Alteration of Flower Color in Viola cornuta cv. “Lutea Splendens” through Metabolic Engineering of Capsanthin/Capsorubin Synthesis
Abstract
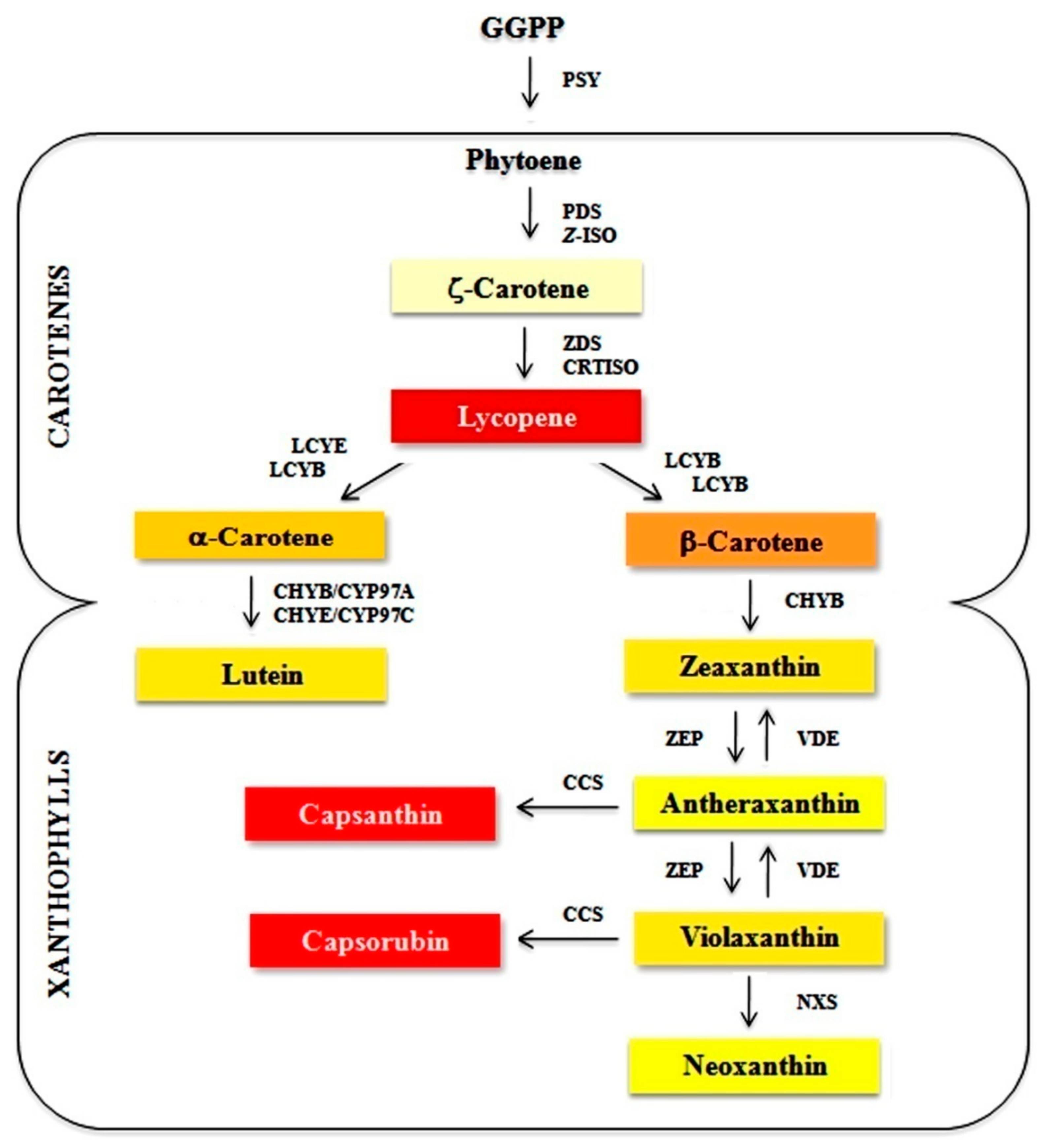
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Agrobacterium-Mediated Genetic Transformation and Regeneration of Transgenic Plants
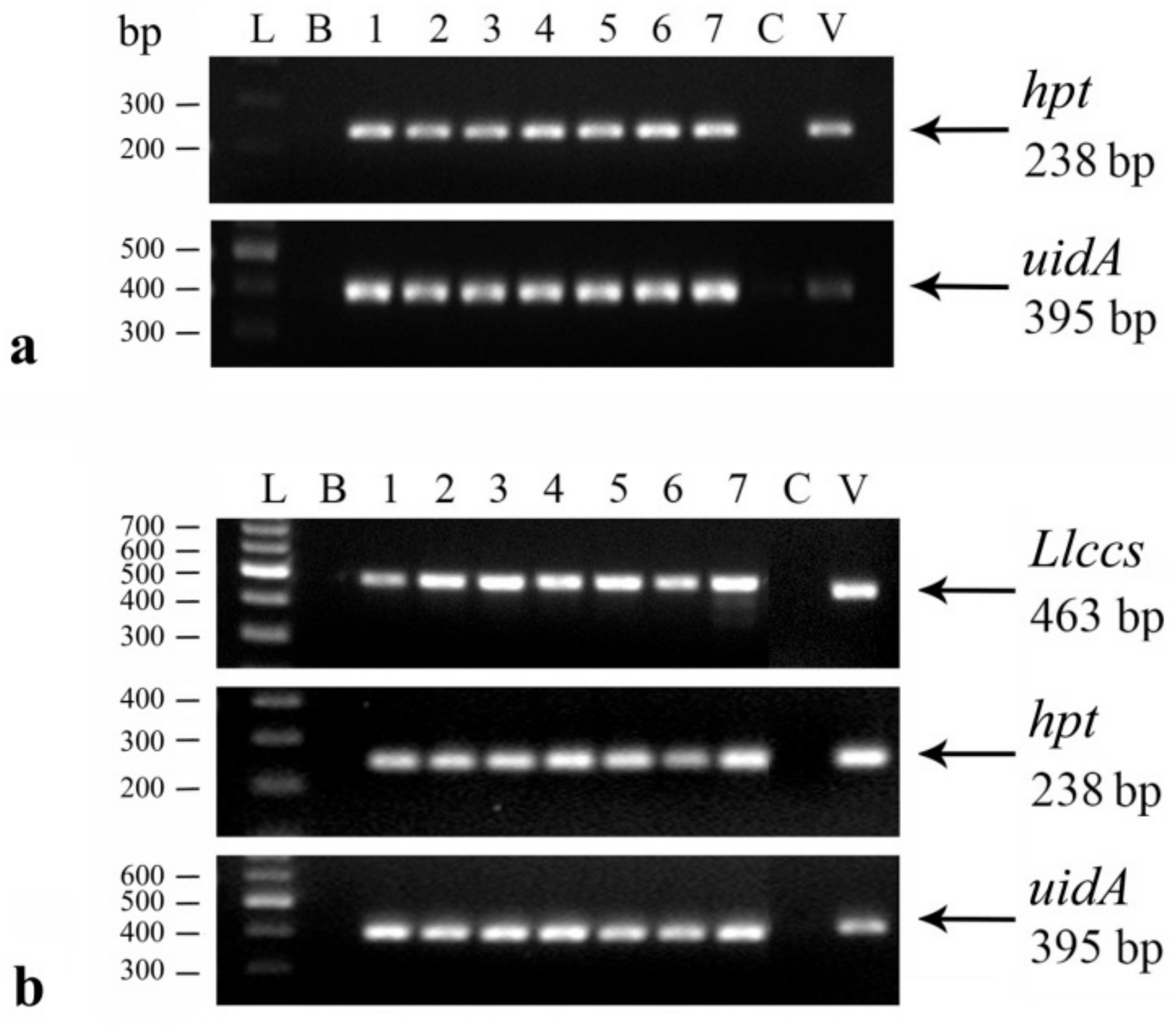
2.2.1. Confirmation of the Genetic Transformation
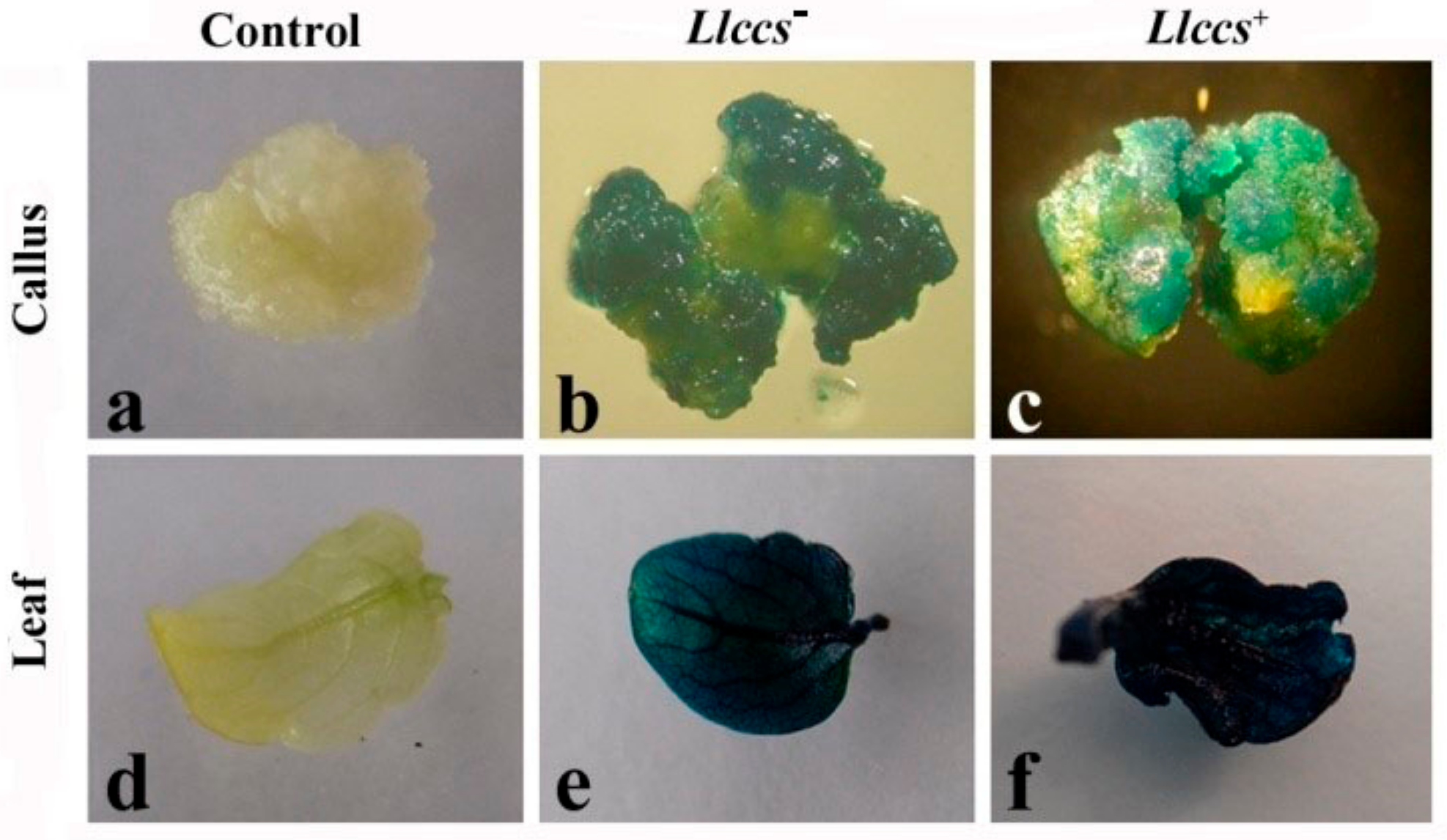
Histochemical GUS Assay
DNA Isolation and Polymerase Chain Reaction (PCR) Analysis
RNA Isolation and Transgene Expression Analysis
2.3. Analysis of Phenotypic Characteristics of Transformed Plants Grown Ex Vitro
2.4. Analysis of Carotenoid Accumulation in Plant Tissue of V. cornuta
2.4.1. Carotenoid Extraction and UHPLC Analysis
2.4.2. Histological Analysis of V. cornuta Tissue
2.4.3. Colorimetric Measurements (CIE L*a*b* System)
2.5. Statistical Analysis
3. Results
3.1. Genetic Transformation and Regeneration of Transgenic V. cornuta Plants
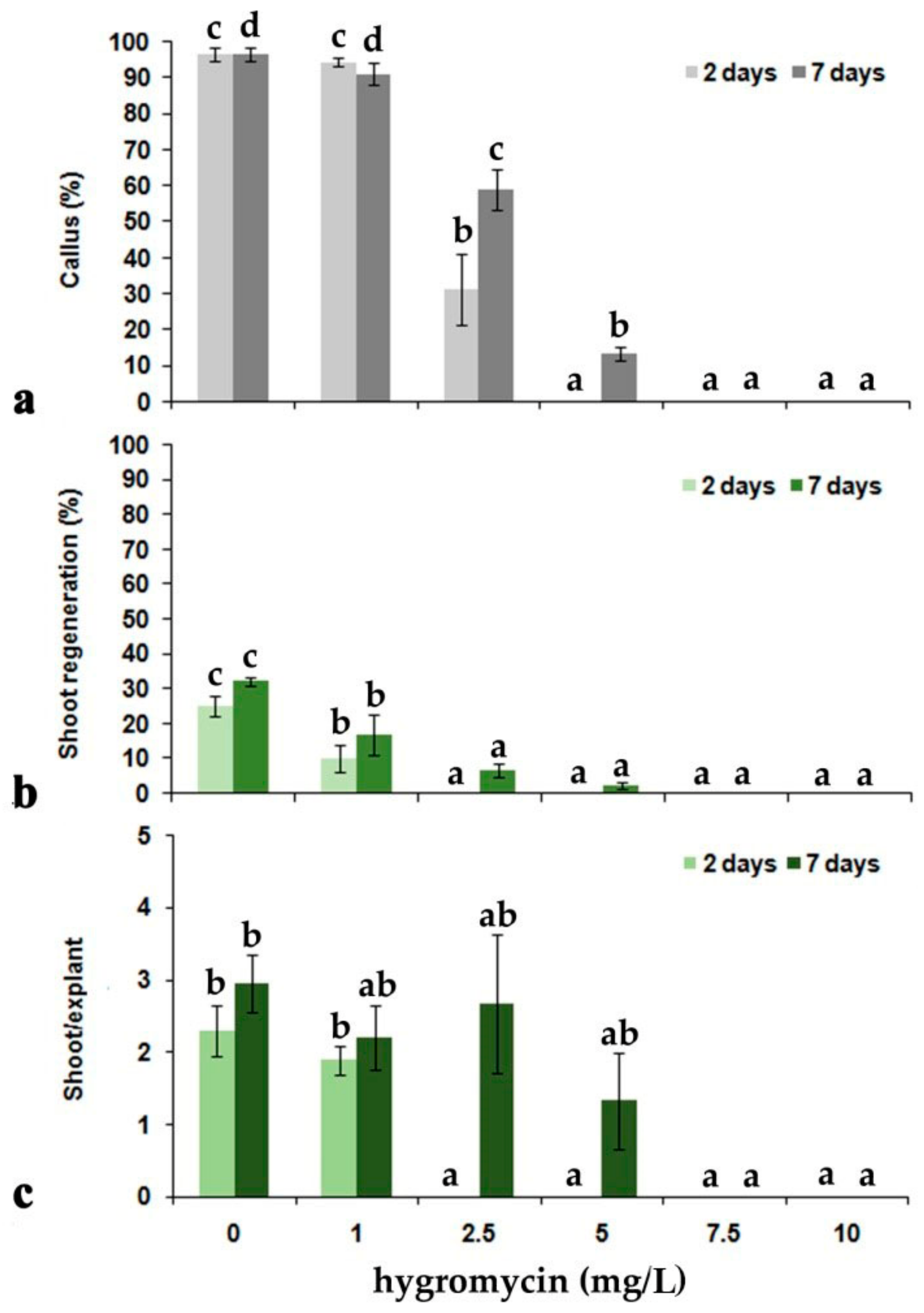
3.1.1. Transformation and Regeneration of V. cornuta Plants from Hypocotyl Explants
3.1.2. Confirmation of Genetic Transformation with Llccs Gene
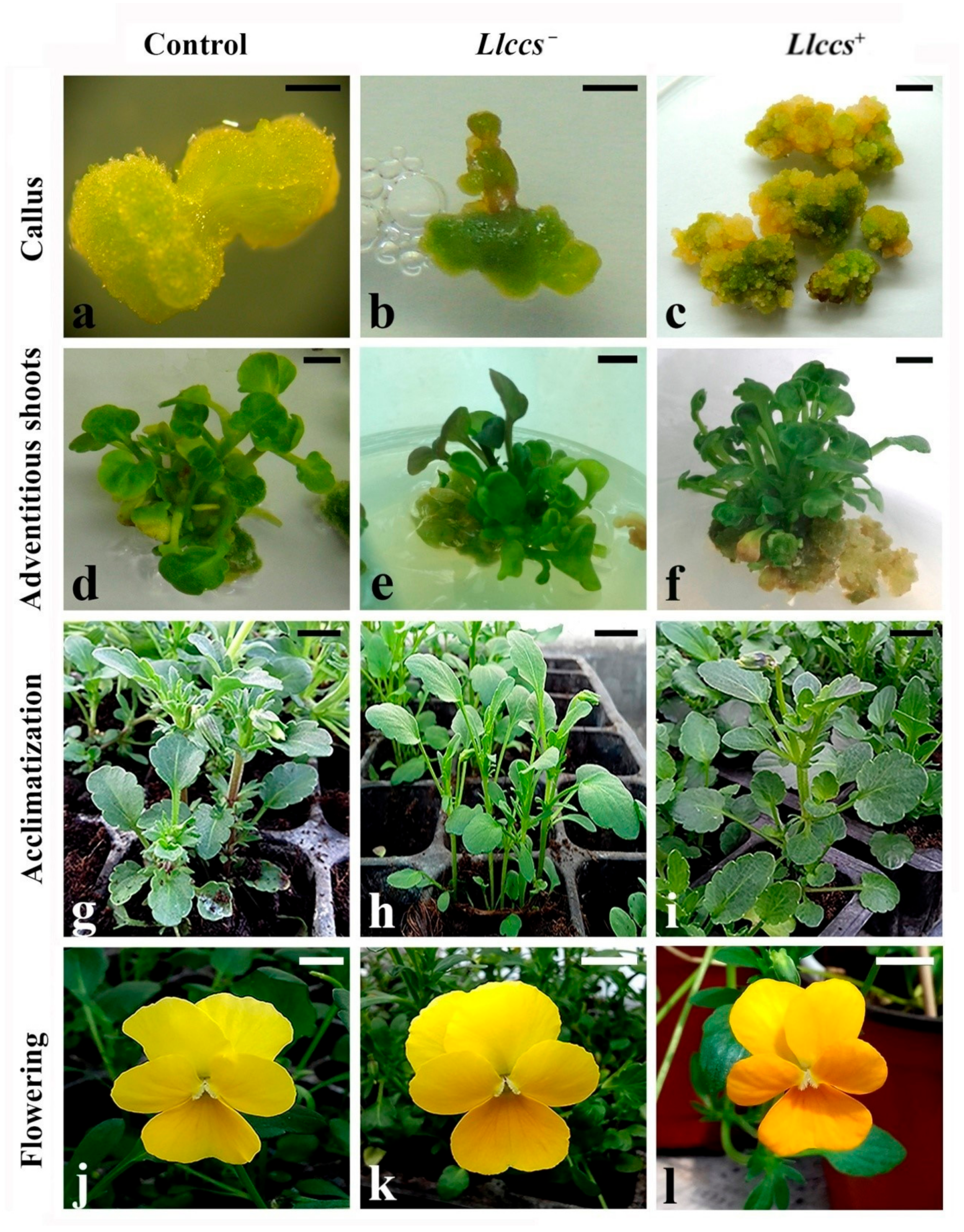
3.1.3. Shoot Multiplication, Rooting and Acclimatization of Transgenic V. cornuta Plants
3.2. Phenotypic Characteristics of Transgenic V. cornuta Plants Grown Ex Vitro
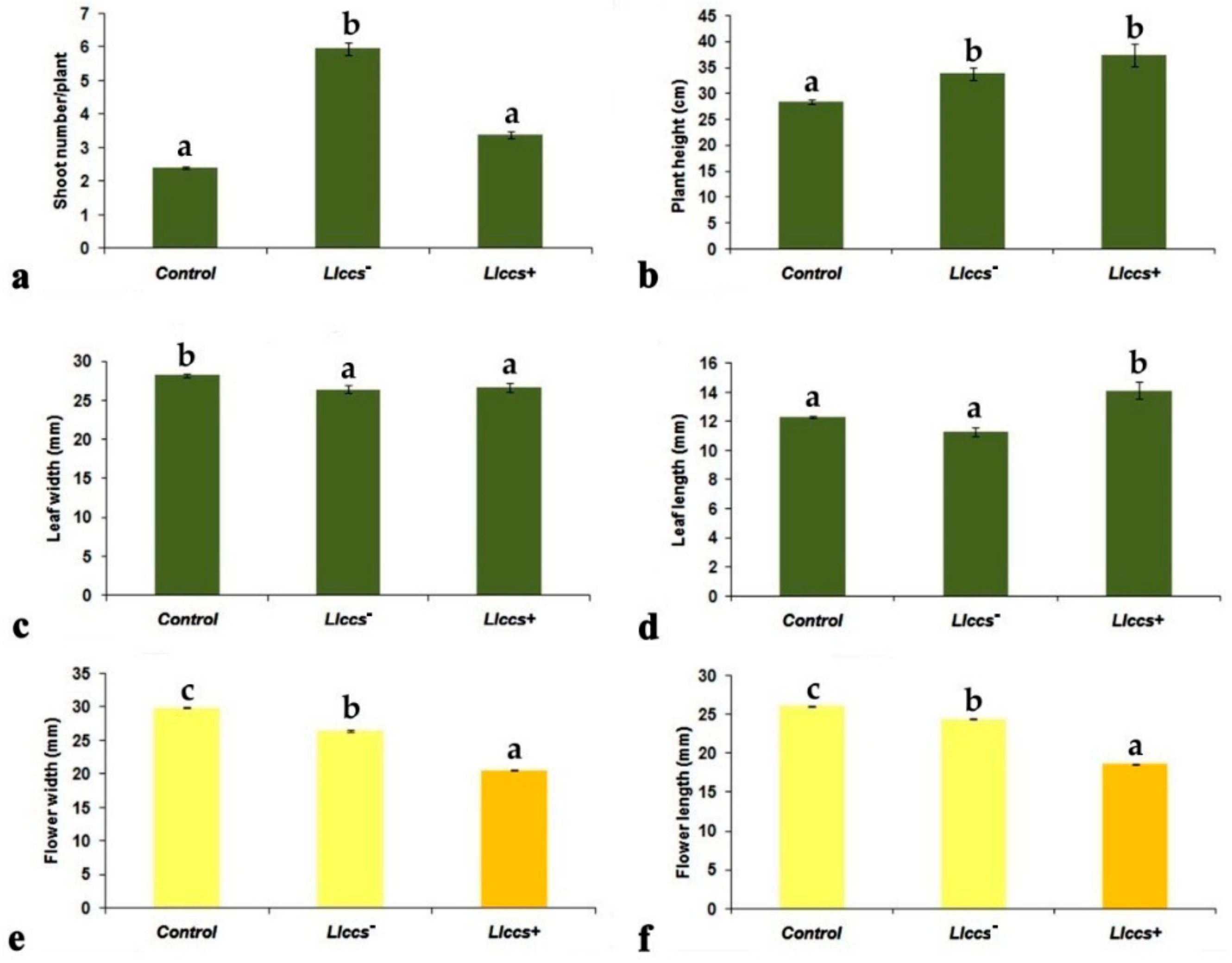
3.2.1. Morphometric Parameters of Transgenic V. cornuta Plants Grown Ex Vitro
3.2.2. Flower Color Alteration in Transgenic V. cornuta Plants Grown Ex Vitro
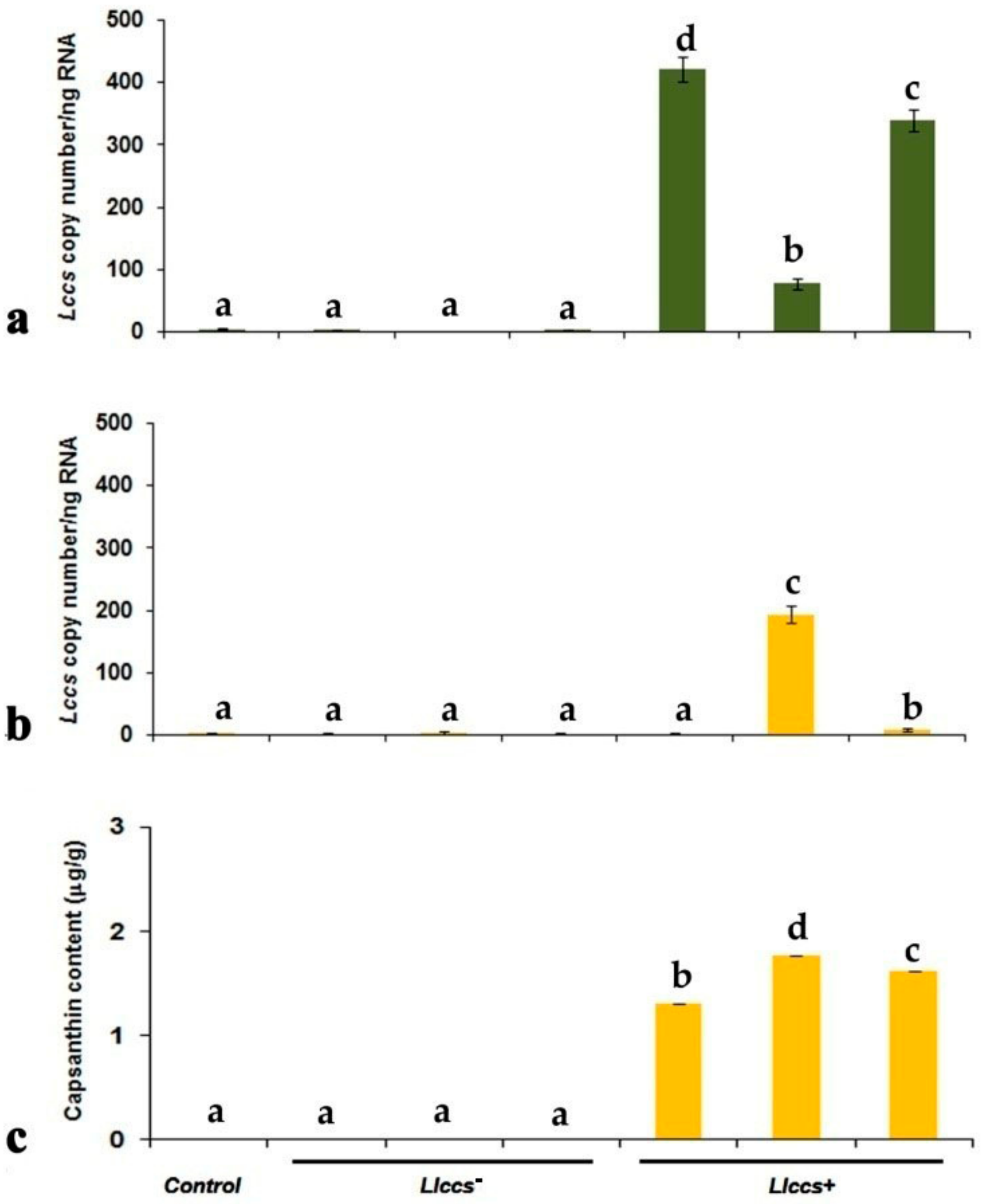
3.2.3. qPCR Quantification of Llccs Expression
3.3. Qualitative and Quantitative Analysis of Newly Synthesized Pigment in Llccs+ Transgenic Plants
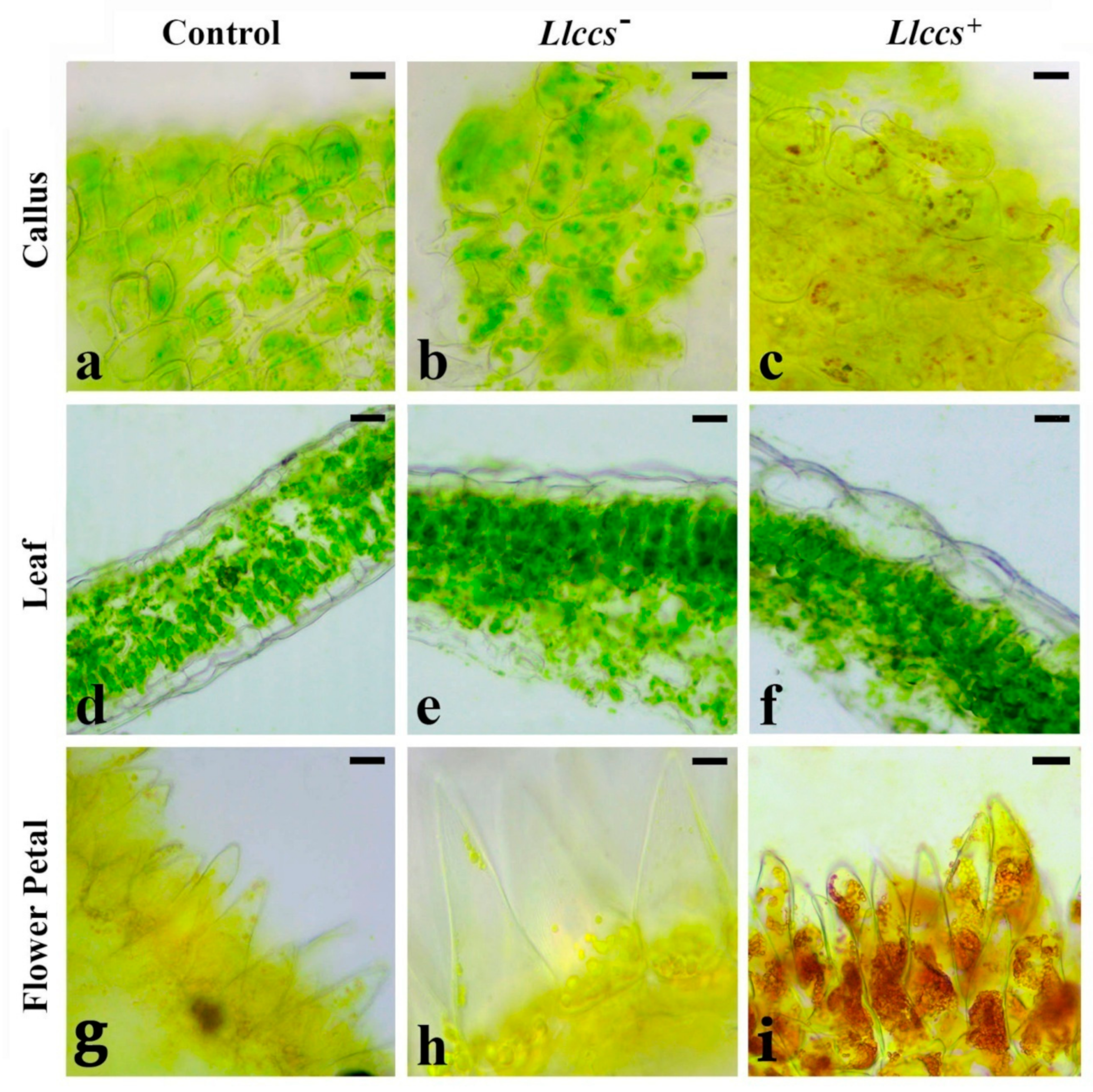
3.3.1. Detection of the Pigment in Plant Tissue
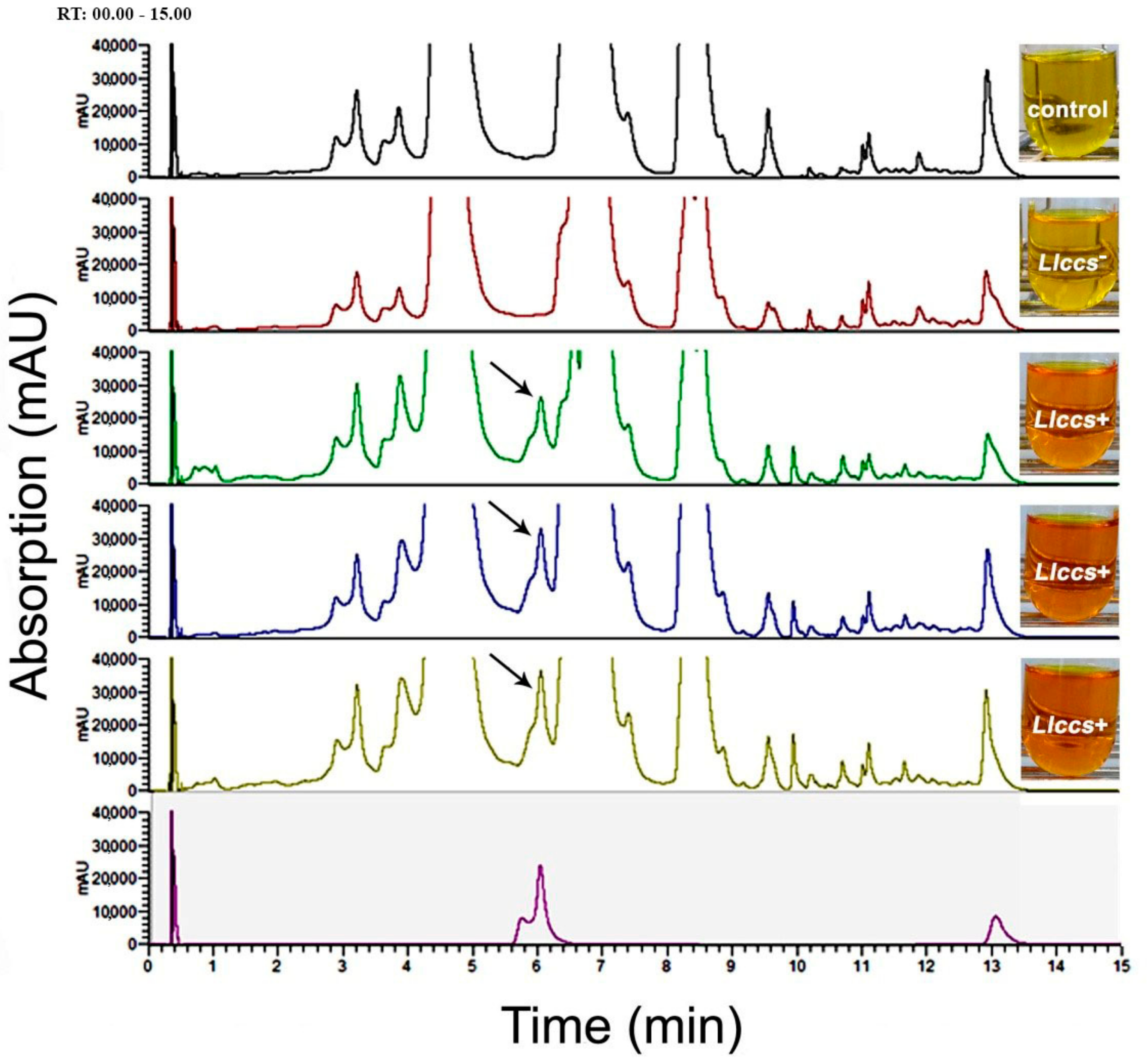
3.3.2. UHPLC Analysis of Carotenoid Pigments in Flower Petals
3.3.3. Chromaticity Changes in the CIE L*a*b* System in Transgenic Flowers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Noman, A.; Ageel, M.; Deng, J.; Khalid, N.; Sanaillah, T.; Shuilin, H. Biotechnological advancements for improving floral attributes in ornamental plants. Front. Plant Sci. 2017, 8, 530. [Google Scholar] [CrossRef] [Green Version]
