Identification and Quantification of Naphthoquinones and Other Phenolic Compounds in Leaves, Petioles, Bark, Roots, and Buds of Juglans regia L., Using HPLC-MS/MS
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Extraction of the Individual Phenolic Compounds
2.3. HPLC–Mass Spectrometry Analysis of the Individual Phenolic Compounds
2.4. Chemicals
2.5. Statistical Analysis
3. Results and Discussion
3.1. Identification of Individual Phenolic Compounds in Walnut
3.2. Quantification of Individual Phenolic Compounds in Walnut
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HHDP | hexahydroxydiphenoyl |
| (U)HPLC | (ultra)high performance liquid chromatography |
| MS(/MS) | (tandem) mass spectrometer |
References
- Medic, A.; Jakopic, J.; Hudina, M.; Solar, A.; Veberic, R. Identification and quantification of the major phenolic constituents in Juglans regia L. peeled kernels and pellicles, using HPLC–MS/MS. Food Chem. 2021, 352, 129404. [Google Scholar] [CrossRef]
- Jahanban-Esfahlan, A.; Ostadrahimi, A.; Tabibiazar, M.; Amarowicz, R. A Comparative review on the extraction, antioxidant content and antioxidant potential of different parts of walnut (Juglans regia L.) fruit and tree. Molecules 2019, 24, 2133. [Google Scholar] [CrossRef] [Green Version]
- Martins, S.; Mussatto, S.I.; Martínez-Avila, G.; Montañez-Saenz, J.; Aguilar, C.N.; Teixeira, J.A. Bioactive phenolic compounds: Production and extraction by solid-state fermentation. A review. Biotechnol. Adv. 2011, 29, 365–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu, D.C.; Vo, P.H.; Coggeshall, M.V.; Lin, C.-H. Identification and characterization of phenolic compounds in black walnut kernels. J. Agric. Food Chem. 2018, 66, 4503–4511. [Google Scholar] [CrossRef]
- Medic, A.; Jakopic, J.; Solar, A.; Hudina, M.; Veberic, R. Walnut (J. regia) agro-residues as a rich source of phenolic compounds. Biology 2021, 10, 535. [Google Scholar] [CrossRef] [PubMed]
- Medic, A.; Solar, A.; Hudina, M.; Veberic, R. Phenolic response to walnut anthracnose (Ophiognomonia leptostyla) infection in different parts of juglans regia husks, using HPLC-MS/MS. Agriculture 2021, 11, 659. [Google Scholar] [CrossRef]
- Nour, V.; Trandafir, I.; Cosmulescu, S. Optimization of ultrasound-assisted hydroalcoholic extraction of phenolic compounds from walnut leaves using response surface methodology. Pharm. Biol. 2016, 54, 2176–2187. [Google Scholar] [CrossRef] [Green Version]
- Bruneton, J. Pharmacognosy, Phytochemistry, Medicinal Plants; Lavoisier Publishing: Paris, France, 1995; p. 15 + 915. [Google Scholar]
- Hadis, E.; Mostafa, H.-Z.S.; Saeedeh, N.; Mehdi, M. Study of the effects of walnut leaf on the levels of a number of Blood Biochemical Factors in normal male rats fed with high cholesterol diet. Clin. Biochem. 2011, 44, S331. [Google Scholar] [CrossRef]
- Hosseini, S.; Huseini, H.F.; Larijani, B.; Mohammad, K.; Najmizadeh, A.; Nourijelyani, K.; Jamshidi, L. The hypoglycemic effect of Juglans regia leaves aqueous extract in diabetic patients: A first human trial. DARU J. Pharm. Sci. 2014, 22, 19. [Google Scholar] [CrossRef] [Green Version]
- Hosseini, S.; Jamshidi, L.; Mehrzadi, S.; Mohammad, K.; Najmizadeh, A.R.; Alimoradi, H.; Huseini, H.F. Effects of Juglans regia L. leaf extract on hyperglycemia and lipid profiles in type two diabetic patients: A randomized double-blind, placebo-controlled clinical trial. J. Ethnopharmacol. 2014, 152, 451–456. [Google Scholar] [CrossRef]
