Transcriptome Profiling Reveals Candidate Key Genes Involved in Sinigrin Biosynthesis in Brassica nigra
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. GSL Extraction and Analysis
2.3. In Silico Identification of BniGSL Genes in B. nigra
2.4. RNA Extraction, Library Construction, Sequencing, and Gene Expression Analysis
3. Results
3.1. Analysis of Glucosinolate Profile in B. nigra
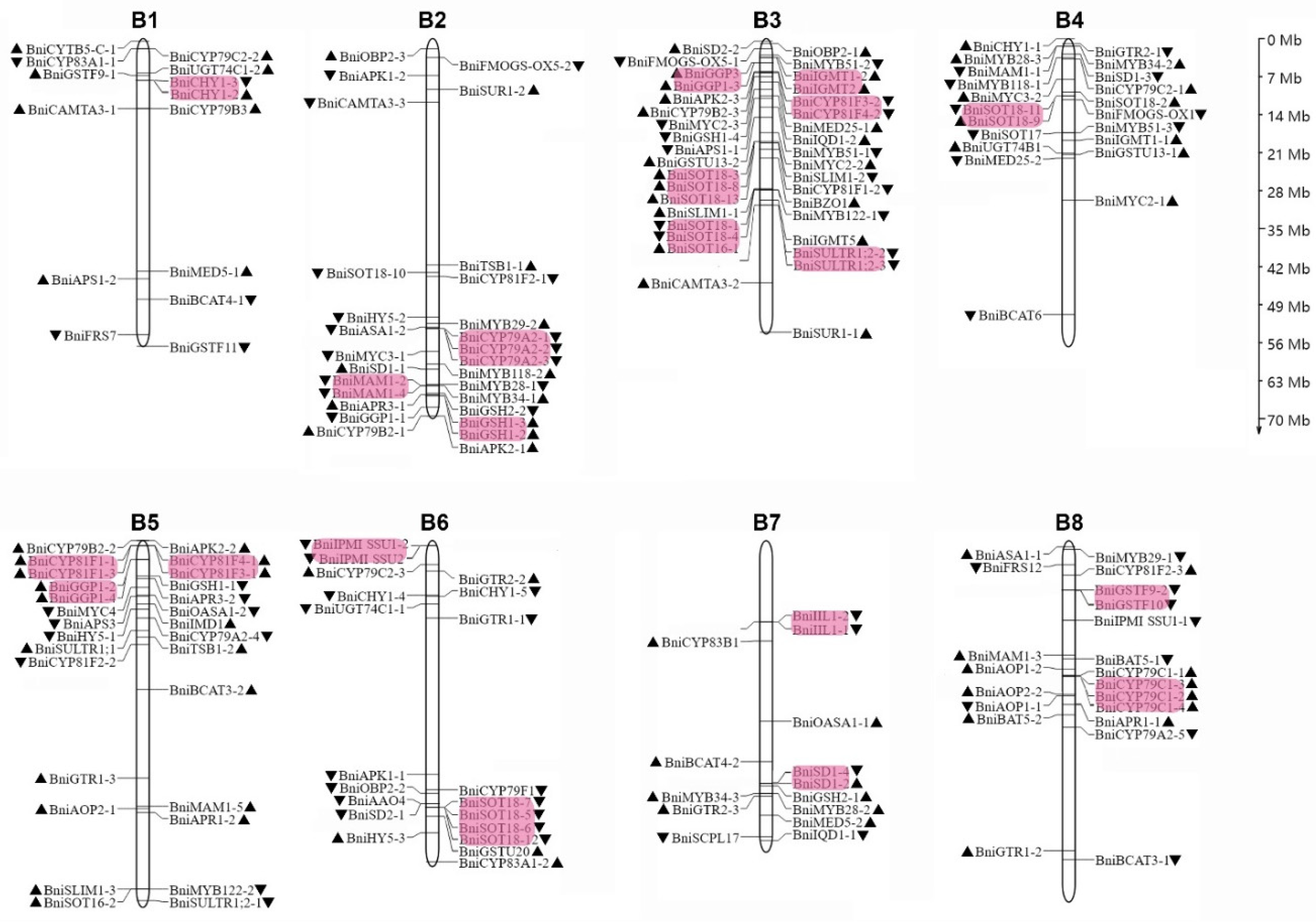
3.2. Identification and Annotation of BniGSL Genes from B. nigra Genome
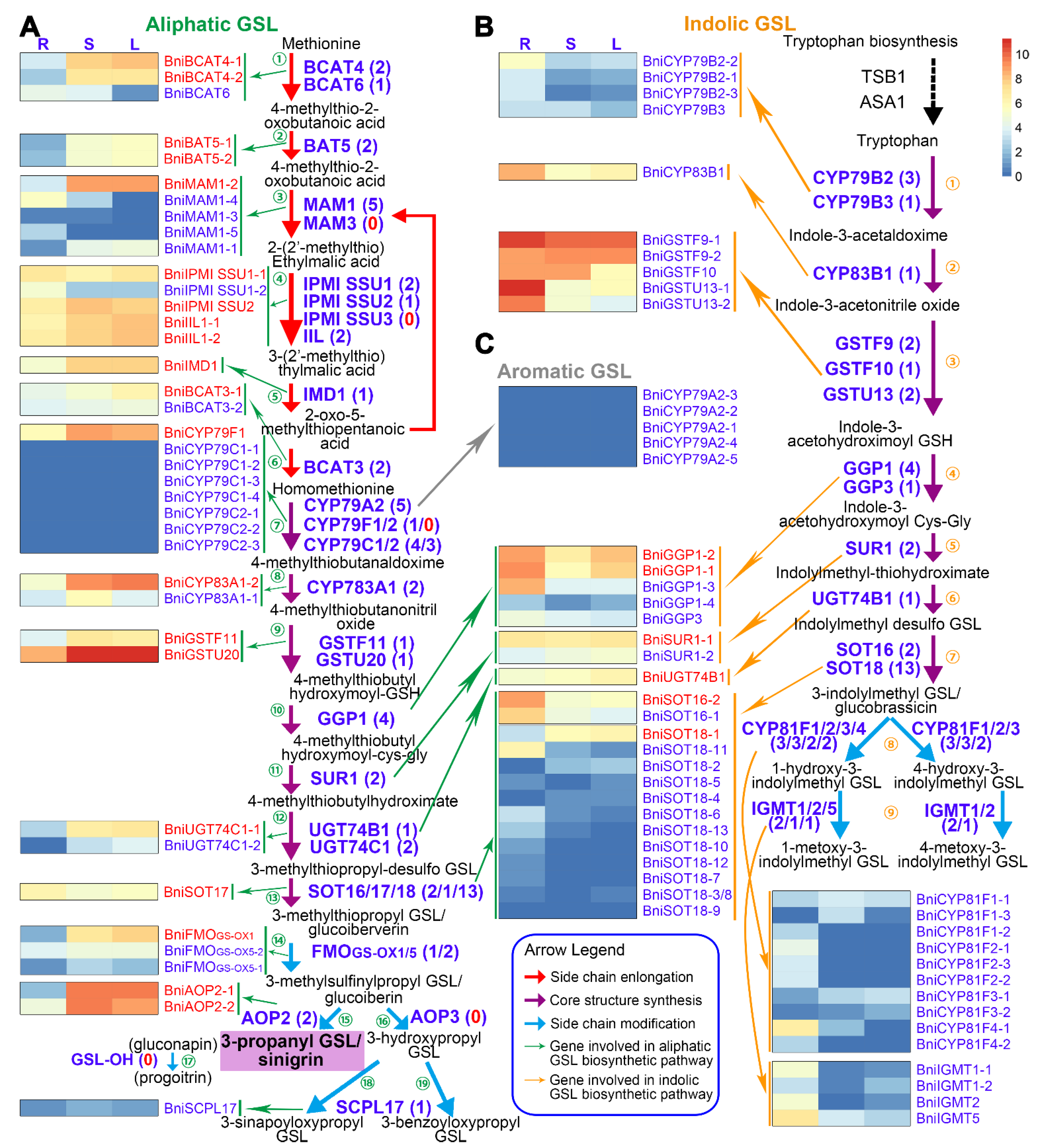
3.3. Expression Patterns of BniGSL Genes Encoding Enzymes in Three Organs of B. nigra
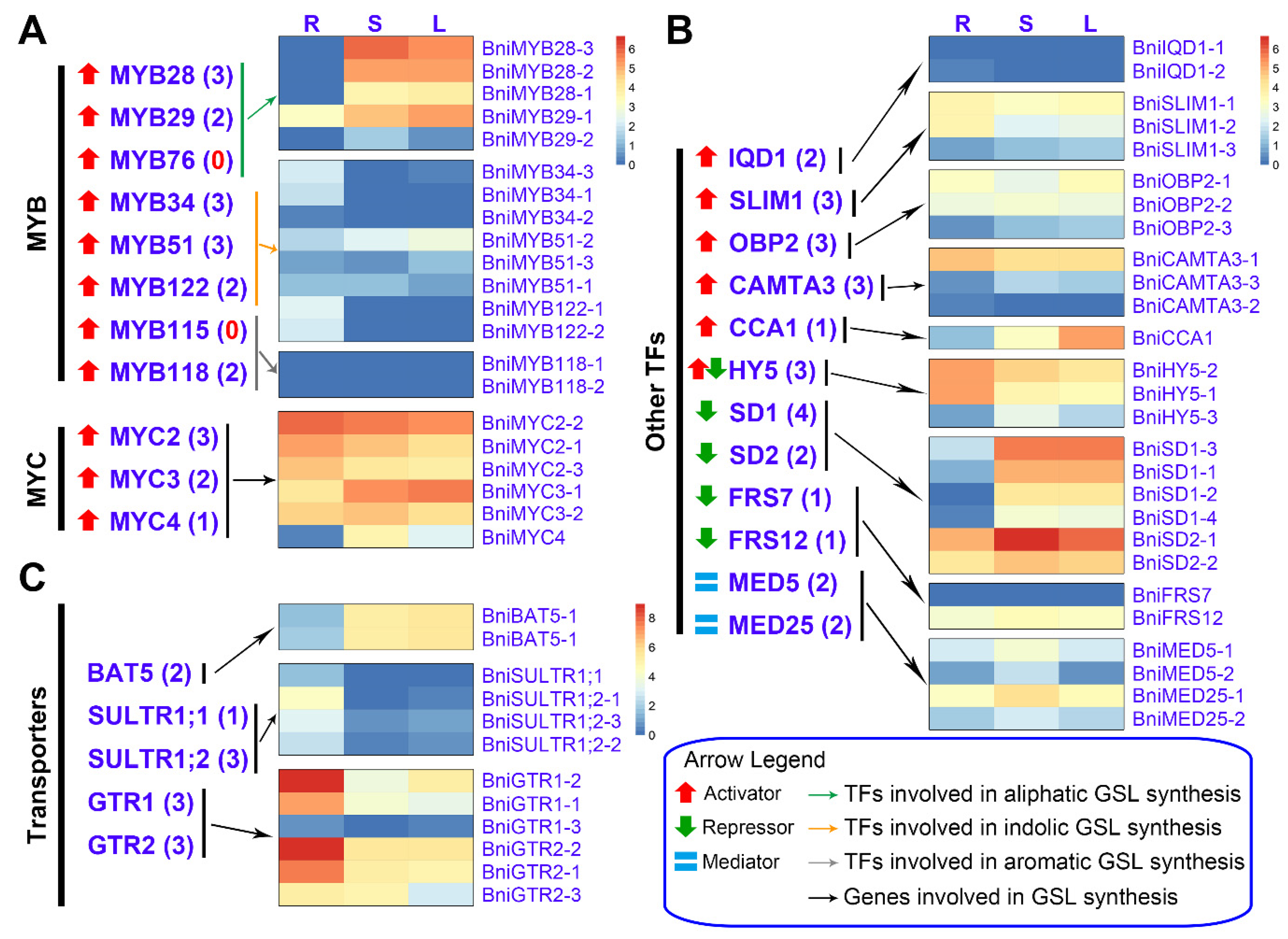
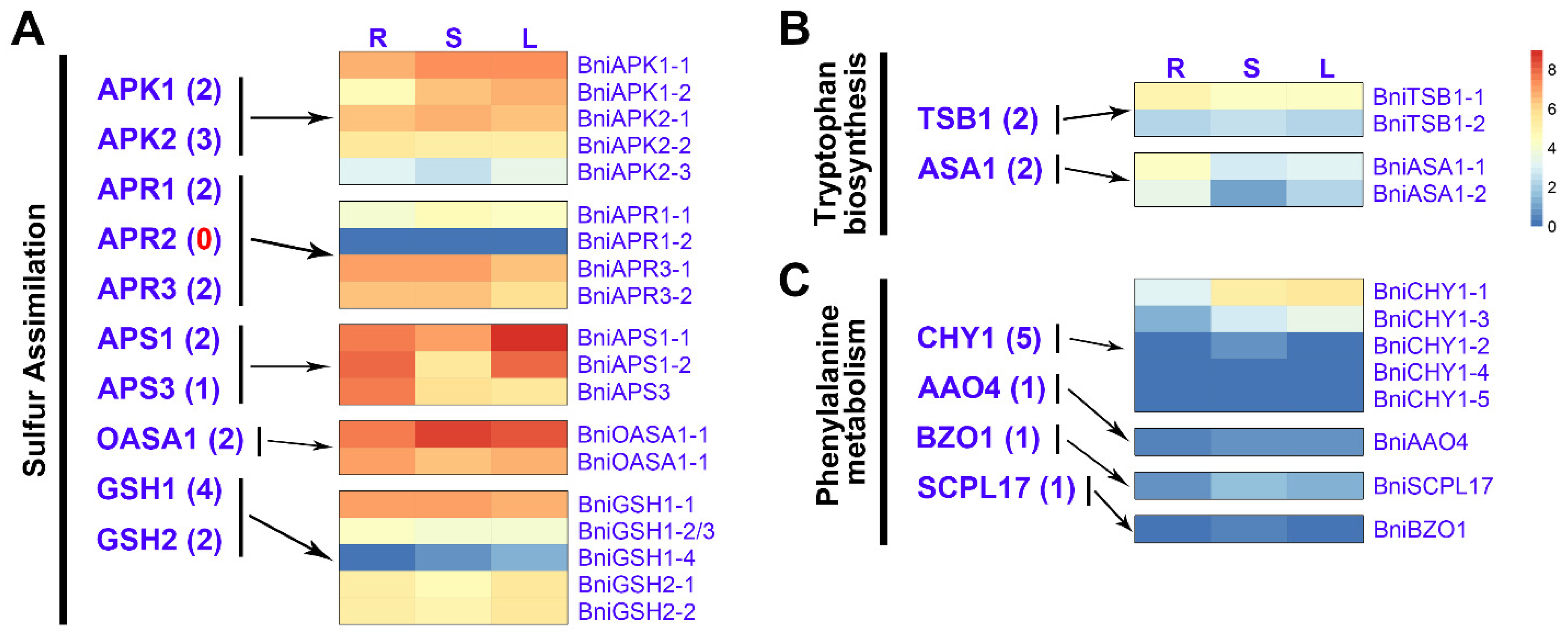
3.4. Expression Patterns of BniGSL Genes Encoding TFs, Transporters and Proteins Involved in Co-Substrate Pathways
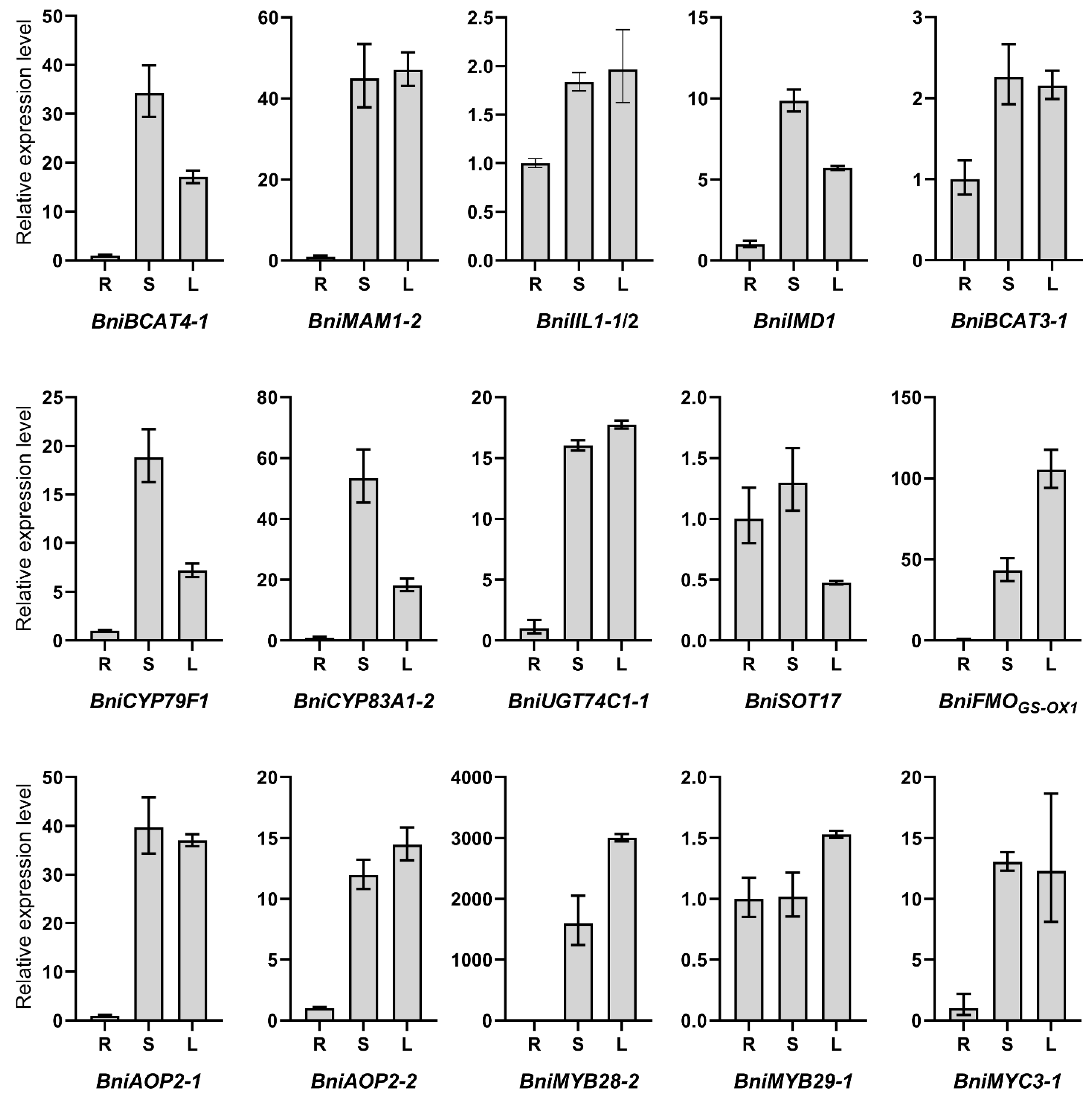
3.5. Expression Patterns of Candidate Key Genes Involved in the Synthesis of Aliphatic GSLs by qRT-PCR
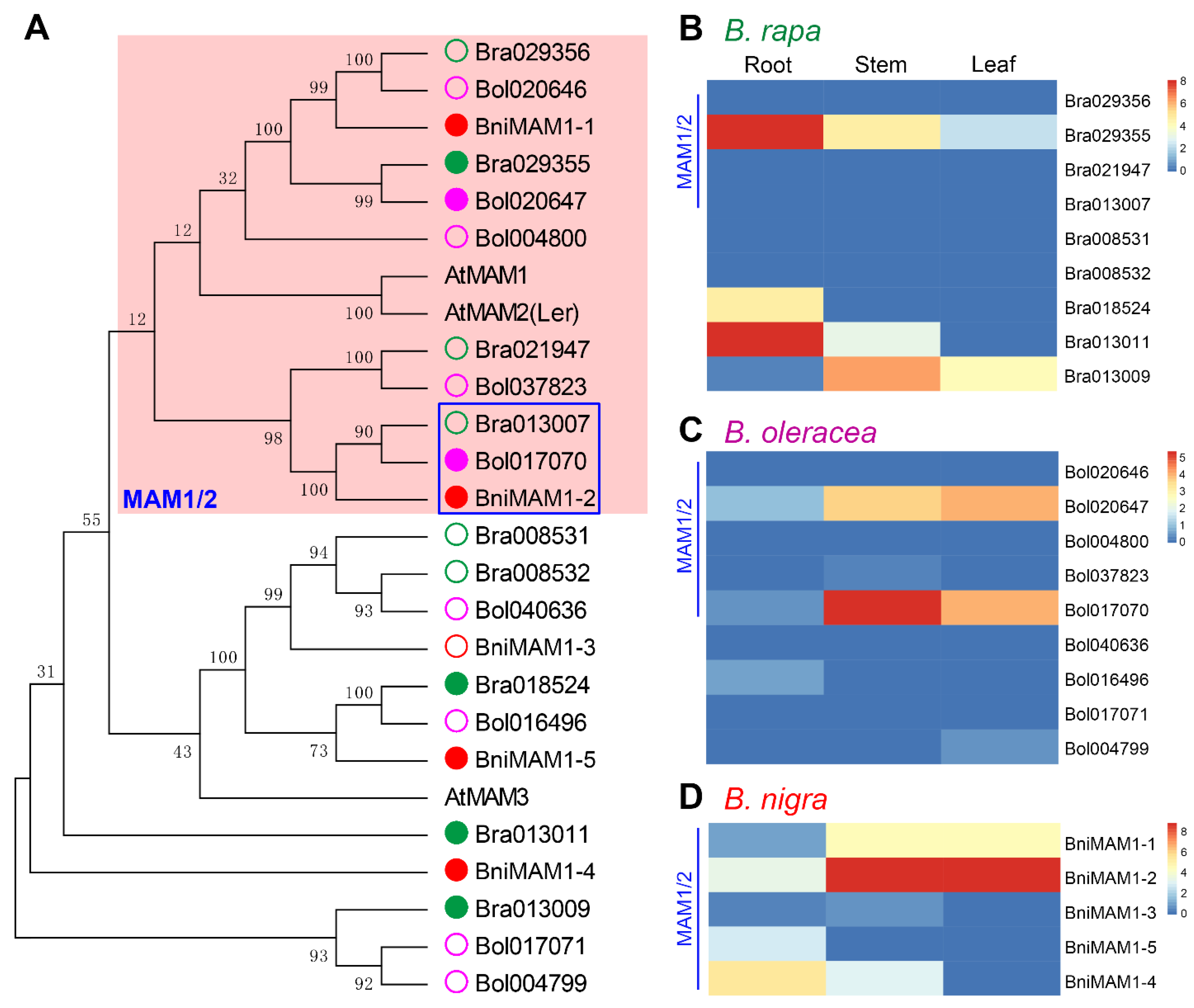
3.6. Key MAM Genes Controlling Side-Chain Elongation in B. nigra
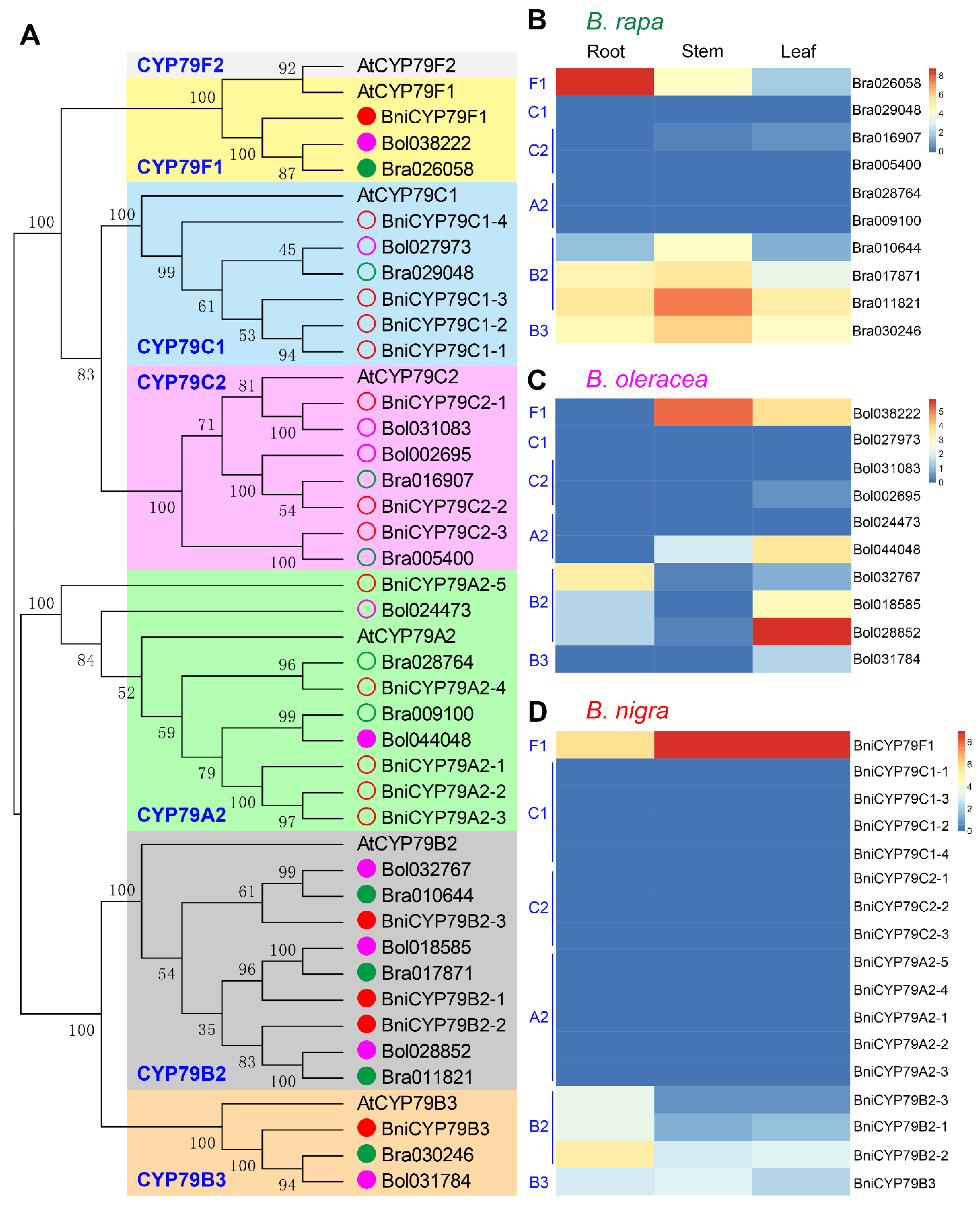
3.7. BniCYP79F1 Was Extremely Highly Expressed in B. nigra
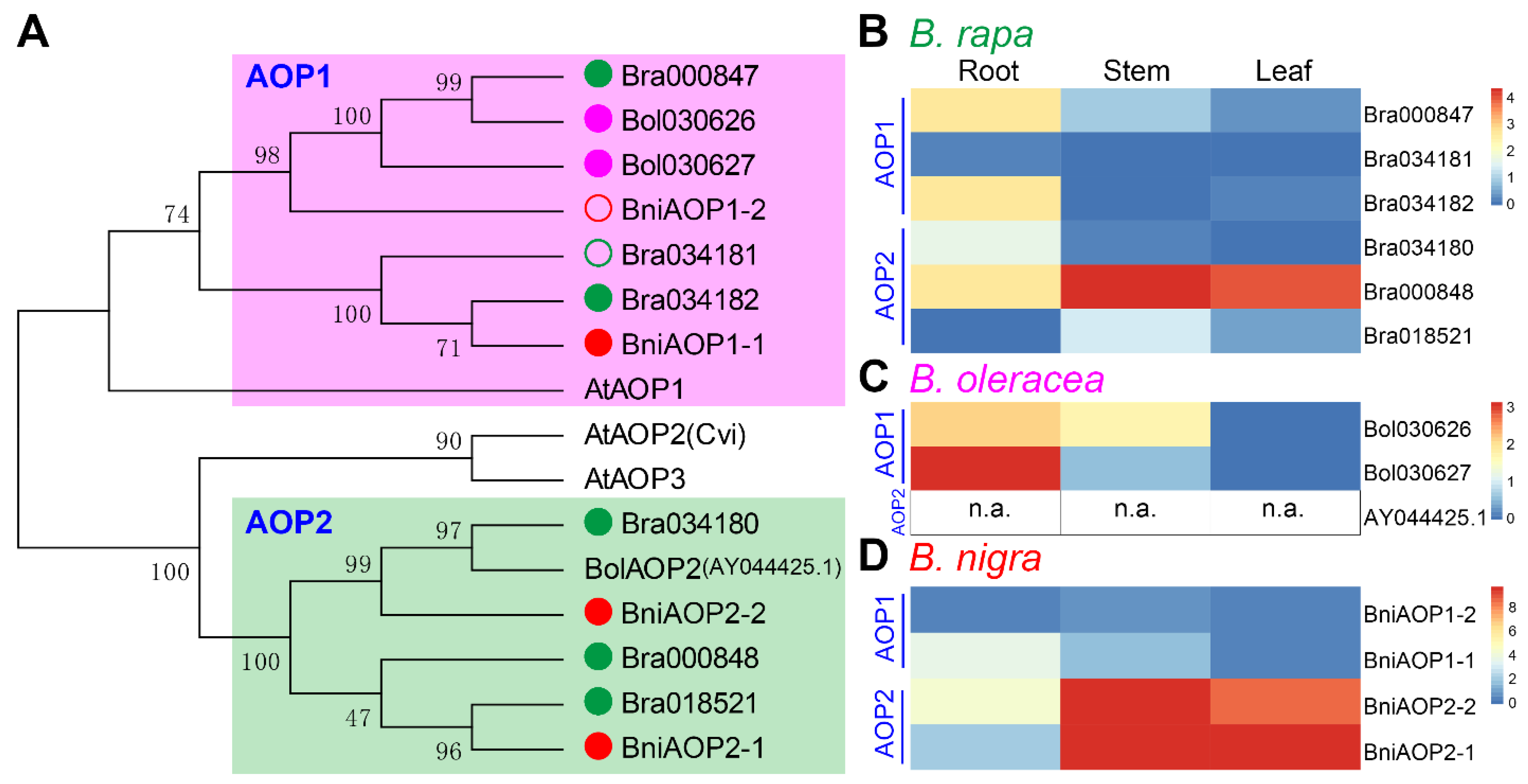
3.8. The Difference in AOP2 Genes Greatly Affect the Diversity of GSLs in Brassica
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blazevic, I.; Montaut, S.; Burcul, F.; Olsen, C.E.; Burow, M.; Rollin, P.; Agerbirk, N. Glucosinolate structural diversity, identification, chemical synthesis and metabolism in plants. Phytochemistry 2019, 169, 112100. [Google Scholar] [CrossRef]
- Agerbirk, N.; Hansen, C.C.; Kiefer, C.; Hauser, T.P.; Ørgaard, M.; Asmussen Lange, C.B.; Cipollini, D.; Koch, M.A. Comparison of glucosinolate diversity in the crucifer tribe Cardamineae and the remaining order Brassicales highlights repetitive evolutionary loss and gain of biosynthetic steps. Phytochemistry 2021, 185, 112668. [Google Scholar] [CrossRef]
- Halkier, B.A.; Gershenzon, J. Biology and Biochemistry of Glucosinolates. Annu. Rev. Plant Biol. 2006, 57, 303–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agerbirk, N.; Olsen, C.E. Glucosinolate structures in evolution. Phytochemistry 2012, 77, 16–45. [Google Scholar] [CrossRef]
- Bones, A.M.; Rossiter, J.T. The enzymic and chemically induced decomposition of glucosinolates. Phytochemistry 2006, 67, 1053–1067. [Google Scholar] [CrossRef]
