accessed on 22 March 2021, 44.84%), 72,868 (KOG, https://www.ncbi.nlm.nih.gov/COG/, accessed on 22 March 2021, 47.55%), 73,836 (KEGG, http://www.genome.jp/kegg, accessed on 22 March 2021, 48.18%), 81,068 (GO, http://geneontology.org, 52.90%), and 60,711 (Pfam, http://pfam.xfam.org, accessed on 22 March 2021, 39.62%) annotated unigenes with at least one sequence read.
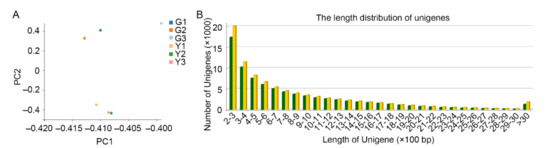
Figure 2.
Summary of paired-end sequencing using DNBSEQ platform (MGI). (A) Principal component analysis (PCA) plot for the expression of unigenes in different samples; (B) the length distribution of unigenes identified in the transcriptomes of green and yellow tissues.
3.3. Unigene Expression Analysis
Unigenes with RPKM value larger than 0 were considered expressed unigenes. Our results showed that 117,000, 116,329, 115,336, 123,027, 122,366, and 119,691 unigenes were detected in the six libraries G1-G3 and Y1-Y3, respectively. Most of the transcripts (>84% for each library) were shared among the three biological replicates (Figure S2a,b). More than 90,000 unigenes were co-expressed in both green and yellow tissues (Figure 3A). In addition, we also found that more than 7000 unigenes were downregulated and more than 13,000 unigenes were upregulated in yellow tissues (Figure 3A). The expression of each unigene was compared among the six libraries. Consistent with PCA results, we found that the unigenes from green samples showed a relative consistent expression pattern, and the expression of these unigenes was largely different from that of the yellow tissues (Figure 3B), indicating a large variance of unigene expression between yellow and green tissues. Taken together, all these results indicated that our RNA-seq data are accurate and robust, and the bud sport mutation leads to chlorophyll deficiency, further altering a large amount of unigene expression.
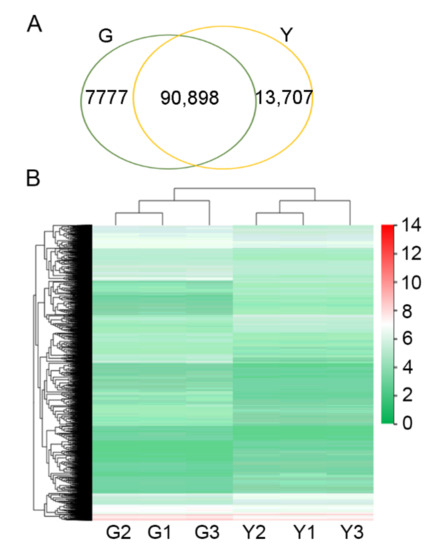
Figure 3.
Unigene expression in green and yellow tissues. (A) Venn diagrams showing genes expressed in green and yellow tissues. G: the shared genes among the three biological replicates for green tissues; Y: the shared genes among the three biological replicates for yellow tissues; (B) heat map showing the expression pattern of genes in green and yellow tissues. Expression level of genes was computed as log10-transformed values of fragments per kilobase per million reads (RPKM).
3.4. Identification and Functional Analysis of Unigenes Differentially Expressed in Green and Yellow Tissues
Upregulated unigenes in the green or yellow tissues were determined using the following criteria: unigenes with fold change (FC, RPKM in yellow tissue/RPKM in green tissue) ≥ 2 and with a Q value ≤ 0.05 are determined as upregulated in yellow tissue. Unigenes with FC ≤ 0.5 and Q value ≤ 0.05 are determined as upregulated in green tissue. Based on these criteria, a total of 4803 and 6846 unigenes were upregulated in yellow and green tissues, respectively (Figure 4A).