- Pitschmann, A.; Zehl, M.; Atanasov, A.G.; Dirsch, V.M.; Heiss, E.; Glasl, S. Walnut leaf extract inhibits PTP1B and enhances glucose-uptake in vitro. J. Ethnopharmacol. 2014, 152, 599–602. [Google Scholar] [CrossRef]
- Seabra, I.J.; Braga, M.E.M.; Oliveira, R.A.; de Sousa, H.C. Two-step high pressure solvent extraction of walnut (Juglans regia L.) husks: scCO2 + CO2/ethanol/H2O. J. CO2 Util. 2019, 34, 375–385. [Google Scholar] [CrossRef]
- Câmara, C.R.S.; Schlegel, V. A review on the potential human health benefits of the black walnut: A comparison with the english walnuts and other tree nuts. Int. J. Food Prop. 2016, 19, 2175–2189. [Google Scholar] [CrossRef]
- Kumagai, Y.; Shinkai, Y.; Miura, T.; Cho, A.K. The chemical biology of naphthoquinones and its environmental implications. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 221–247. [Google Scholar] [CrossRef] [PubMed]
- Wyrepkowski, C.C.; Gomes da Costa, D.L.M.; Sinhorin, A.P.; Vilegas, W.; De Grandis, R.A.; Resende, F.A.; Varanda, E.A.; Dos Santos, L.C. Characterization and quantification of the compounds of the ethanolic extract from Caesalpinia ferrea stem bark and evaluation of their mutagenic activity. Molecules 2014, 19, 16039–16057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huo, J.-H.; Du, X.-W.; Sun, G.-D.; Dong, W.-T.; Wang, W.-M. Identification and characterization of major constituents in Juglans mandshurica using ultra performance liquid chromatography coupled with time-of-flight mass spectrometry (UPLC-ESI-Q-TOF/MS). Chin. J. Nat. Med. 2018, 16, 525–545. [Google Scholar] [CrossRef]
- Ambigaipalan, P.; Oh, W.Y.; Shahidi, F. Epigallocatechin (EGC) esters as potential sources of antioxidants. Food Chem. 2020, 309, 125609. [Google Scholar] [CrossRef]
- Vieira, V.; Pereira, C.; Pires, T.C.S.P.; Calhelha, R.C.; Alves, M.J.; Ferreira, O.; Barros, L.; Ferreira, I.C.F.R. Phenolic profile, antioxidant and antibacterial properties of Juglans regia L. (walnut) leaves from the Northeast of Portugal. Ind. Crops Prod. 2019, 134, 347–355. [Google Scholar] [CrossRef]
- Santos, A.; Barros, L.; Calhelha, R.C.; Dueñas, M.; Carvalho, A.M.; Santos-Buelga, C.; Ferreira, I.C.F.R. Leaves and decoction of Juglans regia L.: Different performances regarding bioactive compounds and in vitro antioxidant and antitumor effects. Ind. Crops Prod. 2013, 51, 430–436. [Google Scholar] [CrossRef]
- Tsugawa, H.; Nakabayashi, R.; Mori, T.; Yamada, Y.; Takahashi, M.; Rai, A.; Sugiyama, R.; Yamamoto, H.; Nakaya, T.; Yamazaki, M.; et al. A cheminformatics approach to characterize metabolomes in stable-isotope-labeled organisms. Nat. Methods 2019, 16, 295–298. [Google Scholar] [CrossRef]
- Sina Niculina, C.; Ion, T.; Gheorghe, A.; Adrian, B. Juglone content in leaf and green husk of five walnut (Juglans regia L.) cultivars. Not. Bot. Horti Agrobot. Cluj-Napoca 2011, 39, 237–240. [Google Scholar] [CrossRef] [Green Version]
- Chong, K.P.; Atong, M.; Rossall, S. The roles of syringic, caffeic and 4-hydroxybenzoicacids in ganoderma-oil palm interaction. Asian J. Microbiol. Biotechnol. Environ. Sci. 2012, 14, 157–166. [Google Scholar]
- Solar, A.; Colarič, M.; Usenik, V.; Stampar, F. Seasonal variations of selected flavonoids, phenolic acids and quinones in annual shoots of common walnut (Juglans regia L.). Plant Sci. 2006, 170, 453–461. [Google Scholar] [CrossRef]