- Padilla, G.; Cartea, M.E.; Velasco, P.; De Haro, A.; Ordás, A. Variation of glucosinolates in vegetable crops of Brassica rapa. Phytochemistry 2007, 68, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, R.J.; van Dam, N.M.; van Loon, J.J.A. Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu. Rev. Entomol. 2009, 54, 57–83. [Google Scholar] [CrossRef]
- Arumugam, A.; Abdull Razis, A.F. Apoptosis as a Mechanism of the Cancer Chemopreventive Activity of Glucosinolates: A Review. Asian Pac. J. Cancer Prev. 2018, 19, 1439–1448. [Google Scholar] [PubMed]
- Mazumder, A.; Dwivedi, A.; Du Plessis, J. Sinigrin and Its Therapeutic Benefits. Molecules 2016, 21, 416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sønderby, I.E.; Geu-Flores, F.; Halkier, B.A. Biosynthesis of glucosinolates—Gene discovery and beyond. Trends Plant Sci. 2010, 15, 283–290. [Google Scholar] [CrossRef]
- Harun, S.; Abdullah-Zawawi, M.-R.; Goh, H.-H.; Mohamed-Hussein, Z.-A. A Comprehensive Gene Inventory for Glucosinolate Biosynthetic Pathway in Arabidopsis thaliana. J. Agric. Food Chem. 2020, 68, 7281–7297. [Google Scholar] [CrossRef] [PubMed]
- Grubb, C.D.; Abel, S. Glucosinolate metabolism and its control. Trends Plant Sci. 2006, 11, 89–100. [Google Scholar] [CrossRef]
- Textor, S.; Kraker, J.-W.; De Hause, B.; Gershenzon, J.; Tokuhisa, J.G. MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol. 2007, 144, 60–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gigolashvili, T.; Yatusevich, R.; Rollwitz, I.; Humphry, M.; Gershenzon, J.; Flügge, U.-I. The plastidic bile acid transporter 5 is required for the biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana. Plant Cell 2009, 21, 1813–1829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Wang, H.; Liu, Z.; Liang, J.; Wu, J.; Cheng, F.; Mei, S.; Wang, X. A naturally occurring variation in the BrMAM-3 gene is associated with aliphatic glucosinolate accumulation in Brassica rapa leaves. Hortic. Res. 2018, 5, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Dissing, M.M.; Agerbirk, N.; Crocoll, C.; Halkier, B.A. Characterization of Arabidopsis CYP79C1 and CYP79C2 by Glucosinolate Pathway Engineering in Nicotiana benthamiana Shows Substrate Specificity Toward a Range of Aliphatic and Aromatic Amino Acids. Front. Recent Dev. Plant Sci. 2020, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Bak, S.; Feyereisen, R. The Involvement of Two P450 Enzymes, CYP83B1 and CYP83A1, in Auxin Homeostasis and Glucosinolate Biosynthesis. Plant Physiol. 2001, 127, 108–118. [Google Scholar] [CrossRef] [Green Version]