- Giovannini, A.; Laura, M.; Nesi, B.; Savona, M.; Cardi, T. Genes and genome editing tools for breeding desirable phenotypes in ornamentals. Plant Cell Rep. 2021, 40, 461–478. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Meyer, P.; Heidemann, I.; Forkmann, G.; Saedler, H. A new petunia flower colour generated by transformation of a mutant with a maize gene. Nature 1987, 330, 677–678. [Google Scholar] [CrossRef]
- Tanaka, Y.; Katsumoto, Y.; Brugliera, F.; Mason, J. Genetic engineering in floriculture. Plant Cell Tissue Organ Cult. 2005, 80, 1–24. [Google Scholar] [CrossRef]
- Nishihara, M.; Nakatsuka, T. Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol. Lett. 2011, 33, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Chandler, S.F.; Sanchez, C. Genetic modification; the development of transgenic ornamental plant varieties. Plant Biotechnol. J. 2012, 10, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Okitsu, N.; Noda, N.; Chandler, S.; Tanaka, Y. Flower Color and Its Engineering by Genetic Modification. In Ornamental Crops, Handbook of Plant Breeding; Van Huylenbroeck, J., Ed.; Springer International Publishing AG: Cham, Switzerland, 2018; Volume 11, pp. 29–62. [Google Scholar] [CrossRef]
- Strack, D.; Vogt, T.; Schliemann, W. Recent advances in betalain research. Phytochemistry 2003, 62, 247–269. [Google Scholar] [CrossRef]
- Polturak, G.; Grossmana, N.; Vela-Corciab, D.; Donga, Y.; Nudelc, A.; Plinera, M.; Levyb, M.; Rogacheva, I.; Aharonia, A. Engineered gray mold resistance, antioxidant capacity, and pigmentation in betalain-producing crops and ornamentals. Proc. Natl. Acad. Sci. USA 2017, 114, 9062–9067. [Google Scholar] [CrossRef] [Green Version]
- Tadmor, Y.; King, S.; Levi, A.; Davis, A.; Meir, A.; Wasserman, B.; Hirschberg, J.; Lewinson, E. Comparative fruit colouration in watermelon and tomato. Food Res. Int. 2005, 38, 837–841. [Google Scholar] [CrossRef]
- Cunnungham, F.X.; Gantt, E. A study in scarlet: Enzymes of ketocarotenoid biosynthesis in the flowers of Adonis aestivalis. Plant J. 2005, 41, 478–492. [Google Scholar] [CrossRef]