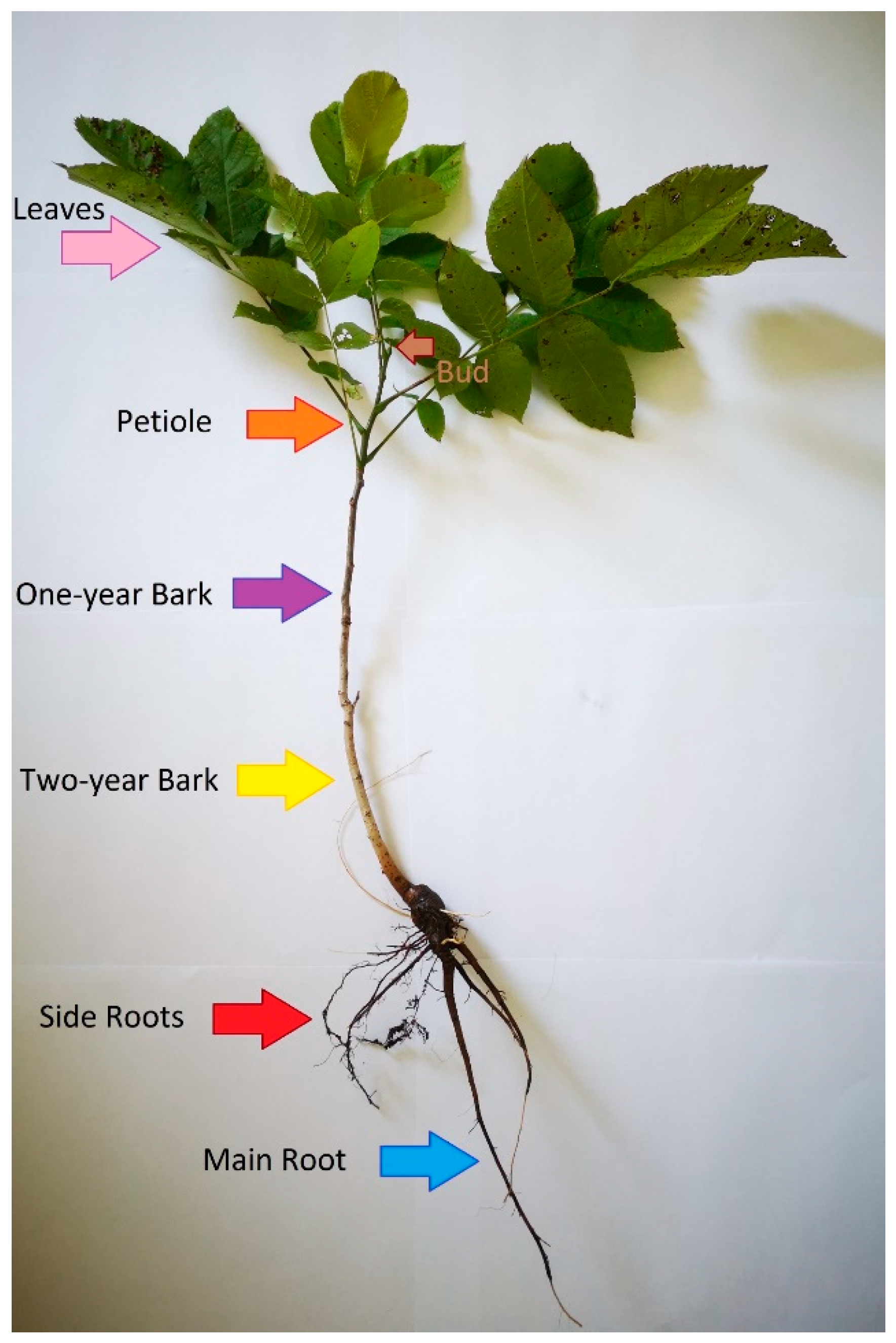
| Compound | Rt | [M-H]− | MS2 | MS3 | MS4 | Plant Tissue | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| (min) | (m/z) | (m/z) | (m/z) | (m/z) | Leaves | Petioles | Bark | Roots | Buds | |
| Bis-HHDP-glucose 1 | 8.45 | 783 | 301, 481, 275, 257 | 257, 229, 185 | x | |||||
| 301, 481, 275, 257 | 257, 231, 203, 247 | 185, 229, 213, 20, 157 | ||||||||
| Epigallocatechin | 8.46 | 305 | 179, 221, 125 | 165, 151, 137, 109 | x | x | x | x | ||
| (epi)Catechin derivative 1 | 9.04 | 593 | 425, 289, 407 | 289 | 245, 205, 179, 125 | x | ||||
| Procyanidin dimer 1 | 9.58 | 577 | 425, 407, 289 | x | x | |||||
| 1-O-(4-Hydroxy-3,5-dimetoxybenzoyl)-D-glucopyranoside | 9.69 | 359 | 197, 239, 299 | 153, 181, 121 | x | x | ||||
| Neochlorogenic acid (3-Caffeoylquinic acid) | 9.74 | 353 | 191, 179, 135 | x | x | x | ||||
| Procyanidin dimer 2 | 10.69 | 577 | 425, 407, 289 | x | x | |||||
| Bis-HHDP-glucose 2 | 10.73 | 783 | 257, 231, 203, 247 | x | x | x | ||||
| (epi)Catechin isomer | 10.76 | 289 | 245, 205, 179, 125 | x | ||||||
| Gallic acid derivative 1 | 11.01 | 483 | 313, 271, 169 | x | ||||||
| Gallic acid derivative 2 | 11.19 | 411 | 169, 241 | 169, 125 | x | |||||
| Gallic acid derivative 3 | 11.56 | 483 | 313, 271, 169 | x | ||||||
| Ellagic acid derivative 1 | 11.68 | 533 | 511, 420, 442 | 502, 275, 301 | 420, 442, 275, 301 | x | ||||
| 502, 275, 301 | 257, 231, 203, 247 | |||||||||
| Tellimagrandin isomer (digalloyl-HHDP-glucose) 1 | 11.95 | 785 | 301, 275 | 257, 229, 185 | x | |||||
| Procyanidin dimer 3 | 12.01 | 577 | 425, 407, 289 | x | x | x | x | |||
| 3-p-Coumaroylquinic acid | 12.36 | 337 | 163, 191, 119, 173 | x | x | x | ||||
| Strictin/isostrictin isomer (galloyl-HHDP-glucose) | 12.47 | 633 | 301, 257, 275, 229, 185 | x | x | |||||
| Cryptochlorogenic acid (4-caffeoylquinic acid) | 12.58 | 353 | 191, 179, 135 | x | x | x | ||||
| (+)Catechin | 12.88 | 289 | 245, 205, 179, 125 | x | x | x | x | x | ||
| Ellagic acid derivative 2 | 13.25 | 467 | 458, 391, 275, 169 | 382, 299, 169 | x | |||||
| 458, 391, 275, 169 | 257, 231, 203, 247 | |||||||||
| Dihydroxytetralone hexoside | 13.27 | 339 | 159, 177 | x | x | x | x | |||
| Hydrojuglone derivative 1 | 13.59 | 401 | 355, 193 | 193 | 175 | x | x | x | x | x |
| Chlorogenic acid (5-caffeoylquinic acid) | 13.86 | 353 | 191, 179, 135 | x | x | |||||
| (epi)Catechin dihexoside | 14.23 | 613 | 603, 458, 301, 289 | 289 | 245, 205, 179, 125 | x | ||||
| Hydrojuglone dihexoside | 14.46 | 499 | 175, 337 | 131, 157, 147, 103 | x | |||||
| Hydrojuglone hexoside derivative | 14.70 | 355 | 193, 319, 175 | 175 | 131, 157, 147, 103 | x | x | |||
| Ellagic acid derivative 3 | 14.75 | 482 | 445, 467, 275, 301 | x | x | |||||
| 5-Hydroxy-2,3-dihydro-1,4-napthalenedione | 14.92 | 175 | 131, 157, 147, 103 | x | x | |||||
| (-)Epicatechin | 15.32 | 289 | 245, 205, 179, 125 | x | ||||||
| Ellagic acid derivative 4 | 15.54 | 661 | 301, 275 | 257, 229, 185 | x | |||||
| Ellagic acid derivative 5 | 15.63 | 467 | 391, 301 | 301, 275 | 257, 229, 185 | x | x | x | ||
| Procyanidin dimer 4 | 15.64 | 577 | 425, 407, 289 | x | ||||||
| Tellimagrandin isomer (digalloyl-HHDP-glucose) 2 | 15.88 | 785 | 301, 275 | 257, 229, 185 | x | |||||