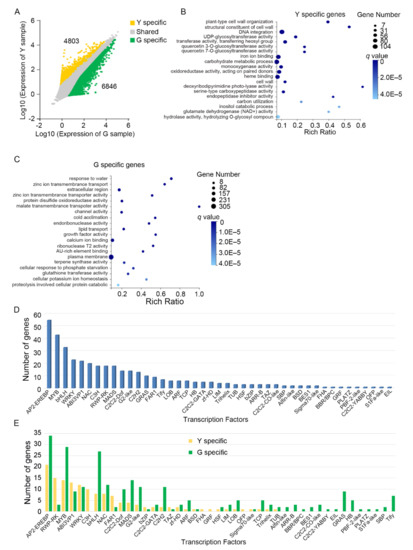
Figure 4.
Unigenes that are differentially expressed between green and yellow leaf tissues. (A) Scatter plot showing the expression pattern of genes in green and yellow tissues. Yellow color indicates the upregulated unigenes in yellow tissues, green color indicates the upregulated unigenes in green tissues, gray color indicates genes that are expressed in both green and yellow tissues. The tissue-enriched unigenes were identified by q ≤ 0.05, with |Log2(RPKM_Y/RPKM_G)| ≥ 1; (B–C) gene ontology (GO) analysis of differentially expressed unigenes in yellow tissues (B) or green tissues (C). G indicates green leaf tissue; Y indicates yellow leaf tissue; (D–E) number of tissue-enriched unigenes from different transcription factor family; (D) all unigenes that are differentially expressed in both green and yellow tissues were analyzed; (E) the unigenes that are differentially expressed in green and yellow tissues were analyzed.
To obtain a functional insight into the tissue-enriched unigenes in both types of leaf tissues, gene ontology (GO) analysis was performed and we found that GO terms for unigenes upregulated in yellow tissues are enriched in the plant development and metabolic pathways, such as carbohydrate metabolism, monooxygenase and glutamate dehydrogenase (NAD+) activity, which are associated with photosynthesis (Figure 4B). Additionally, GO terms identified in green tissues are enriched in multiple biological processes, including ion/water transport and response, stress response, and enzyme activity, which may be involved in physiological activities and responses to environmental stimuli (Figure 4C). Taken together, these results indicated that the bud sport mutation could alter the expression of unigenes that regulate the developmental processes, particularly the photosynthesis that is strictly chlorophyll dependent.
Given that transcription factors (TFs) are proteins that can bind to specific DNA sequences and regulate transcriptional cascades of thousands of genes, it is vital to gain insight into the effects of chlorophyll deficiency on these TFs in plants. Here, we examined all the TFs amongst the unigenes and found that most of the unigenes belonged to AP2-EREBP, MYB, bHLH, WRKY, ABI3VP1, and NAC families (Figure 4D). We also performed TFs enrichment analysis and found that the TFs from S1Fa-like, ABI3VP1, HB, AP2-EREBP, and zf-DB families are highly enriched (Figure S3A), indicating their important roles in response to chlorophyll deficiency in plants. We further found that the TFs from ABI3VP1, NAC, RWP-RK, and FAR1 families are highly enriched in the unigenes upregulated in yellow tissues, while bHLH, HB, C2C2-Dof, MADS, G2-like, and C2H2 families are highly enriched in green tissues (Figure 4E, Figure S3B). These TFs have been reported to bind to cis-acting elements of plant growth, developmental processes, biotic and abiotic stress responses, including nitrogen response, chloroplast division, and chlorophyll biosynthesis [28,29,30,31,32], indicating crucial and pleiotropic effects of chlorophyll deficiency in plants.