- Yamagishi, M.; Kishimoto, S.; Nakayama, M. Carotenoid composition and changes in expression of carotenoid biosynthetic genes in tepals of Asiatic hybrid lily. Plant Breed. 2010, 129, 100–107. [Google Scholar] [CrossRef]
- Wang, H.M.; To, K.Y.; Lai, H.M.; Jeng, S.T. Modification of flower colour by supressing β-ring carotene hydroxylase in Oncidium. Plant Biol. 2016, 18, 220–229. [Google Scholar] [CrossRef]
- Niyogi, K. Safety valves for photosynthesis. Curr. Opin. Plant Biol. 2000, 3, 455–460. [Google Scholar] [CrossRef]
- Howitt, C.A.; Pogson, B.J. Carotenoid accumulation and function in seeds and non-green tissues. Plant Cell Environ. 2006, 29, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, S.; Oda-Yamamizo, C.; Ohmiya, A. Regulation of carotenoid pigmentation in corollas of petunias. Plant Mol. Biol. Report. 2018, 36, 632–642. [Google Scholar] [CrossRef]
- Hirschberg, J. Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 2001, 4, 210–218. [Google Scholar] [CrossRef]
- Zhu, C.; Bai, C.; Sanahuja, G.; Yuan, D.; Farré, G.; Naqvu, S.; Shi, L.; Capell, T.; Christou, P. The regulation of carotenoid pigmentation in flowers. Arch. Biochem. Biophys. 2010, 504, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Zhang, J.; Nageswaran, D.; Li, L. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2015, 2, 15036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohmiya, A.; Kato, M.; Shimada, T.; Nashima, K.; Kishimoto, S.; Nagata, M. Molecular basis of carotenoid accumulation in horticultural crops. Hortic. J. 2019, 88, 135–149. [Google Scholar] [CrossRef] [Green Version]
- Camara, B. Plant phytoene synthase complex:component enzymes immunology, and biogenesis. Methods Enzymol. 1993, 214, 352–365. [Google Scholar] [CrossRef]
- Ohmiya, A. Diversity of carotenoid composition in flower petals. Jpn. Agric. Res. Q. JARQ 2011, 45, 163–171. [Google Scholar] [CrossRef] [Green Version]
- Giuliano, G.; Bartley, G.E.; Scolnik, P.A. Regulation of carotenoid biosynthesis during tomato development. Plant Cell 1993, 5, 379–387. [Google Scholar] [CrossRef] [Green Version]
- Welsch, R.; Beyer, P.; Hugueney, P.; Kleining, H.; von Lintig, J. Regulation and activation of phytoene synthase, a key enzyme in carotenoid biosynthesis, during photomorphogenesis. Planta 2000, 211, 846–854. [Google Scholar] [CrossRef]