| p-Coumaroylquinic acid | 16.01 | 337 | 163, 191, 119, 173 | x | x | x | ||||
| Myricetin galloyl hexoside | 16.25 | 631 | 479 | 316, 317 | x | |||||
| Trigalloyl-glucose isomer | 16.47 | 635 | 465, 313 | 313, 295, 169, 271 | x | |||||
| Hydrojuglone-β-D-glucopyranoside | 17.39 | 337 | 175 | 131, 157, 147, 103 | x | x | x | x | x | |
| Myricetin-3-galactoside | 17.73 | 479 | 316, 317 | 179, 151 | x | x | x | |||
| Hydrojuglone derivative 2 | 18.00 | 451 | 301, 325, 319, 193 | 215, 257, 283, 175 | 147, 131, 103, 157 | x | x | x | ||
| 301, 325, 319, 193 | 192, 235, 177 | |||||||||
| Ellagic acid pentoside | 18.08 | 433 | 301 | 257, 229, 185 | x | |||||
| Hydrojuglone derivative 3 | 18.17 | 465 | 301, 193, 151, 319 | 215, 257, 283, 175 | 186, 171, 143 | x | x | |||
| 215, 257, 283, 175 | 147, 131, 103, 157 | |||||||||
| Myricetin-3-glucoside | 18.21 | 479 | 316, 317 | 179, 151 | x | |||||
| Gallic acid derivative 4 | 18.50 | 491 | 271, 331 | x | ||||||
| Gallic acid derivative 5 | 18.50 | 469 | 393, 169, 301, 275 | x | ||||||
| Gallic acid derivative 6 | 18.51 | 475 | 313, 271, 169 | 169, 125 | x | |||||
| Hydrojuglone derivative pentoside 1 | 18.66 | 435 | 303, 285 | x | x | |||||
| Quercetin galloyl hexoside | 18.93 | 615 | 463, 301 | 301 | 179, 151 | x | ||||
| Hydrojuglone derivative 4 | 19.21 | 465 | 301, 193, 151, 319 | x | ||||||
| Tetralone hexoside | 19.48 | 491 | 271, 331 | x | x | x | x | x | ||
| Gallic acid derivative 7 | 19.94 | 475 | 313, 271, 169 | x | x | x | ||||
| Myricetin pentoside | 19.96 | 449 | 317, 316 | 179, 151 | x | |||||
| Myricetin-3-rhamnoside | 20.12 | 463 | 316, 317 | 179, 151 | x | x | x | x | ||
| Quercetin-3-galactoside | 20.36 | 463 | 301 | 179, 151 | x | x | x | x | ||
| Ellagic acid | 20.54 | 301 | 257, 229, 185 | x | x | |||||
| Quercetin-3-glucoside | 20.59 | 463 | 301 | 179, 151 | x | x | x | |||
| (epi)Catechin galloyl | 20.73 | 441 | 289 | 245, 205, 179, 125 | x | x | ||||
| Gallic acid derivative 8 | 20.94 | 469 | 393, 169 | 317, 169, 125 | x | |||||
| Trihydroxytetralone galloyl hexoside | 20.97 | 507 | 331, 271 | x | x | |||||
| Digalloylgallate | 21.18 | 473 | 313, 271, 169 | 169, 125 | x | |||||
| Gallic acid derivative 9 | 21.23 | 489 | 271, 313, 169 | x | ||||||
| Hydrojuglone derivative rhamnoside | 21.26 | 449 | 303, 285 | x | x | x | ||||
| Gallic acid derivative 10 | 21.43 | 489 | 271, 313, 169 | x | x | x | ||||
| Quercetin-3-xyloside | 21.43 | 433 | 301 | 179, 151 | x | |||||
| Hydrojuglone derivative pentoside 2 | 21.56 | 435 | 303, 285 | x | x | x | x | x | ||
| Kaempferol-3-glucoside | 21.67 | 447 | 284, 285 | 255, 227, 151 | x | x | ||||
| Quercetin-3-arabinopyranoside | 21.77 | 433 | 301 | 179, 151 | x | x | x | x | ||
| 3-O-Methylellagic acid-4-O-β-D-arabinopyranoside | 22.01 | 447 | 315 | 300 | x | x | ||||
| Quercetin-3-arabinofuranoside | 22.18 | 433 | 301 | 179, 151 | x | x | ||||
| Gallic acid derivative 11 | 22.28 | 489 | 271, 313, 169 | x | x | x | ||||
| Quercetin-3-rhamnoside | 22.42 | 447 | 301 | 179, 151 | x | x | x | x | ||
| Kaempferol-7-hexoside 1 | 22.78 | 447 | 285 | 165, 119 | x | |||||
| Kaempferol-pentoside 1 | 22.91 | 417 | 284, 285 | 255, 227, 151 | x | x | ||||
| Kaempferol-7-hexoside 2 | 23.12 | 447 | 285 | 165, 119 | x | |||||
| p-Coumaric acid hexoside derivative 1 | 23.13 | 487 | 325 | 307 | 145, 163, 235 | x | x | |||