- Naur, P.; Petersen, B.L.; Mikkelsen, M.D.; Bak, S.; Rasmussen, H.; Olsen, C.E.; Halkier, B.A. CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol. 2003, 133, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Mikkelsen, M.D.; Naury, P.; Halkier, B.A. Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 2004, 37, 770–777. [Google Scholar] [CrossRef]
- Grubb, C.D.; Zipp, B.J.; Kopycki, J.; Schubert, M.; Quint, M.; Lim, E.-K.; Bowles, D.J.; Pedras, M.S.C.; Abel, S. Comparative analysis of Arabidopsis UGT74 glucosyltransferases reveals a special role of UGT74C1 in glucosinolate biosynthesis. Plant J. 2014, 79, 92–105. [Google Scholar] [CrossRef]
- Piotrowski, M.; Schemenewitz, A.; Lopukhina, A.; Müller, A.; Janowitz, T.; Weiler, E.W.; Oecking, C. Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J. Biol. Chem. 2004, 279, 50717–50725. [Google Scholar] [CrossRef] [Green Version]
- Hirschmann, F.; Krause, F.; Baruch, P.; Chizhov, I.; Mueller, J.W.; Manstein, D.J.; Papenbrock, J.; Fedorov, R. Structural and biochemical studies of sulphotransferase 18 from Arabidopsis thaliana explain its substrate specificity and reaction mechanism. Sci. Rep. 2017, 7, 4160. [Google Scholar] [CrossRef] [PubMed]
- Burow, M.; Atwell, S.; Francisco, M.; Kerwin, R.E.; Halkier, B.A.; Kliebenstein, D.J. The Glucosinolate Biosynthetic Gene AOP2 Mediates Feed-back Regulation of Jasmonic Acid Signaling in Arabidopsis. Mol. Plant 2015, 8, 1201–1212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, W.; Li, J.; Yu, Q.; Cang, W.; Xu, R.; Wang, Y.; Ji, W. Two Novel Flavin-Containing Monooxygenases Involved in Biosynthesis of Aliphatic Glucosinolates. Front. Plant Sci. 2016, 7, 1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gigolashvili, T.; Engqvist, M.; Yatusevich, R.; Müller, C.; Flügge, U.-I. HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 2008, 177, 627–642. [Google Scholar] [CrossRef]
- Sønderby, I.E.; Burow, M.; Rowe, H.C.; Kliebenstein, D.J.; Halkier, B.A. A complex interplay of three R2R3 MYB transcription factors determines the profile of aliphatic glucosinolates in Arabidopsis. Plant Physiol. 2010, 153, 348–363. [Google Scholar] [CrossRef] [Green Version]
- Frerigmann, H.; Gigolashvili, T. MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol. Plant 2014, 7, 814–828. [Google Scholar] [CrossRef] [Green Version]
- Mitreiter, S.; Gigolashvili, T. Regulation of glucosinolate biosynthesis. J. Exp. Bot. 2021, 72, 70–91. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, B.; Huai, D.; Zhou, Y.; Kliebenstein, D.J. The conserved transcription factors, MYB115 and MYB118, control expression of the newly evolved benzoyloxy glucosinolate pathway in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 343. [Google Scholar] [CrossRef] [Green Version]