- Jeknić, Z.; Jeknić, S.; Jevremović, S.; Subotić, A.; Chen, T.H. Alteration of flower color in Iris germanica L. ’Fire Bride’ through ectopic expression of phytoene synthase gene (crtB) from Pantoeaagglomerans. Plant Cell Rep. 2014, 33, 1307–1321. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Wang, J.; Dong, X.; Han, Y.; Ma, Q.; Ding, Y.; Zhao, F.; Zhang, J.; Chen, H.; Xu, Q.; et al. Carotenoid accumulation affects redox status, starch metabolism, and flavonoid/anthocyanin accumulation in citrus. BCM Plant Biol. 2015, 15, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umemoto, N.; Takano, M.; Shimada, H.; Mamiya, K.; Toguri, T. Flower color modification by xanthophyll biosynthetic genes in petunia. Plant Cell Physiol. 2006, 47, S110. [Google Scholar]
- Umemoto, N.; Toguri, T. Peptide Transporting to Chromoplasts in Petals and Method of Constructing Plant Having Yellowish Petals by Using the Same. U.S. Patent 8143478 B2, 27 March 2012. [Google Scholar]
- Suzuki, S.; Nishihara, M.; Nakatsuka, T.; Misawa, N.; Ogiwara, I.; Yamamura, S. Flower color alteration in Lotus japonicusby modification of the carotenoid biosynthetic pathway. Plant Cell Rep. 2007, 26, 951–959. [Google Scholar] [CrossRef]
- Mortimer, C.L.; Misawa, N.; Perez-Fons, L.; Robertson, F.P.; Harada, H.; Bramley, P.M.; Fraser, P.D. The formation and sequestration of nonendogenousketocarotenoids in transgenic Nicotianaglauca. Plant Physiol. 2017, 173, 1617–1635. [Google Scholar] [CrossRef] [Green Version]
- Neamtu, G.; Illyds, G.; Bodea, C. Biosynthesis of carotenoids. IV Carotenoid biosynthesis in Aesculus hippocastanum. Stud. Cercet. Biochim. 1969, 12, 77–82. [Google Scholar]
- Neamtu, G.; Bodea, C. Chemotaxonomic investigations on higher plants. VII. Carotenoid pigments in some species of Aesculus genus. Stud. Cercet. Biochim. 1974, 17, 41–46. [Google Scholar]
- Neamtu, G.; Bodea, C. Biosynthesis of carotenoids V Carotenoid biosynthesis in Aesculus Rubicunda. Stud. Cercet. Biochim. 1973, 16, 35–43. [Google Scholar]
- Deli, J.; Molnár, P.; Ősz, E.; Zsila, F.; Simonyi, M.; Tóth, G. Analysis of carotenoids in the fruits of Asparagus falcatus: Isolation of 5,6-Diepikarpoxanthin. Chromatografia 2000, 51, S183–S187. [Google Scholar] [CrossRef]