| Gallic acid derivative 12 | 23.31 | 489 | 337, 301, 313, 271 | 317, 229, 187, 247 | x | |||||
| Dihydrokaempferol pentoside 1 | 23.21 | 419 | 287, 269, 259, 179 | 259, 243, 201, 125 | x | |||||
| Dihydrokaempferol pentoside 2 | 23.56 | 419 | 287, 269, 259, 179 | 259, 243, 201, 125 | x | |||||
| 1-β-D-Glucopyranosyloxy-4,8-dihydroxy-2-napthoic acid | 23.64 | 381 | 218, 229, 247, 175 | 173 | x | |||||
| 218, 229, 247, 175 | 131, 103, 147, 157 | |||||||||
| Kaempferol-pentoside 2 | 23.84 | 417 | 285, 284 | 257, 267, 241, 229, 151 | x | x | ||||
| Kaempferol-3-rhamnoside | 24.15 | 431 | 285, 284 | 257, 267, 241, 229, 151 | x | |||||
| 1,4,8-trihydroxynapthalene-1-D-glucopyranoside | 24.34 | 503 | 327, 285, 217, 229, 175 | 131, 103, 147, 157 | x | |||||
| Isofraxidin | 24.50 | 221 | 206, 191 | 191, 177 | 163, 135 | x | ||||
| Isofraxidin derivative | 24.64 | 265 | 221 | 206, 191 | x | |||||
| Caffeic acid hexoside derivative | 25.06 | 517 | 341, 371, 281, 209, 251 | x | x | |||||
| Hydrojuglone derivative 5 | 25.51 | 601 | 285, 303 | 241, 175 | x | x | ||||
| p-Coumaric acid hexoside derivative 2 | 26.25 | 485 | 325 | 235, 163 | x | |||||
| Hydrojuglone derivative 6 | 26.73 | 517 | 175 | 131, 157, 103, 147 | x | x | ||||
| Hydrojuglone dihexoside derivative | 27.08 | 515 | 355, 193 | 193 | 175 | x | ||||
| 1,4-Napthoquinone | 28.33 | 173 | 111, 155, 129, 145 | x | x | x | x | x | ||
| Hydrojuglone | 28.33 | 175 | 131, 147, 157, 115, 103 | x | x | x | x | x | ||
| Juglone (5-hydroxy-1,4-napthoquinone) | 29.58 | 189 | 161 | 117, 133 | x | x | x | x | x | |
| Compound | Content in Plant Tissue (g/kg Dry Weight) | ||||||
|---|---|---|---|---|---|---|---|
| Leaves | Petioles | Bark | Roots | Bud | |||
| One-year | Two-Year | Side | Main | ||||
| Hydroxycinnamic acids | |||||||
| Neochlorogenic acid (3-caffeoylquinic acid) 1 | 2.01 ± 0.14 b | 0.41 ± 0.03 a | nd | nd | nd | nd | 0.76 ± 0.05 a |
| Cryptochlorogenic acid (4-caffeoylquinic acid) 2 | 1.56 ± 0.13 b | 0.23 ± 0.01 a | 4.81 ± 0.11 d | 3.48 ± 0.22 c | nd | nd | 1.67 ± 0.07 b |
| Chlorogenic acid (5-caffeoylquinic acid) 3 | 0.17 ± 0.01 | nd | nd | nd | nd | nd | nd |
| 3-p-Coumaroylquinic acid 4 | 0.48 ± 0.04 b | 0.12 ± 0.01 a | nd | nd | nd | nd | 0.39 ± 0.03 c |
| p-Coumaroylquinic acid 4 | nd | 0.12 ± 0.01 a | 0.16 ± 0.04 a | 0.09 ± 0.01 a | nd | nd | 0.38 ± 0.02 b |
| p-Coumaric acid hexoside derivative 1 4 | nd | nd | nd | nd | 0.26 ± 0.01 b | 0.28 ± 0.03 b | 0.09 ± 0.01 a |
| p-Coumaric acid hexoside derivative 2 4 | 0.11 ± 0.00 | nd | nd | nd | nd | nd | nd |
| Caffeic acid hexoside derivative 5 | 0.16 ± 0.01 b | 0.01 ± 0.00 a | nd | nd | nd | nd | nd |
| Hydroxybenzoic acids | |||||||
| bis-HHDP-glucose 1 6 | nd | nd | nd | nd | 3.20 ± 0.06 a | 3.54 ± 0.24 a | nd |
| bis-HHDP-glucose 2 6 | nd | nd | 0.57 ± 0.01 ab | 0.35 ± 0.02 a | 4.57 ± 0.06 c | 4.80 ± 0.18 c | 0.82 ± 0.03 b |
| Tellimagrandin isomer (digalloyl-HHDP-glucose) 1 6 | nd | nd | nd | nd | 2.37 ± 0.03 a | 3.77 ± 0.27 b | nd |
| Tellimagrandin isomer (digalloyl-HHDP-glucose) 2 6 | nd | nd | nd | nd | 2.03 ± 0.09 a | 5.39 ± 0.46 b | nd |
| Strictin/isostrictin isomer (galloyl-HHDP-glucose) 6 | nd | nd | 0.35 ± 0.05 a | 0.23 ± 0.02 a | 1.22 ± 0.03 b | 1.80 ± 0.03 c | nd |
| Digalloylgallate 6 | nd | nd | nd | nd | 0.87 ± 0.04 a | 1.16 ± 0.17 a | nd |