3.5. Unigenes That Respond to Chlorophyll Deficiency
In the bud sport mutation, the expression of 10 unigenes from the GRAS family was altered, of which nine unigenes showed lower expression in the yellow tissues, including the unigenes encoding scarecrow-like proteins (Figure 5A). SCARECROW has been shown to be involved in the positioning of stem cell niche in roots [33], accumulates in the ground meristem cells of developing leaf primordia, and regulates cell division in the leaves [34,35]. Thus, the decline in the average size of yellow leaves could in part be due to the lower transcriptional level of the unigenes in GRAS family (Figure 1 and Figure 5A).
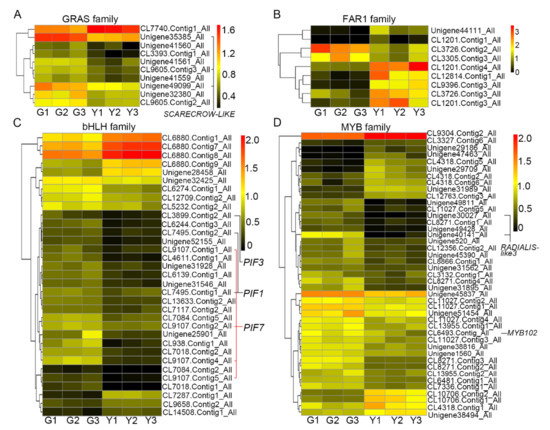
Figure 5.
Summary of unigenes that may contribute to chlorophyll deficiency. Heat map showing the expression pattern of unigenes from the GRAS (A), FAR1 (B), bHLH (C), and MYB (D) families in green and yellow tissues. G1–G3: three biological replicates for green tissues; Y1–Y3: three biological replicates for yellow tissues.
In total, nine unigenes from FAR1 TF family are differentially expressed in green or yellow leaves (Figure 5B). Candidate unigenes such as FAR-RED ELONGATED HYPOCOTYL 3 have been reported to regulate light-mediated floral meristem determinacy and shoot apical meristem maintenance in Arabidopsis [36]. Consistent with this report, our results showed that the expression of FAR-RED ELONGATED HYPOCOTYL 3 (CL3305.Contig3_All) unigene in yellow tissues was lower than that in green tissues (Figure 5B). In addition, FAR1-RELATED SEQUENCE 7, as a transcriptional repressor, negatively regulates plant development, including flowering timer determination, hypocotyl elongation, and plant immunity [37,38]. In line with these reports, our results showed that the expression of FAR1-RELATED SEQUENCE 7 (CL9396.Contig3_All) gene in yellow tissues was higher than that in green tissues (Figure 5B).
Many unigenes from the bHLH and MYB families are essential factors involved in plant growth and response to environmental stimuli, and these unigenes are highly enriched in the unigenes differentially expressed in green and yellow tissues (Figure 4D,E and Figure 5C,D). Moreover, the unigenes from both of these two TF families are also involved in the regulation of chlorophyll deficiency. Interestingly, our results showed that the expression level of PIF1/3/7 from the bHLH family (Figure 5C) and both of RADIALIS-like3 and MYB102 from the MYB family (Figure 5D) were differentially regulated, indicating that these genes could be functionally associated with the regulation of chlorophyll deficiency in chrysanthemum, or possibly affected by the chlorophyll deficiency.
To validate the expression of unigenes in green and yellow tissues, a subset of the eight tissue enriched unigenes were selected and quantified by using qRT-PCR (Figure 6A and Supplementary Figure S4). We compared the transcription profiles obtained from qRT-PCR to those generated from the RNA-seq analysis on the green and yellow tissues of chrysanthemum (Figure 6A,B). Figure 6 shows that most of these eight candidate unigenes demonstrated expression patterns consistent with the expression levels obtained from RNA-seq analysis.