- Simpson, D.J.; Baqar, M.R.; Lee, T.H. Fine-structure and catotenoid composition of fibrillar chromoplasts of Asparagus officinalis L. Ann. Bot. 1977, 41, 1101–1108. [Google Scholar] [CrossRef]
- Seybold, A. Undersuchungenüber Den Farbwechsel Von Blumenblattern, Früchten Und Samenschalen. Sitzungsberichte der Heidelberger Akademie der Wissenschaften, Mathematisch-Naturwissenschaftliche Klasse 1953, 4, 31–124. [Google Scholar]
- Wierzchowski, Z.; Bubicz, M. Carotenoids of the berries of Berberis vulgaris. Annales Universitatis Mariae Curie-Skłodowska Lublin-Polonia 1959, XIV, 383–405. [Google Scholar]
- Bubicz, M. Occurrence of carotenoids in fruits of the genus Berberis. Bulletin de l’Academie Polonaise des Sciences Serie des Sciences Biologiques 1965, 13, 251–255. [Google Scholar]
- Zechmeister, L.; Cholnoky, L. Untersuchungenüber den Paprika-Farbstoff. Justus Liebigs Annalen der Chemie 1927, 454, 70–84. [Google Scholar] [CrossRef]
- Zechmeister, L.; Cholnoky, L. Untersuchungentiber den Paprika-Farbstoff. V. Naturliche und synthetische Ester des Capsanthins. Justus Liebigs Annalen der Chemie 1931, 487, 197–213. [Google Scholar] [CrossRef]
- Bouvier, F.; Hugueney, P.; d’Harlingue, A.; Kuntz, M.; Camara, B. Xanthophyll biosynthesis in chromoplasts: Isolation and molecular cloning of an enzyme catalyzing the coversion of 5,6-epoxycarotenoid into ketocarotenoid. Plant J. 1994, 6, 45–54. [Google Scholar] [CrossRef]
- Murillo, E.; Watts, M.; Reyna, G.; Giuffida, D.; Durant-Archibold, A.A. Carotenoid composition of Cionosiyos macranthus fruit. Nat. Prod. Commun. 2019, 14, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Murillo, E.; Deli, J.; Nagy, V.; Molinar-Toribo, E.; Sándor, V.; Marton, K.; Agócs, A. Carotenoid profile of two capsorubin-rich tropical plants. J. Food Compos. Anal. 2021, 97, 103798. [Google Scholar] [CrossRef]
- Valadon, L.R.G.; Mummery, R.S. Carotenoids of lilies and of red pepper: Biogenesis of capsanthin and capsorubin. Zeitschrift für Pflanzenphysiologie 1977, 82, 407–410. [Google Scholar] [CrossRef]