| Trigalloyl-glucose isomer 6 | nd | nd | nd | nd | 1.90 ± 0.07 a | 4.28 ± 0.51 b | nd |
| Gallic acid derivative 1 6 | nd | nd | nd | nd | 2.27 ± 0.05 b | 1.88 ± 0.04 a | nd |
| Gallic acid derivative 2 6 | nd | nd | nd | nd | 4.88 ± 0.07 b | 4.04 ± 0.09 a | nd |
| Gallic acid derivative 3 6 | nd | nd | 0.78 ± 0.04 b | 0.54 ± 0.04 a | nd | nd | nd |
| Gallic acid derivative 4 6 | nd | nd | nd | nd | nd | nd | 1.08 ± 0.11 |
| Gallic acid derivative 5 6 | nd | nd | 1.22 ± 0.13 b | 0.73 ± 0.04 a | nd | nd | nd |
| Gallic acid derivative 6 6 | nd | nd | nd | nd | 2.19 ± 0.04 a | 3.65 ± 0.12 b | nd |
| Gallic acid derivative 7 6 | nd | nd | 1.40 ± 0.03 bc | 0.69 ± 0.05 a | 4.05 ± 0.08 d | 1.64 ± 0.12 c | 1.11 ± 0.02 b |
| Gallic acid derivative 8 6 | nd | nd | nd | nd | 0.65 ± 0.03 a | 0.82 ± 0.06 b | nd |
| Gallic acid derivative 9 6 | nd | nd | nd | nd | 7.29 ± 0.18 a | 8.96 ± 0.43 b | nd |
| Gallic acid derivative 10 6 | nd | nd | 12.49 ± 6.51 d | 9.04 ± 0.57 d | 1.77 ± 0.05 b | 3.38 ± 0.10 c | 0.47 ± 0.02 a |
| Gallic acid derivative 11 6 | nd | nd | 2.86 ± 0.08 a | 1.52 ± 0.09 a | 5.60 ± 0.06 b | 16.95 ± 0.95 d | 13.30 ± 0.36 c |
| Gallic acid derivative 12 6 | nd | nd | nd | nd | 1.57 ± 0.06 a | 2.68 ± 0.03 b | nd |
| Ellagic acid 7 | nd | nd | 1.02 ± 0.14 a | 0.45 ± 0.05 a | 6.13 ± 0.07 b | 8.66 ± 0.74 c | nd |
| Ellagic acid pentoside 7 | nd | nd | nd | nd | 13.65 ± 0.11 a | 33.66 ± 2.14 b | nd |
| Ellagic acid derivative 1 7 | nd | nd | nd | nd | 6.68 ± 0.10 b | 5.22 ± 0.16 a | nd |
| Ellagic acid derivative 2 7 | nd | nd | nd | nd | 5.03 ± 0.09 a | 5.57 ± 0.37 a | nd |
| Ellagic acid derivative 3 7 | nd | nd | 1.97 ± 0.20 a | 0.93 ± 0.09 a | 5.41 ± 0.30 b | 9.53 ± 0.74 c | nd |
| Ellagic acid derivative 4 7 | nd | nd | nd | nd | 4.95 ± 0.19 a | 10.59 ± 0.59 b | nd |
| Ellagic acid derivative 5 7 | nd | nd | 2.06 ± 0.28 a | 0.88 ± 0.10 a | 5.01 ± 0.23 b | 11.82 ± 0.99 c | 2.24 ± 0.03 a |
| 3-O-Methylellagic acid-4-O-β-D-arabinopyranoside 7 | nd | nd | 1.54 ± 0.12 a | 0.99 ± 0.06 a | 3.20 ± 0.05 b | 7.40 ± 0.63 c | nd |
| 1-O-(4-Hydroxy-3,5-dimetoxybenzoyl)-D-glucopyranoside 6 | nd | nd | 0.78 ± 0.07 a | 0.51 ± 0.03 a | 4.18 ± 0.10 b | 5.58 ± 0.69 b | nd |
| Flavanols | |||||||
| (+)Catechin 8 | 3.81 ± 0.26 a | 3.98 ± 0.28 a | nd | nd | 11.73 ± 0.23 c | 11.06 ± 0.33 c | 9.30 ± 0.08 b |
| (-)Epicatechin 9 | nd | 0.71 ± 0.06 | nd | nd | nd | nd | nd |
| (epi)Catechin galloyl 8 | nd | nd | 2.58 ± 0.08 b | 1.49 ± 0.07 a | nd | nd | 1.74 ± 0.06 a |
| (epi)Catechin dihexoside 8 | nd | nd | 2.55 ± 0.20 b | 1.58 ± 0.20 a | nd | nd | nd |
| (epi)Catechin isomer 8 | nd | nd | nd | nd | nd | nd | 1.14 ± 0.02 |
| (epi)Catechin derivative 1 8 | nd | nd | 1.69 ± 0.23 b | 1.06 ± 0.07 a | nd | nd | nd |
| Epigallocatechin 8 | 0.48 ± 0.04 c | 0.29 ± 0.02 ab | 0.38 ± 0.01 ac | 0.27 ± 0.02 a | nd | nd | 0.40 ± 0.02 bc |
| Procyanidin dimer 1 10 | 10.73 ± 0.70 b | 1.81 ± 0.13 a | nd | nd | nd | nd | nd |
| Procyanidin dimer 2 10 | 2.14 ± 0.18 a | 2.92 ± 0.19 a | nd | nd | nd | nd | nd |
| Procyanidin dimer 3 10 | 5.33 ± 0.35 c | 2.24 ± 0.22 a | 1.95 ± 0.04 a | 1.57 ± 0.10 a | nd | nd | 3.90 ± 0.40 b |
| Procyanidin dimer 4 10 | nd | 1.80 ± 0.21 | nd | nd | nd | nd | nd |
| Flavonols | |||||||
| Myricetin-3-galactoside 11 | 0.19 ± 0.02 a | 0.81 ± 0.08 b | nd | nd | nd | nd | 2.64 ± 0.07 c |
| Myricetin-3-glucoside 11 | nd | 0.26 ± 0.03 | nd | nd | nd | nd | nd |