- Bischoff, A.; Trémulot, S. Differentiation and adaptation in Brassica nigra populations: Interactions with related herbivores. Oecologia 2011, 165, 971–981. [Google Scholar] [CrossRef] [PubMed]
- Obi, R.K.; Nwanebu, F.C.; Ndubuisi, U.U.; Orji, N.M. Antibacterial qualities and phytochemical screening of the oils of Curcubita pepo and Brassica nigra. J. Med. Plants Res. 2009, 3, 429–432. [Google Scholar]
- Alam, M.B.; Hossain, M.S.; Haque, M.E. Antioxidant and anti-inflammatory activities of the leaf extract of Brassica nigra. Int. J. Pharmaceut. Sci. Res. 2011, 2, 303–310. [Google Scholar]
- Song, X.; Wei, Y.; Xiao, D.; Gong, K.; Sun, P.; Ren, Y.; Yuan, J.; Wu, T.; Yang, Q.; Li, X.; et al. Brassica carinata genome characterization clarifies U’s triangle model of evolution and polyploidy in Brassica. Plant Physiol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Liang, J.; Cai, C.; Cai, X.; Wu, J.; Wang, X. Genome sequencing supports a multi-vertex model for Brassiceae species. Curr. Opin. Plant Biol. 2017, 36, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Padmaja, K.L.; Gupta, V.; Paritosh, K.; Pradhan, A.K.; Pental, D. Two plastid DNA lineages—Rapa/Oleracea and Nigra--within the tribe Brassiceae can be best explained by reciprocal crosses at hexaploidy: Evidence from divergence times of the plastid genomes and R-block genes of the A and B genomes of Brassica juncea. PLoS ONE 2014, 9, e93260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volden, J.; Borge, G.I.A.; Hansen, M.; Wicklund, T.; Bengtsson, G.B. Processing (blanching, boiling, steaming) effects on the content of glucosinolates and antioxidant-related parameters in cauliflower (Brassica oleracea L. ssp. botrytis). LWT Food Sci. Technol. 2009, 42, 63–73. [Google Scholar] [CrossRef]
- Wang, J.; Yu, H.; Zhao, Z.; Sheng, X.; Shen, Y.; Gu, H. Natural Variation of Glucosinolates and Their Breakdown Products in Broccoli (Brassica oleracea var. italica) Seeds. J. Agric. Food Chem. 2019, 67, 12528–12537. [Google Scholar] [CrossRef]
- Yang, B.; Quiros, C.F. Survey of glucosinolate variation in leaves of Brassica rapa crops. Genet. Resour. Crop Evol. 2010, 57, 1079–1089. [Google Scholar] [CrossRef] [Green Version]
- Mnzava, N.A.; Olson, K. Studies on tropical vegetables. Part 1: Seed amino, fatty acid and glucosinolate profile of Ethiopian mustards (Brassica carinata Braun). Food Chem. 1990, 35, 229–235. [Google Scholar] [CrossRef]