- Karrer, P.; Oswald, A. Carotinoideaus den staubbeuteln von Lilium tigrinum Einneues carotenoid: Anthetaxanthin. Helv. Chim. Acta 1935, 13, 1303–1305. [Google Scholar] [CrossRef]
- Marki-Fisher, E.; Eugster, C.H. Das carotenoid spektrum der antheren und petalen von Lilium tigrinum cv. ‘Red Night’. Helv. Chim. Acta 1985, 68, 1708–1715. [Google Scholar] [CrossRef]
- Jeknić, Z.; Morré, J.T.; Jeknić, S.; Jevremović, S.; Subotić, A.; Chen, T.H.H. Cloning and functional characterization of a gene for capsanthin-capsorubin synthase from Tiger Lily (Lilium lancifolium Thunb. ’Splendens’). Plant Cell Physiol. 2012, 53, 1899–1912. [Google Scholar] [CrossRef] [PubMed]
- Partali, V.; Liaaen-Jensen, S.; Huneck, S.; Khaidav, T. Carotenoids from the flowers of Lilium pumilum. Die Pharmazie 1987, 42, 208. [Google Scholar]
- Heriyanto, I.A.G.; Fujii, R.; Maoka, T.; Shiosi, Y.; Kameubun, K.M.B.; Limantara, L.; Brotosudarmo, T.H.P. Carotenoid composition in buahmerah (Pandanus conoideus Lam.) an indigenous red fruit of the papua Islands. J. Food Compos. Anal. 2021, 96, 103722. [Google Scholar] [CrossRef]
- Gulyás-Fekete, G.; Murillo, E.; Kurtán, T.; Papp, T.; Illyes, T.Z.; Drahos, I.; Visy, J.; Agócs, A.; Turcsi, E.; Deli, J. Cryptocapsinepoxide-type carotenoids from red mamey, Pouteria sapota. J. Nat. Prod. 2013, 76, 607–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumagai, M.H.; Keller, Y.; Bouvier, F.; Clry, D.; Camara, B. Functional integration of non-native carotenoids into chloroplasts by viral-derived expression of capsanthin–capsorubin synthase in Nicotiana benthamiana. Plant J. 1998, 14, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Van der Meer, I.M.; Brouwer, M.; Spelt, C.E.; Mol, J.N.; Stuitje, A.R. The TACPyAT repeats in the chalcone synthase promoter of Petunia hybrida act as a dominant negative cis-acting module in the control of organ-specific expression. Plant J. 1992, 2, 525–535. [Google Scholar] [CrossRef]
- Jeknić, S. Alteration of Flower Color in Solanum Lycopersicum through Ectopic Expression of a Gene for Capsanthin-Capsorubin Synthase from Lilium Lancifolium. Bachelor’s Thesis, Oregon State University, Corvallis, OR, USA, 2015; pp. 1–37. Available online: https://ir.library.oregonstate.edu/concern/honors_college_theses/st74cs45q (accessed on 10 September 2021).