| Myricetin-3-rhamnoside 12 | 0.57 ± 0.03 bc | 0.41 ± 0.03 a | 0.65 ± 0.02 c | 0.46 ± 0.04 ab | nd | nd | 3.90 ± 0.06 d |
| Myricetin pentoside 11 | nd | 0.15 ± 0.01 | nd | nd | nd | nd | nd |
| Myricetin galloyl hexoside 11 | nd | nd | nd | nd | nd | nd | 3.44 ± 0.09 |
| Quercetin-3-galactoside 13 | 0.66 ± 0.02 a | 0.70 ± 0.05 a | 1.47 ± 0.04 b | 0.53 ± 0.08 a | nd | nd | 2.08 ± 0.05 c |
| Quercetin-3-glucoside 14 | 0.49 ± 0.01 b | 0.29 ± 0.02 a | nd | nd | nd | nd | 0.75 ± 0.01 c |
| Quercetin-3-xyloside 15 | nd | 0.23 ± 0.01 | nd | nd | nd | nd | nd |
| Quercetin-3-arabinopyranoside 16 | 0.24 ± 0.01 a | 0.20 ± 0.01 a | 1.20 ± 0.10 c | 0.62 ± 0.02 b | nd | nd | 0.90 ± 0.19 bc |
| Quercetin-3-arabinofuranoside 17 | 0.64 ± 0.01 a | 0.59 ± 0.04 a | nd | nd | nd | nd | nd |
| Quercetin-3-rhamnoside 18 | 0.64 ± 0.02 b | 0.54 ± 0.04 b | 0.68 ± 0.04 b | 0.34 ± 0.02 a | nd | nd | 4.03 ± 0.08 c |
| Quercetin galloyl hexoside 14 | nd | nd | nd | nd | nd | nd | 1.87 ± 0.05 |
| Kaempferol-3-glucoside 19 | 0.24 ± 0.02 a | 0.25 ± 0.02 a | nd | nd | nd | nd | nd |
| Kaempferol-3-rhamnoside 19 | 0.28 ± 0.01 | nd | nd | nd | nd | nd | nd |
| Kaempferol-pentoside 1 19 | 0.58 ± 0.04 b | 0.11 ± 0.01 a | nd | nd | nd | nd | nd |
| Kaempferol-pentoside 2 19 | 0.54 ± 0.06 b | 0.08 ± 0.01 a | nd | nd | nd | nd | nd |
| Kaempferol-7-hexoside 1 19 | nd | nd | 1.47 ± 0.05 b | 0.92 ± 0.05 a | nd | nd | nd |
| Kaempferol-7-hexoside 2 19 | nd | nd | 0.33 ± 0.02 b | 0.17 ± 0.01 a | nd | nd | nd |
| Dihydrokaempferol pentoside 1 19 | 1.30 ± 0.09 | nd | nd | nd | nd | nd | nd |
| Dihydrokaempferol pentoside 2 19 | 1.02 ± 0.25 | nd | nd | nd | nd | nd | nd |
| Napthoquinones | |||||||
| Juglone (5-hydroxy-1,4-napthoquinone) 20 | 0.13 ± 0.06 a | 0.70 ± 0.02 c | 0.32 ± 0.02 b | 0.65 ± 0.01 c | 0.20 ± 0.00 ab | 0.14 ± 0.01 a | 0.22 ± 0.01 ab |
| 1,4-Napthoquinone 21 | 0.06 ± 0.01 a | 0.03 ± 0.00 a | 0.01 ± 0.00 a | 0.02 ± 0.00 a | 0.23 ± 0.02 c | 0.15 ± 0.01 b | 0.02 ± 0.00 a |
| Hydrojuglone 20 | 0.14 ± 0.01 c | 0.01 ± 0.00 a | 0.02 ± 0.00 ab | 0.02 ± 0.00 ab | 0.03 ± 0.00 b | 0.02 ± 0.00 a | 0.01 ± 0.00 a |
| Hydrojuglone-β-D-glucopyranoside 20 | 74.72 ± 2.86 e | 19.63 ± 0.28 ab | 43.89 ± 0.87 c | 27.68 ± 0.59 b | 15.84 ± 0.82 a | 54.00 ± 1.26 d | 54.88 ± 4.81 d |
| Hydrojuglone dihexoside 20 | nd | nd | nd | nd | nd | nd | 13.48 ± 0.21 |
| Hydrojuglone dihexoside derivative 20 | 0.64 ± 0.04 | nd | nd | nd | nd | nd | nd |
| Hydrojuglone derivative rhamnoside 20 | nd | nd | 22.59 ± 0.32 d | 12.81 ± 0.67 c | 3.39 ± 0.09 a | 10.76 ± 0.88 bc | 9.88 ± 0.17 b |
| Hydrojuglone derivative pentoside 1 20 | 2.89 ± 0.17 b | 1.87 ± 0.12 a | nd | nd | nd | nd | nd |
| Hydrojuglone derivative pentoside 2 20 | 31.90 ± 1.84 e | 5.08 ± 0.33 cd | 0.76 ± 0.13 ab | 0.35 ± 0.03 a | 3.94 ± 0.29 bc | 7.66 ± 0.30 d | 8.97 ± 0.36 d |
| Hydrojuglone hexoside derivative 20 | 2.18 ± 0.09 b | 0.98 ± 0.10 a | nd | nd | nd | nd | nd |
| Hydrojuglone derivative 1 20 | 1.59 ± 0.08 a | 1.45 ± 0.11 a | 3.32 ± 0.27 a | 1.82 ± 0.12 a | 16.14 ± 0.51 b | 25.78 ± 1.90 c | 2.46 ± 0.12 a |
| Hydrojuglone derivative 2 20 | 1.02 ± 0.05 a | 2.60 ± 0.21 b | nd | nd | nd | nd | 2.92 ± 0.21 b |
| Hydrojuglone derivative 3 20 | nd | nd | 7.16 ± 0.79 b | 3.82 ± 0.33 a | nd | nd | 10.16 ± 0.45 c |
| Hydrojuglone derivative 4 20 | nd | nd | 7.36 ± 1.45 a | 9.27 ± 0.47 a | nd | nd | nd |