- Rangkadilok, N.; Nicolas, M.E.; Richard, B.N.; Premier, R.R.; Eagling, D.R.; Taylor, P.W.J. Developmental changes of sinigrin and glucoraphanin in threee Brassica species (Brassica nigra, Brassica juncea and Brassica oleracea var. italica). Sci. Hortic. 2002, 96, 11–26. [Google Scholar] [CrossRef]
- Bellostas, N.; Sørensen, J.C.; Sørensen, H. Profiling glucosinolates in vegetative and reproductive tissues of four Brassica species of the U-triangle for their biofumigation potential. J. Sci. Food Agric. 2007, 87, 1586–1594. [Google Scholar] [CrossRef]
- Wang, H.; Wu, J.; Sun, S.; Liu, B.; Cheng, F.; Sun, R.; Wang, X. Glucosinolate biosynthetic genes in Brassica rapa. Gene 2011, 487, 135–142. [Google Scholar] [CrossRef]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.P.; Zhao, M.; Ma, J.; Yu, J.; Huang, S.; et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, D.; Wang, X.; Ji, C.; Cheng, F.; Liu, B.; Hu, Z.; Chen, S.; Pental, D.; Ju, Y.; et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016, 48, 1225–1232. [Google Scholar] [CrossRef]
- Zhu, B.; Yang, J.; He, Y.; Zang, Y.; Zhu, Z. Glucosinolate Accumulation and Related Gene Expression in Pak Choi (Brassica rapa L. ssp. chinensis var. communis N. Tsen & S.H. Lee Hanelt) in Response to Insecticide Application. J. Agric. Food Chem. 2015, 63, 9683–9689. [Google Scholar]
- Yi, G.-E.; Robin, A.H.K.; Yang, K.; Park, J.-I.; Kang, J.-G.; Yang, T.-J.; Nou, I.-S. Identification and expression analysis of glucosinolate biosynthetic genes and estimation of glucosinolate contents in edible organs of Brassica oleracea subspecies. Molecules 2015, 20, 13089–13111. [Google Scholar] [CrossRef]
- Roshan, K.; Arya, G.C.; Bisht, N.C. Differential expression and interaction specificity of the heterotrimeric G-protein family in Brassica nigra reveal their development- and condition-specific role. Plant Cell Physiol. 2014, 11, 1954–1968. [Google Scholar]
- Tong, C.; Wang, X.; Yu, J.; Wu, J.; Li, W.; Huang, J.; Dong, C.; Hua, W.; Liu, S. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genomics 2013, 14, 689. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Tehrim, S.; Zhang, F.; Tong, C.; Huang, J.; Cheng, X.; Dong, C.; Zhou, Y.; Qin, R.; Hua, W.; et al. Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genomics 2014, 15, 3. [Google Scholar] [CrossRef] [Green Version]
- Nour-Eldin, H.H.; Andersen, T.G.; Burow, M.; Madsen, S.R.; Jørgensen, M.E.; Olsen, C.E.; Dreyer, I.; Hedrich, R.; Geiger, D.; Halkier, B.A. NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 2012, 488, 531–534. [Google Scholar] [CrossRef]