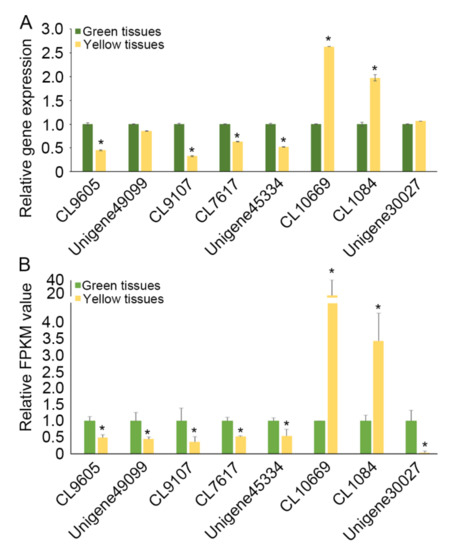
Figure 6.
Validation of unigenes that respond to chlorophyll deficiency. (A) Relative unigene expression values detected by qPCR. Relative transcript levels were normalized with CmEF1α as the standard. The green and yellow tissues from three leaves in a single plant were bulked for qRT-PCR test. Error bars indicate the standard error of the mean of three technical replicates. (B) Relative unigene expression values detected by RNA-seq. Error bars indicate the standard error of the mean of three biological replicates. * p < 0.05 when compared to the control by one-tailed unpaired t-test. Error bars represent SD.
4. Discussion
In this study, we performed transcriptome analysis to reveal the unigene expression dynamics in response to chlorophyll deficiency in chrysanthemum. Our analysis of RNA-seq data generated from the yellow and green tissues not only identified the unigenes that respond to chlorophyll deficiency, but also provided valuable knowledge on the underlying biological and physiological effects of chlorophyll deficiency in chrysanthemum.
Chlorophyll-deficient mutations are common in a wide variety of cultivated and natural plant species; a large number of chlorophyll deficient mutants have been identified in brassica napus, barley, maize, rice, and soybean [8,39,40,41,42,43]. It is of vital importance to determine the genetic loci that contribute to the formation of abnormal phenotype to fully understand the underlying mechanisms. Most of the researchers combined the quantitative trait locus (QTL) mapping or genome-wide association studies (GWAS) technique with the next generation sequencing to identify the genetic loci that contribute to the phenotype [44,45,46,47], which may be applied to identify the locus containing the bud sport mutation. Another approach to identify the somatic mutation sites is by comparison the SNPs/Indels between mutation and wild type tissues [48]. We are working on the mutation sites identification using somatic mutation mining approaches, and several candidate sites have been determined. Further research to verify the putative genetic locus that is linked to the bud sport mutation by engineered transgenes or genetic research shall be conducted in our future study.
A recent study using transcriptome analysis detected that MYB, NAC, bHLH, AP2-EREBP, WRKY, G2-like, GRAS and C2H2 TF families were differentially expressed between chlorophyll b-deficient mutant and wild type control in rice [41]. In Areca catechu L. trees, bHLH, WRKY, and NAC TFs were found to be differentially expressed in response to iron deficiency induced chlorosis [49]. In addition to these findings, we also found that RWP-RKs and C3Hs TFs are specifically enriched in the yellow or green tissues. RWP-RKs are plant-specific TFs and are key regulators that bind to cis-acting elements of nitrogen use efficiency-related genes and genes responsible for gametogenesis and embryogenesis [29,31]. C3Hs are characterized by their signature motif of three cysteine and one histidine residue, and play important roles in regulation of plant growth, developmental processes, and environmental responses [30]. Due to their vital roles in maintaining plant growth, it is reasonable to observe that these TFs are highly enriched in response to chlorophyll deficiency. In the future, experiments related to cloning and functional verification of these TFs in response to chrysanthemum chlorophyll deficiency are needed to further identify the underlying molecular mechanism.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/horticulturae8010014/s1, Figure S1: Number of raw reads and clean reads for each library, Figure S2: Unigene expression in green and yellow tissues. (A,B) Venn diagrams showing unigenes expressed in green (A) and yellow (B) tissues. G1-G3: three biological replicates for green leaf; Y1-Y3: three biological replicates for yellow leaf, Figure S3: TFs enrichment analysis. (A) The TFs enrichments in all unigenes that differentially expressed in both green and yellow tissues. (B) The TFs enrichments in the unigenes that differentially expressed in green and yellow tissues, separately. TFs enrichments were calculated by the (number of TFs in the unigenes)/(number of TFs in one TF family), Figure S4: qPCR Validation of unigenes that respond to chlorophyll deficiency using CmUbituitin as control. * p < 0.05 when compare to the control by one-tailed unpaired t-test. Error bars represent SD. Table S1: Primers used for qRT-PCR, Table S2: Sequence information of unigenes examined by qRT-PCR. Primers were highlighted. Table S3: The original Fv/Fm values were obtained by PAM fluorimeter (Imaging-PAM; WALZ, Effeltrich, Germany).