- González-Barrio, R.; Periago, M.J.; Luna-Recio, C.; Garcia-Alonso, F.J.; Navarro González, I. Chemical composition of the edible flowers, pansy (Viola wittrockiana) and snapdragon (Antirrhinum majus) as new sources of bioactive compounds. Food Chem. 2018, 252, 373–380. [Google Scholar] [CrossRef]
- Trajković, M.; Antonić, D.; Cingel, A.; Ghalawenji, N.; Subotić, A.; Jevremović, S. Advancement in protocol for in vitro seed germination, plant regeneration and cryopreservation of Viola cornuta. 3 Biotech 2019, 9, 17. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Haymes, K.M. Mini-prep method suitable for a plant breeding program. Plant Mol. Biol. Report. 1996, 14, 280–284. [Google Scholar] [CrossRef]
- Gašić, K.; Hernandez, A.; Korban, S.S. RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol. Biol. Report. 2004, 22, 437. [Google Scholar] [CrossRef]
- Bogdanović, M.D.; Dragićević, M.B.; Tanić, N.T.; Todorović, S.I.; Mišić, D.M.; Živković, S.T.; Tissier, A.; Simonović, A.D. Reverse transcription of 18S rRNA with poly (dT) 18 and other homopolymers. Plant Mol. Biol. Report. 2013, 31, 55–63. [Google Scholar] [CrossRef]
- Lee, H.S.; Castle, W.S.; Coates, G.A. High-performance liquid chromatography for the characterization of carotenoids in the sweet orange (Earlygold) grown in Florida, USA. J. Chromatogr. A 2001, 913, 371–377. [Google Scholar] [CrossRef]
- Chandler, S.F.; Brugliera, F. Genetic modification in floriculture. Biotechnol. Lett. 2011, 33, 207–214. [Google Scholar] [CrossRef]
- Boutigny, A.L.; Dohin, N.; Pornin, D.; Rolland, M. Overview and detectability of the genetic modifications in ornamental plants. Hortic. Res. 2020, 7, 11. [Google Scholar] [CrossRef] [Green Version]
- Azadi, P.; Bagheri, H.; Nalousi, A.M.; Nazari, F.; Chandler, S.F. Current status and biotechnological advances in genetic engineering of ornamental plants. Biotechnol. Adv. 2016, 34, 1073–1090. [Google Scholar] [CrossRef] [PubMed]
- Ralley, L.; Enfissi, E.M.; Misawa, N.; Schuch, W.; Bramly, P.M.; Fraser, P.D. Metabolic engineering of ketocarotenoid formation in higher plants. Plant J. 2004, 39, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Gerjets, T.; Sandmann, G. Nicotianaglauca engineered for the production of ketocarotenoids in flowers and leaves by expressing the cyanobacterial crtO ketolase gene. Transgenic Res. 2007, 16, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Mann, V.; Harker, M.; Pecker, I.; Hirschberg, J. Metabolic engineering of astaxanthin production in tobacco flowers. Nat. Biotechnol. 2000, 18, 888–892. [Google Scholar] [CrossRef]
- Watanabe, K.; Oda-Yamamizo, C.; Sage-Ono, K.; Ohmiya, A.; Ono, M. Overexpression of carotenogenic genes in the Japanese morning glory Ipomoea (Pharbitis) nil. Plant Biotechnol. 2017, 34, 177–185. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.Y.; Nugent, G.; Wardley-Richardson, T.; Chandler, S.F.; Young, R.; Dalling, M.J. Agrobacterium mediated transformation of carnation (Dianthus caryophyllus L.). Nat. Biotechnol. 1991, 9, 864–868. [Google Scholar] [CrossRef]
- Park, E.J.; Jeknić, Z.; Sakamoto, A.; DeNoma, J.; Yuwansiri, R.; Murata, N.; Chen, T.H.H. Genetic engineering of glycinebetaine synthesis in tomato protects seeds, plants, and flowers from chilling damage. Plant J. 2004, 40, 474–487. [Google Scholar] [CrossRef]
- Antonić, D.; Trajković, M.; Cingel, A.; Subotić, A.; Jevremović, S. Plant regenerationfrom in vitro-derived leaf and petiole explants of Viola cornuta L. ’Lutea Splendens’. Propag. Ornam. Plants 2017, 17, 95–102. [Google Scholar]
- Dai, R.C.; Lin, R.H.; He, W.J.; Ma, Z.W.; Chen, Y.Q. Studies on resveratrol synthase gene transformation for Viola diffusaGing. mediated by Agrobacterium tumefaciens. Biotechnology 2009, 4, 16–17. [Google Scholar]
- Wu, J.; Liu, C.; Seng, S.; Khan, M.A.; Sui, J.; Gong, B.; Liu, C.; Wu, C.; Zhong, X.; He, J.; et al. Somatic embryogenesis and Agrobacterium-mediated transformation of Gladiolus hybridus cv. ‘Advance red’. Plant Cell Tussue Oragan Cult. 2015, 120, 717–728. [Google Scholar] [CrossRef]
- Maheshwari, P.; Kovalchuk, I. Agrobacterium-mediated stable genetic transformation of Populus angustifolia and Populus balsamifera. Front. Plant Sci. 2016, 7, 296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, R.; Wang, Z.; Ren, Y.; Li, H.; Li, N.; Sun, H. Establishment of efficient genetic transformation systems and application of CRISPR/Cas9 genome editing technology in Lilium pumilum DC. Fisch. And Lilium longiflorum White Heaven. Int. J. Mol. Sci. 2019, 20, 2920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miki, B.; McHugh, S. Selectable marker genes in transgenic plants: Applications, alternatives and biosafety. J. Biotechnol. 2004, 107, 193–232. [Google Scholar] [CrossRef] [PubMed]
- Boszoradova, E.; Libantova, J.; Matusikova, I.; Poloniova, Z.; Jopick, M.; Berenyi, M.; Moravickova, J. Agrobacterium-mediated genetic transformation of economically important oilseed rape cultivars. Plant Cell Tissue Organ Cult. 2011, 107, 317–323. [Google Scholar] [CrossRef]
- Abou-Alaiwi, W.A.; Potlakayala, S.D.; Goldman, S.L.; Josekutty, P.C.; Karelia, D.N.; Rudrabhatla, S.V. Agrobacterium-mediated transformation of the medicinal plant Centaurea montana. Plant Cell Tissue Organ Cult. 2012, 109, 1–8. [Google Scholar] [CrossRef]
- Qin, Y.H.; Teixeira da Silva, J.A.; Bi, J.H.; Zhang, S.L.; Hu, G.B. Response of in vitro strawberry to antibiotics. Plant Growth Regul. 2011, 65, 183–193. [Google Scholar] [CrossRef]
- Cellini, F.; Chesson, A.; Colquhoun, I.; Constable, A.; Davies, H.V.; Engel, K.H.; Gatehouse, A.M.; Kärenlampi, S.; Kok, E.J.; Leguay, J.J.; et al. Unintended effects and their detection in genetically modified crops. Food Chem. Toxicol. 2004, 42, 1089–1125. [Google Scholar] [CrossRef]
- Simó, C.; Ibáez, C.; Valdés, A.; Cifuentes, A.; García-Cañas, V. Metabolomics of genetically modified crops. Int. J. Mol. Sci. 2014, 15, 18941–18966. [Google Scholar] [CrossRef] [Green Version]
- Ladics, G.S.; Bartholomaeus, A.; Bregitzer, P.; Doerrer, N.G.; Gray, A.; Holzhauser, T.; Jordan, M.; Keese, P.; Kok, E.; MacDonald, P.; et al. Genetic basis and detection of unintended effects in genetically modified crop plants. Transgenic Res. 2015, 24, 587–603. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, P.; Dabi, M.; More, P.; Patel, K.; Jana, K.; Agarwal, P.K. Improved shoot regeneration, salinity tolerance and reduced fungal susceptibility in transgenic tobacco constitutively expressing PR-10a gene. Front. Plant Sci. 2016, 7, 217. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.H.; Kim, J.K.; Jeong, Y.S.; You, M.-K.; Lim, S.H.; Kim, J.K. Stepwise pathway engineering to the biosynthesis of zeaxanthin, astaxanthin and capsanthin in rice endosperm. Metab. Eng. 2019, 52, 178–189. [Google Scholar] [CrossRef]