| Hydrojuglone derivative 5 20 | nd | nd | 1.48 ± 0.29 b | 0.66 ± 0.09 a | 3.44 ± 0.05 c | 5.52 ± 0.24 d | nd |
| Hydrojuglone derivative 6 20 | nd | nd | 1.82 ± 0.17 a | 0.63 ± 0.13 a | 17.7 ± 0.21 b | 66.34 ± 1.75 c | nd |
| Tetralone hexoside 20 | 1.89 ± 0.16 a | 2.06 ± 0.15 a | 2.96 ± 0.25 a | 2.01 ± 0.14 a | 8.02 ± 0.22 b | 14.25 ± 1.79 c | 24.82 ± 0.33 d |
| Dihydroxytetralone hexoside 20 | 1.91 ± 0.10 c | 1.33 ± 0.12 b | 1.25 ± 0.04 b | 0.74 ± 0.04 a | nd | nd | 3.58 ± 0.27 d |
| Trihydroxytetralone galloyl hexoside 20 | nd | 0.19 ± 0.03 a | nd | nd | nd | nd | 4.83 ± 0.10 b |
| 5-Hydroxy-2,3-dihydro-1,4-napthalenedione 20 | 3.16 ± 0.16 b | 2.03 ± 0.16 a | nd | nd | nd | nd | nd |
| 1-β-D-Glucopyranosyloxy-4,8-dihydroxy-2-napthoic acid 20 | nd | nd | nd | nd | 4.69 ± 0.27 a | 7.55 ± 0.82 b | nd |
| 1,4,8-trihydroxynapthalene-1-D-glucopyranoside 20 | nd | nd | nd | nd | 4.81 ± 0.35 a | 4.85 ± 0.53 a | nd |
| Coumarins | |||||||
| Isofraxidin 6 | nd | nd | nd | nd | 1.61 ± 0.05 a | 4.23 ± 0.05 b | nd |
| Isofraxidin derivative 6 | nd | nd | nd | nd | 0.92 ± 0.02 a | 1.72 ± 0.15 b | nd |
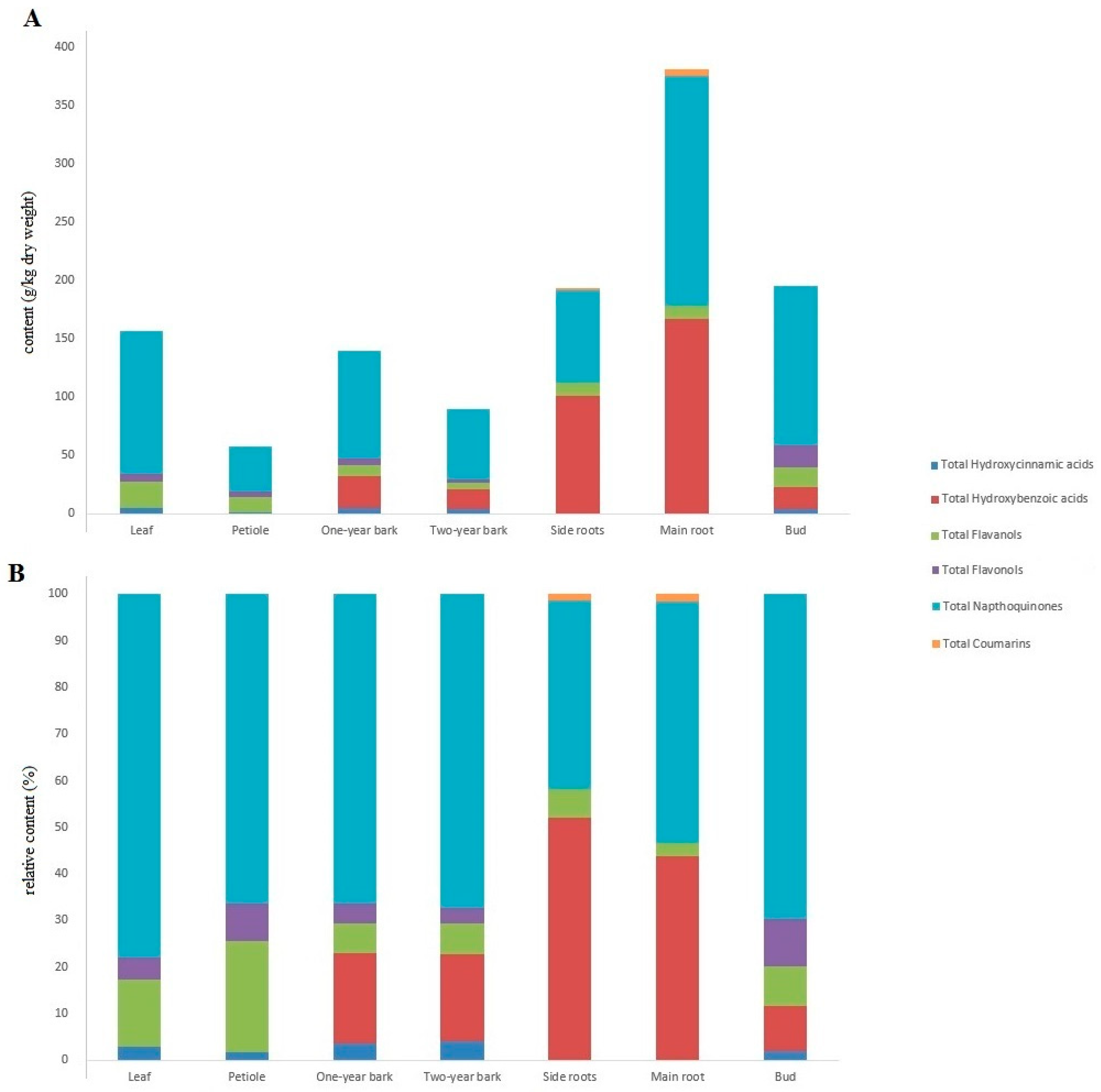
| Total Hydroxycinnamic acids | 4.49 ± 0.30 cd | 0.89 ± 0.05 a | 4.97 ± 0.14 d | 3.57 ± 0.23 b | 0.26 ± 0.01 a | 0.28 ± 0.03 a | 3.84 ± 0.08 bc |
| Total Hydroxybenzoic acids | nd | nd | 27.05 ± 6.09 a | 16.86 ± 1.10 a | 100.67 ± 1.24 b | 166.79 ± 9.93 d | 19.02 ± 0.46 a |
| Total Flavanols | 22.49 ± 1.39 e | 13.75 ± 1.09 cd | 9.14 ± 0.43 a | 5.97 ± 0.43 b | 11.73 ± 0.23 bc | 11.06 ± 0.33 bc | 16.49 ± 0.44 d |
| Total Flavonols | 7.40 ± 0.29 d | 4.61 ± 0.34 b | 5.81 ± 0.17 c | 3.04 ± 0.21 a | nd | nd | 19.60 ± 0.44 e |
| Total Napthoquinones | 122.22 ± 4.60 d | 37.96 ± 1.57 a | 92.92 ± 3.77 c | 60.46 ± 1.33 b | 78.41 ± 1.16 c | 197.01 ± 5.83 e | 136.22 ± 5.71 d |
| Total Coumarins | nd | nd | nd | nd | 2.53 ± 0.06 a | 5.94 ± 0.17 b | nd |
| Total Analysed Phenolic Content | 156.60 ± 4.69 bc | 57.21 ± 3.03 a | 139.89 ± 2.38 b | 89.90 ± 3.22 a | 193.60 ± 2.61 d | 381.08 ± 16.21 e | 195.16 ± 7.08 cd |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Medic, A.; Zamljen, T.; Hudina, M.; Veberic, R. Identification and Quantification of Naphthoquinones and Other Phenolic Compounds in Leaves, Petioles, Bark, Roots, and Buds of Juglans regia L., Using HPLC-MS/MS. Horticulturae 2021, 7, 326. https://doi.org/10.3390/horticulturae7090326
Medic A, Zamljen T, Hudina M, Veberic R. Identification and Quantification of Naphthoquinones and Other Phenolic Compounds in Leaves, Petioles, Bark, Roots, and Buds of Juglans regia L., Using HPLC-MS/MS. Horticulturae. 2021; 7(9):326. https://doi.org/10.3390/horticulturae7090326
Chicago/Turabian StyleMedic, Aljaz, Tilen Zamljen, Metka Hudina, and Robert Veberic. 2021. "Identification and Quantification of Naphthoquinones and Other Phenolic Compounds in Leaves, Petioles, Bark, Roots, and Buds of Juglans regia L., Using HPLC-MS/MS" Horticulturae 7, no. 9: 326. https://doi.org/10.3390/horticulturae7090326
APA StyleMedic, A., Zamljen, T., Hudina, M., & Veberic, R. (2021). Identification and Quantification of Naphthoquinones and Other Phenolic Compounds in Leaves, Petioles, Bark, Roots, and Buds of Juglans regia L., Using HPLC-MS/MS. Horticulturae, 7(9), 326. https://doi.org/10.3390/horticulturae7090326