Author Contributions
Conceptualization, S.C., F.C., L.W. and J.J.; investigation, G.S., R.L., Z.Q., H.Z., Q.H. and Y.Z.; supervision, S.C., F.C., L.W. and J.J.; writing—original draft, H.Z.; writing—review and editing, L.W. and J.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Acknowledgments
We thank Zhengyao Shao (Department of Molecular Biosciences, The University of Texas at Austin) for comments and critical reading. We thank Yuehua Ma (Central laboratory of College of Horticulture, Nanjing Agricultural University) for assistance in using pulse amplitude modulated (PAM) fluorimeter (Imaging-PAM; WALZ, Germany).
Conflicts of Interest
The authors declare no competing interests.
References
- Chang, Q.; Chen, S.; Chen, Y.; Deng, Y.; Chen, F.; Zhang, F.; Wang, S. Anatomical and physiological differences and differentially expressed genes between the green and yellow leaf tissue in a variegated chrysanthemum variety. Mol. Biotechnol. 2013, 54, 393–411. [Google Scholar] [CrossRef] [PubMed]
- Imsande, J. Iron, sulfur, and chlorophyll deficiencies: A need for an integrative approach in plant physiology. Physiol. Plant. 1998, 103, 139–144. [Google Scholar] [CrossRef]
- Li, Z.; Peng, J.; Wen, X.; Guo, H. Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 2013, 25, 3311–3328. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Yang, C.; Gao, S.; Zhang, W.; Li, L.; Kuai, B. Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 2014, 7, 1776–1787. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Dai, S.; Zhang, Z.L.; Lao, W.; Wang, R.; Meng, X.; Zhou, X. Ethylene and salicylic acid synergistically accelerate leaf senescence in Arabidopsis. J. Integr. Plant Biol. 2021, 63, 828–833. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Xu, Y.; Yan, S. Salicylic acid and ethylene coordinately promote leaf senescence. J. Integr. Plant Biol. 2021, 63, 823–827. [Google Scholar] [CrossRef]
- Li, N.; Jia, J.; Xia, C.; Liu, X.; Kong, X. Characterization and mapping of novel chlorophyll deficient mutant genes in durum wheat. Breed. Sci. 2013, 63, 169–175. [Google Scholar] [CrossRef]
- Shi, D.; Zheng, X.; Li, L.; Lin, W.; Xie, W.; Yang, J.; Chen, S.; Jin, W. Chlorophyll deficiency in the maize elongated mesocotyl2 mutant is caused by a defective heme oxygenase and delaying grana stacking. PLoS ONE 2013, 8, e80107. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Gao, J.; Yao, L.; Ren, G.; Zhu, X.; Gao, S.; Qiu, K.; Zhou, X.; Kuai, B. The role of ANAC072 in the regulation of chlorophyll degradation during age- and dark-induced leaf senescence. Plant Cell Rep. 2016, 35, 1729–1741. [Google Scholar] [CrossRef]
- Niu, F.; Cui, X.; Zhao, P.; Sun, M.; Yang, B.; Deyholos, M.K.; Li, Y.; Zhao, X.; Jiang, Y.Q. WRKY42 transcription factor positively regulates leaf senescence through modulating SA and ROS synthesis in Arabidopsis thaliana. Plant J. 2020, 104, 171–184. [Google Scholar] [CrossRef]