- Nambiar, D.M.; Kumari, J.; Augustine, R.; Kumar, P.; Bajpai, P.K.; Bisht, N. GTR1 and GTR2 transporters differentially regulate tissue-specific glucosinolate contents and defence responses in the oilseed crop Brassica juncea. Plant Cell Environ. 2021. [Google Scholar] [CrossRef] [PubMed]
- Benderoth, M.; Textor, S.; Windsor, A.J.; Mitchell-Olds, T.; Gershenzon, J.; Kroymann, J. Positive selection driving diversification in plant secondary metabolism. Proc. Natl. Acad. Sci. USA 2006, 103, 9118–9123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benderoth, M.; Pfalz, M.; Kroymann, J. Methylthioalkylmalate synthases: Genetics, ecology and evolution. Phytochem. Rev. 2009, 8, 255–268. [Google Scholar] [CrossRef] [Green Version]
- Hansen, N.; Ostermeier, A. Completely Derandomized Self-Adaptation in Evolution Strategies. Evol. Comput. 2001, 9, 159–195. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Glawischnig, E.; Jørgensen, K.; Naur, P.; Jørgensen, B.; Olsen, C.-E.; Hansen, C.H.; Rasmussen, H.; Pickett, J.A.; Halkier, B.A. CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J. 2003, 33, 923–937. [Google Scholar] [CrossRef] [Green Version]
- Miao, H.; Wei, J.; Zhao, Y.; Yan, H.; Sun, B.; Huang, J.; Wang, Q. Glucose signalling positively regulates aliphatic glucosinolate biosynthesis. J. Exp. Bot. 2013, 64, 1097–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikkelsen, M.D.; Halkier, B.A. Metabolic engineering of valine- and isoleucine-derived glucosinolates in Arabidopsis expressing CYP79D2 from Cassava. Plant Physiol. 2003, 131, 773–779. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Hull, A.K.; Gupta, N.R.; Goss, K.A.; Alonso, J.; Ecker, J.R.; Normanly, J.; Chory, J.; Celenza, J.L. Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 2003, 16, 3100–3112. [Google Scholar] [CrossRef] [Green Version]
- Zang, Y.-X.; Kim, D.-H.; Park, B.-S.; Hong, S.-B. Metabolic engineering of indole glucosinolates in Chinese cabbage hairy roots expressing Arabidopsis CYP79B2, CYP79B3, and CYP83B1. Biotechnol. Bioprocess Eng. 2009, 14, 467–473. [Google Scholar] [CrossRef]
- Kliebenstein, D.J.; Kroymann, J.; Brown, P.; Figuth, A.; Pedersen, D.; Gershenzon, J.; Mitchell-Olds, T. Genetic Control of Natural Variation in Arabidopsis Glucosinolate Accumulation. Plant Physiol. 2001, 126, 811–825. [Google Scholar] [CrossRef] [Green Version]