- Furubayashi, M.; Kubo, A.; Takemura, M.; Otani, Y.; Maoka, Y.; Terada, Y.; Yaoi, K.; Ohdan, K.; Misawa, N.; Mitani, Y. Capsanthin production in Escherichia coli by overexpression of capsanthin/capsorubin synthase from Capsicum Annuum. J. Agric. Food Chem. 2021, 69, 5070–5085. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, W.; Ruttink, T.; Borst-Vrenssen, A.W.M.; van der Plas, L.H.W.; van der Krol, A.R. Characterization of position-induced spatial and temporal regulation of transgene promoter activity in plants. J. Exp. Bot. 2001, 52, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Sunilkumar, G.; Mohr, L.A.; Lopata-Finch, E.; Emani, C.; Rathore, K.S. Developmental and tissue-specific expression of CaMV 35S promoter in cotton as revealed by GFP. Plant Mol. Biol. 2002, 50, 463–479. [Google Scholar] [CrossRef]
- Pretóvá, A.; Obert, B.; Wetzstein, H.Y. Leaf developmental stage and tissue location affect the detection of β-glucuronidase in transgenic tobacco plants. Biotechnol. Lett. 2001, 23, 555–558. [Google Scholar] [CrossRef]
- Schnurr, J.A.; Guerra, D.J. The CaMV-35S promoter is sensitive to shortened photoperiod in transgenic tobacco. Plant Cell Rep. 2000, 19, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Windels, P.; De Buck, S.; Depicker, A. Agrobacterium tumefaciens-mediated transformation: Patterns of T-DNA integration into the host genome. In Agrobacterium: From Biology to Biotechnology; Tzfira, T., Citovsky, V., Eds.; Springer: New York, NY, USA, 2008; pp. 441–481. [Google Scholar] [CrossRef]
- Muskens, M.W.M.; Vissers, A.P.A.; Mol, J.N.M.; Kooter, J.M. Role ofinverted DNA repeats in transcriptional and post-transcriptional gene silencing. Plant Mol. Biol. 2000, 43, 243–260. [Google Scholar] [CrossRef]
- Jin, Y.; Guo, H.S. Transgene-induced gene silencing in plants. In Plant Gene Silencing; Humana Press: New York, USA, 2015; pp. 105–117. [Google Scholar] [CrossRef]
- Ohmiya, A. Qualitative and quantitative control of carotenoid accumulation in flower petals. Sci. Hortic. 2013, 163, 10–19. [Google Scholar] [CrossRef]
- Egea, I.; Barsan, C.; Bian, W.; Purgatto, E.; Latché, A.; Chervin, C.; Bouyayen, M.; Pech, J.C. Chromoplast differentiation: Current status and perspectives. Plant Cell Physiol. 2010, 51, 1601–1611. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Bai, J.; Duan, X.; Wang, J. Accumulation characteristics of carotenoids and adaptive fruit color variations in ornamental peper. Sci. Hortic. 2021, 275, 109699. [Google Scholar] [CrossRef]
- Pathare, P.B.; Opara, U.L.; Al-Said, F.A.J. Colour Measurement and Analysis in Fresh and Processed Foods: A Review. Food Bioprocess. Technol. 2013, 6, 36–60. [Google Scholar] [CrossRef]

| Plant Species | Capsanthin | Capsorubin | Plant Part | Reference |
|---|---|---|---|---|
| Aesculus hippocastanum | + | + | anthers | [32] |
| Aesculus parviflora | + | + | anthers | [33] |
| Aesculus rubicunda | + | + | anthers | [34] |
| Asparagus falcatus | + | + | fruit | [35] |
| Asparagus officinalis | + | + | fruit | [36] |
| Asparagus tenuifolia | + | n.d. | fruit | [37] |
| Berberis s sp. (9 species) | + | n.d. | fruit | [38,39] |
| Capsicum annuum | + | + | fruit | [40,41,42] * |
| Cajophoralateritia | n.d. | + | petals | [37] |
| Cionosicyos macranthus | + | n.d. | fruit | [43] |
| Cardulovica palmata | + | + | fruit | [44] |
| Encephalartos altensteinii | + | + | fruit | [37] |
| Encephalartos villosus | + | n.d. | fruit | [37] |
| Lilium amabile | + | + | petals | [37] |
| Lilium davidii | + | + | petals | [45] |
| Lilium lancifolium (formerly L. tigrinum) | + | + | anthers, petals | [46,47,48] ** |
| Lilium leichtinii | + | + | petals | [45] |
| Lilium Maxwill (L. davidii × L. leichtinii) | + | + | petals, anthers, styles | [45] |
| Lilium pumilum | + | + | petals | [49] *** |
| Lilium sp.′Saija′ | + | n.d. | red tepals | [13] |
| Pandanus conoideus | + | + | fruit | [50] |
| Pouteria sapota | + | n.d. | fruit | [51] |
| Zamia dressleri | + | + | leaf, seed | [44] |
| Gene * | GenBankTM Accession Number | Primer Sequence | Product Size (bp) |
|---|---|---|---|
| uidA | A00196.1 | F: 5′-GTGACAAAAACCACCCAAGC-3′ | 395 |
| R: 5′-CAGCCATGCACACTGATAGT-3′ | |||
| hpt | FJ457012.1 | F: 5′-GATGTTGGCGACCTCGTATT-3′ | 238 |
| R: 5′-GATTGCTGATCCCCATGTGT-3′ | |||
| Llccs (PCR) | JF304153.1 | F: 5′-GTCAGATTCCACCCCTCCAA-3′ | 463 |
| R: 5′-AACACTTCTCCTCCTCCAGC-3′ | |||
| Llccs (qPCR) | JF304153.1 | F:5′-GTACGACAGACCGAGAAACC-3′ | 164 |
| R: 5′-TTGGAATAGCAGCGTTGTGA-3′ | |||
| 18S | 18S consensus: X16077 | F:5′-TGACGGAGAATTAGGGTTCG-3′ | 190–191 |
| R:5′-CAATGGATCCTCGTTAAGGG-3′ |
| Treatment | Callus Formation (%) | Shoot Regeneration (%) |
|---|---|---|
| Control | 100 | 32.00 ± 4.90 * |
| Llccs− | 21.00 ± 1.00 | 2.00 ± 1.15 |
| Llccs+ | 15.40 ± 2.35 | 0.20 ± 0.20 |
| Line | Shoot Multiplication | Rooting | ||
|---|---|---|---|---|
| N° of Shoots | Frequency (%) | N° of Roots/Shoot | Length of Longest Root (mm) | |
| Control | 1.95 ± 0.04 c | 100 a | 5.51 ± 0.11 b | 2.75 ± 0.10 b |
| Llccs− | 1.52 ± 0.20 b | 90.73 ± 2.59 a | 9.28 ± 0.90 c | 8.50 ± 0.64 c |
| Llccs+ | 1.03 ± 0.09 a | 28.65 ± 6.45 b | 2.08 ± 0.21 a | 0.85 ± 0.18 a |
| Sample | L* | a* | b* | C* | h* |
|---|---|---|---|---|---|
| Control | 78.32 ± 0.51 b | −4.02 ± 0.83 a | 71.06 ± 2.73 b | 71.38 ± 2.71 a | 93.83 ± 0.74 b |
| Llccs− | 78.54 ± 0.51 b | −4.15 ± 1.07 a | 70.88 ± 2.40 b | 71.35 ± 2.34 a | 94.26 ± 1.03 b |
| Llccs+ | 69.26 ± 0.29 a | 13.22 ± 0.98 b | 64.66 ± 0.65 a | 66.40 ± 0.58 a | 78.34 ± 0.89 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trajković, M.; Jevremović, S.; Dragićević, M.; Simonović, A.D.; Subotić, A.R.; Milošević, S.; Cingel, A. Alteration of Flower Color in Viola cornuta cv. “Lutea Splendens” through Metabolic Engineering of Capsanthin/Capsorubin Synthesis. Horticulturae 2021, 7, 324. https://doi.org/10.3390/horticulturae7090324
Trajković M, Jevremović S, Dragićević M, Simonović AD, Subotić AR, Milošević S, Cingel A. Alteration of Flower Color in Viola cornuta cv. “Lutea Splendens” through Metabolic Engineering of Capsanthin/Capsorubin Synthesis. Horticulturae. 2021; 7(9):324. https://doi.org/10.3390/horticulturae7090324
Chicago/Turabian StyleTrajković, Milena, Slađana Jevremović, Milan Dragićević, Ana D. Simonović, Angelina R. Subotić, Snežana Milošević, and Aleksandar Cingel. 2021. "Alteration of Flower Color in Viola cornuta cv. “Lutea Splendens” through Metabolic Engineering of Capsanthin/Capsorubin Synthesis" Horticulturae 7, no. 9: 324. https://doi.org/10.3390/horticulturae7090324
APA StyleTrajković, M., Jevremović, S., Dragićević, M., Simonović, A. D., Subotić, A. R., Milošević, S., & Cingel, A. (2021). Alteration of Flower Color in Viola cornuta cv. “Lutea Splendens” through Metabolic Engineering of Capsanthin/Capsorubin Synthesis. Horticulturae, 7(9), 324. https://doi.org/10.3390/horticulturae7090324