- Piao, W.; Kim, S.H.; Lee, B.D.; An, G.; Sakuraba, Y.; Paek, N.C. Rice transcription factor OsMYB102 delays leaf senescence by down-regulating abscisic acid accumulation and signaling. J. Exp. Bot. 2019, 70, 2699–2715. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xin, J.; Liu, L.; Song, A.; Guan, Z.; Fang, W.; Chen, F. A temporal gene expression map of Chrysanthemum leaves infected with Alternaria alternata reveals different stages of defense mechanisms. Hortic. Res. 2020, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Guan, Y.; Liu, Y.; Zhang, Z.; Jaffar, M.A.; Song, A.; Chen, S.; Jiang, J.; Chen, F. Regulation of flowering time in chrysanthemum by the R2R3 MYB transcription factor CmMYB2 is associated with changes in gibberellin metabolism. Hortic. Res. 2020, 7, 96. [Google Scholar] [CrossRef] [PubMed]
- Doi, M.; Nakagawa, Y.; Watabe, S.; Aoe, K.; Inamoto, K.; Imanishi, H. Ethylene-induced Leaf Yellowing in Cut Chrysanthemums (Dendranthema grandiflora Kitamura). J. Jpn. Soc. Hortic. Sci. 2003, 72, 533–535. [Google Scholar] [CrossRef][Green Version]
- Doi, M.; Aoe, K.; Watabe, S.; Inamoto, K.; Imanishi, H. Leaf Yellowing of Cut Standard Chrysanthemum (Dendranthema grandiflora Kitamura) ‘Shuho-no-chikara’ Induced by Ethylene and the Postharvest Increase in Ethylene Sensitivity. J. Jpn. Soc. Hortic. Sci. 2004, 73, 229–234. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef]
- Ahammed, G.J.; Ruan, Y.-P.; Zhou, J.; Xia, X.-J.; Shi, K.; Zhou, Y.-H.; Yu, J.-Q. Brassinosteroid alleviates polychlorinated biphenyls-induced oxidative stress by enhancing antioxidant enzymes activity in tomato. Chemosphere 2013, 90, 2645–2653. [Google Scholar] [CrossRef] [PubMed]
- Kramer, D.M.; Johnson, G.; Kiirats, O.; Edwards, G.E. New Fluorescence Parameters for the Determination of QA Redox State and Excitation Energy Fluxes. Photosynth. Res. 2004, 79, 209. [Google Scholar] [CrossRef]
- Cheng, P.; Liu, Y.; Yang, Y.; Chen, H.; Cheng, H.; Hu, Q.; Zhang, Z.; Gao, J.; Zhang, J.; Ding, L.; et al. CmBES1 is a regulator of boundary formation in chrysanthemum ray florets. Hortic. Res. 2020, 7, 129. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gao, T.; Li, P.; Tian, C.; Song, A.; Jiang, J.; Guan, Z.; Fang, W.; Chen, F.; Chen, S. Chrysanthemum CmWRKY53 negatively regulates the resistance of chrysanthemum to the aphid Macrosiphoniella sanborni. Hortic. Res. 2020, 7, 109. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Zhao, K.; Zhang, X.; Song, A.; Su, J.; Hu, Y.; Zhao, W.; Jiang, J.; Chen, F. Comprehensive characterization of a floral mutant reveals the mechanism of hooked petal morphogenesis in Chrysanthemum morifolium. Plant Biotechnol. J. 2019, 17, 2325–2340. [Google Scholar] [CrossRef] [PubMed]
- Sipka, G.B.; Magyar, M.; Mezzetti, A.; Akhtar, P.; Zhu, Q.; Xiao, Y.; Han, G.; Santabarbara, S.; Shen, J.R.; Lambrev, P.H.; et al. Light-adapted charge-separated state of photosystem II: Structural and functional dynamics of the closed reaction center. Plant Cell 2021, 33, 1286–1302. [Google Scholar] [CrossRef]