- Jensen, L.M.; Kliebenstein, D.J.; Burow, M. Investigation of the multifunctional gene AOP3 expands the regulatory network fine-tuning glucosinolate production in Arabidopsis. Front. Plant Sci. 2015, 6, 762. [Google Scholar] [CrossRef] [Green Version]
- Vallejo, F.; Tomás-Barberán, F.A.; Benavente-García, A.G.; García-Viguera, C. Total and individual glucosinolate contents in inflorescences of eight broccoli cultivars grown under various climatic and fertilisation conditions. J. Sci. Food Agric. 2003, 83, 307–313. [Google Scholar] [CrossRef]
- Schonhof, I.; Krumbein, A.; Brückner, B. Genotypic effects on glucosinolates and sensory properties of broccoli and cauliflower. Nahrung/Food 2004, 48, 25–33. [Google Scholar] [CrossRef]
- Farnham, M.W.; Wilson, P.E.; Stephenson, K.K.; Fahey, J.W. Genetic and environmental effects on glucosinolate content and chemoprotective potency of broccoli. Plant Breed. 2004, 123, 60–65. [Google Scholar] [CrossRef]
- Zang, Y.-X.; Kim, H.U.; Kim, J.A.; Lim, M.-H.; Jin, M.; Lee, S.C.; Kwon, S.-J.; Lee, S.-I.; Hong, J.K.; Park, T.-H.; et al. Genome-wide identification of glucosinolate synthesis genes in Brassica rapa. FEBS J. 2010, 276, 3559–3574. [Google Scholar] [CrossRef]
- Lysak, M.A.; Koch, M.A.; Pecinka, A.; Schubert, I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005, 15, 516–525. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.-H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Hansen, B.G.; Ober, J.A.; Kliebenstein, D.J.; Halkier, B.A. Subclade of flavin-monooxygenases involved in aliphatic glucosinolate biosynthesis. Plant Physiol. 2008, 148, 1721–1733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edger, P.P.; Pires, J.C. Gene and genome duplications: The impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 2009, 17, 699–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef]
- Wang, J.; Qiu, Y.; Wang, X.; Yue, Z.; Yang, X.; Chen, X.; Zhang, X.; Shen, D.; Wang, H.; Song, J.; et al. Insights into the species-specific metabolic engineering of glucosinolates in radish (Raphanus sativus L.) based on comparative genomic analysis. Sci. Rep. 2017, 7, 16040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X. Glucosinolates in Chinese Brassica campestris Vegetables: Chinese Cabbage, Purple Cai-tai, Choysum, Pakchoi, and Turnip. Hortscience 2008, 43, 571–574. [Google Scholar] [CrossRef] [Green Version]
| Organ | Aliphatic | Indolic | Aromatic | ||||
|---|---|---|---|---|---|---|---|
| SIN | GNA | 4OHI3M | I3M | 4MOI3M | 1MOI3M | 2PHET | |
| St | 82.35 ± 2.12d | 0.52 ± 0.3c | 0.25 ± 0.02a | 0.12 ± 0.02b | 0.05 ± 0.02ab | 0.09 ± 0.02b | 0.25 ± 0.01a |