- Wu, H.; Roy, S.; Alami, M.; Green, B.R.; Campbell, D.A. Photosystem II photoinactivation, repair, and protection in marine centric diatoms. Plant Physiol. 2012, 160, 464–476. [Google Scholar] [CrossRef]
- Son, K.; Yu, S.; Shin, W.; Han, K.; Kang, K. A Simple Guideline to Assess the Characteristics of RNA-Seq Data. Biomed. Res. Int. 2018, 2018, 2906292. [Google Scholar] [CrossRef]
- Zhang, N.; Zhou, S.; Yang, D.; Fan, Z. Revealing Shared and Distinct Genes Responding to JA and SA Signaling in Arabidopsis by Meta-Analysis. Front. Plant Sci. 2020, 11, 908. [Google Scholar] [CrossRef]
- Dietz, K.J.; Vogel, M.O.; Viehhauser, A. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 2010, 245, 3–14. [Google Scholar] [CrossRef]
- Chardin, C.; Girin, T.; Roudier, F.; Meyer, C.; Krapp, A. The plant RWP-RK transcription factors: Key regulators of nitrogen responses and of gametophyte development. J. Exp. Bot. 2014, 65, 5577–5587. [Google Scholar] [CrossRef]
- Liu, C.; Xu, X.; Kan, J.; Cheng, Z.M.; Chang, Y.; Lin, J.; Li, H. Genome-wide analysis of the C3H zinc finger family reveals its functions in salt stress responses of Pyrus betulaefolia. PeerJ 2020, 8, e9328. [Google Scholar] [CrossRef]
- Yin, M.; Zhang, Z.; Xuan, M.; Feng, H.; Ye, W.; Zheng, X.; Wang, Y. Conserved Subgroups of the Plant-Specific RWP-RK Transcription Factor Family Are Present in Oomycete Pathogens. Front. Microbiol. 2020, 11, 1724. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.; Fan, C.; Wei, Y.; Meng, J.; Li, Z.; Zhong, C. Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia. BMC Plant Biol. 2021, 21, 328. [Google Scholar] [CrossRef]
- Sabatini, S.; Heidstra, R.; Wildwater, M.; Scheres, B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003, 17, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Dhondt, S.; Coppens, F.; De Winter, F.; Swarup, K.; Merks, R.M.; Inze, D.; Bennett, M.J.; Beemster, G.T. SHORT-ROOT and SCARECROW regulate leaf growth in Arabidopsis by stimulating S-phase progression of the cell cycle. Plant Physiol. 2010, 154, 1183–1195. [Google Scholar] [CrossRef] [PubMed]
- Hughes, T.E.; Sedelnikova, O.V.; Wu, H.; Becraft, P.W.; Langdale, J.A. Redundant SCARECROW genes pattern distinct cell layers in roots and leaves of maize. Development 2019, 146, dev177543. [Google Scholar] [CrossRef]
- Li, D.; Fu, X.; Guo, L.; Huang, Z.; Li, Y.; Liu, Y.; He, Z.; Cao, X.; Ma, X.; Zhao, M.; et al. FAR-RED ELONGATED HYPOCOTYL3 activates SEPALLATA2 but inhibits CLAVATA3 to regulate meristem determinacy and maintenance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 9375–9380. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Tang, W.; Ma, T.; Niu, D.; Jin, J.B.; Wang, H.; Lin, R. A pair of light signaling factors FHY3 and FAR1 regulates plant immunity by modulating chlorophyll biosynthesis. J. Integr. Plant Biol. 2016, 58, 91–103. [Google Scholar] [CrossRef]
- Ritter, A.; Inigo, S.; Fernandez-Calvo, P.; Heyndrickx, K.S.; Dhondt, S.; Shi, H.; De Milde, L.; Vanden Bossche, R.; De Clercq, R.; Eeckhout, D.; et al. The transcriptional repressor complex FRS7-FRS12 regulates flowering time and growth in Arabidopsis. Nat. Commun. 2017, 8, 15235. [Google Scholar] [CrossRef]
- Falbel, T.G.; Staehelin, L.A. Characterization of a Family of Chlorophyll-Deficient Wheat (Triticum) and Barley (Hordeum vulgare) Mutants with Defects in the Magnesium-Insertion Step of Chlorophyll Biosynthesis. Plant Physrol. 1994, 104, 639–648. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, D.; Han, S.; Zhang, X.; Yu, D. Identification and gene mapping of a soybean chlorophyll-deficient mutant. Plant Breed. 2011, 130, 133–138. [Google Scholar] [CrossRef]
- Nguyen, M.K.; Shih, T.-H.; Lin, S.-H.; Lin, J.-W.; Nguyen, H.C.; Yang, Z.-W.; Yang, C.-M. Transcription Profile Analysis of Chlorophyll Biosynthesis in Leaves of Wild-Type and Chlorophyll b-Deficient Rice (Oryza sativa L.). Agriculture 2021, 11, 401. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, W.; Xie, Y.; Lu, W.; Zhang, R. Comparative proteomics of thylakoid membrane from a chlorophyll b-less rice mutant and its wild type. Plant Sci. 2007, 173, 397–407. [Google Scholar] [CrossRef]
- Chu, P.; Yan, G.X.; Yang, Q.; Zhai, L.N.; Zhang, C.; Zhang, F.Q.; Guan, R.Z. iTRAQ-based quantitative proteomics analysis of Brassica napus leaves reveals pathways associated with chlorophyll deficiency. J. Proteom. 2015, 113, 244–259. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Yang, X.; Zhang, F.; Wu, S.; Xiong, S.; Shi, L.; Guan, Z.; Fang, W.; Chen, F. Dynamic and epistatic QTL mapping reveals the complex genetic architecture of waterlogging tolerance in chrysanthemum. Planta 2018, 247, 899–924. [Google Scholar] [CrossRef]
- Chong, X.; Su, J.; Wang, F.; Wang, H.; Song, A.; Guan, Z.; Fang, W.; Jiang, J.; Chen, S.; Chen, F.; et al. Identification of favorable SNP alleles and candidate genes responsible for inflorescence-related traits via GWAS in chrysanthemum. Plant Mol. Biol. 2019, 99, 407–420. [Google Scholar] [CrossRef]
- Su, J.; Zhang, F.; Chong, X.; Song, A.; Guan, Z.; Fang, W.; Chen, F. Genome-wide association study identifies favorable SNP alleles and candidate genes for waterlogging tolerance in chrysanthemums. Hortic. Res. 2019, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Fang, X.; Su, J.; Ding, L.; Guan, Z.; Jiang, J.; Chen, S.; Chen, F.; Fang, W.; Zhang, F. Genetic dissection of floral traits in anemone-type chrysanthemum by QTL mapping. Mol. Breed. 2019, 39, 136. [Google Scholar] [CrossRef]
- Liu, Y.; Loewer, M.; Aluru, S.; Schmidt, B. SNVSniffer: An integrated caller for germline and somatic single-nucleotide and indel mutations. BMC Syst. Biol. 2016, 10, 47. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, J.; Cao, X.; Jia, X.; Liu, L.; Cao, H.; Qin, W.; Li, M. Iron Deficiency Leads to Chlorosis Through Impacting Chlorophyll Synthesis and Nitrogen Metabolism in Areca catechu L. Front. Plant Sci. 2021, 12, 710093. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).