| RL | 13.20 ± 1.08a | - | 0.50 ± 0.14b | 0.04 ± 0.01a | 0.04 ± 0.01a | 0.03 ± 0.01a | 0.78 ± 0.10b |
| CL | 53.80 ± 2.52b | 0.32 ± 0.05b | 0.32 ± 0.01ab | 0.13 ± 0.02b | 0.05 ± 0.00ab | 0.08 ± 0.01b | - |
| In | 59.14 ± 2.39c | 0.32 ± 0.25b | 1.50 ± 0.17c | - | - | - | 0.34 ± 0.01a |
| Si | 51.31 ± 0.25b | 0.35 ± 0.11a | 0.37 ± 0.07ab | 0.13 ± 0.03b | 0.06 ± 0.01b | 0.08 ± 0.01b | - |
| Name1 | Name2 | Gene ID | Chromosome Location | AA | Identity/% | AGI ID | |
|---|---|---|---|---|---|---|---|
| Side-chain elongation | |||||||
| BCAT4 | BniBCAT4-1 | BniB01g044210 | B1 (+) | 48,590,123–48,592,361 | 356 | 82.3 | AT3G19710 |
| BniBCAT4-2 | BniB07g030730 | B7 (−) | 41,353,700–41,356,224 | 306 | 62.7 | ||
| BCAT6 | BniBCAT6 | BniB04g060340 | B4 (+) | 51,535,934–51,537,864 | 359 | 86.7 | AT1G50110 |
| MAM1 | BniMAM1-1 | BniB04g003060 | B4 (+) | 1,498,531–1,501,807 | 505 | 79.8 | AT5G23010 |
| BniMAM1-2 | BniB02g076300 | B2 (+) | 64,723,274–64,726,274 | 504 | 79.1 | ||
| BniMAM1-3 | BniB08g036280 | B8 (−) | 21,370,428–21,376,553 | 495 | 74.8 | ||
| BniMAM1-4 | BniB02g076310 | B2 (+) | 64,728,011–64,730,517 | 397 | 59.6 | ||
| BniMAM1-5 | BniB05g055400 | B5 (−) | 49,610,184–49,616,319 | 388 | 56.6 | ||
| MAM3 | * | AT5G23020 | |||||
| IPMI SSU1 | BniIPMI SSU1-1 | BniB08g028010 | B8 (+) | 14,699,124–14,699,873 | 249 | 87.7 | AT2G43090 |
| BniIPMI SSU1-2 | BniB06g001990 | B6 (+) | 917,687–917,463 | 258 | 79.6 | ||
| IPMI SSU2 | BniIPMI SSU2 | BniB06g002000 | B6 (+) | 919,426–920,271 | 281 | 72.7 | AT2G43100 |
| IPMI SSU3 | * | AT3G58990 | |||||
| IIL1 | BniIIL1-1 | BniB07g011950 | B7 (+) | 15,245,075–15,248,497 | 505 | 94.5 | AT4G13430 |
| BniIIL1-2 | BniB07g011910 | B7 (+) | 15,114,671–15,118,112 | 505 | 94.3 | ||
| IMD1 | BniIMD1 | BniB05g024330 | B5 (−) | 11,915,893–11,917,777 | 408 | 91.7 | AT5G14200 |
| BCAT3 | BniBCAT3-1 | BniB08g063340 | B8 (+) | 59,567,292–59,569,478 | 419 | 87.1 | AT3G49680 |
| BniBCAT3-2 | BniB05g048020 | B5 (−) | 27,787,734–27,790,073 | 419 | 84.5 | ||
| Core structure formation | |||||||
| CYP79F1 | BniCYP79F1 | BniB06g041700 | B6 (+) | 46,406,179–46,408,341 | 541 | 82.1 | AT1G16410 |
| CYP79F2 | * | AT1G16400 | |||||
| CYP79A2 | BniCYP79A2-1 | BniB02g055340 | B2 (+) | 53,932,413–53,934,250 | 532 | 77.6 | AT5G05260 |
| BniCYP79A2-2 | BniB02g055360 | B2 (+) | 53,941,026–53,942,864 | 532 | 77.6 | ||
| BniCYP79A2-3 | BniB02g055420 | B2 (+) | 53,958,847–53,960,685 | 532 | 76.3 | ||
| BniCYP79A2-4 | BniB05g029220 | B5 (+) | 14,358,148–14,360,431 | 562 | 73.1 | ||
| BniCYP79A2-5 | BniB08g051390 | B8 (+) | 34,728,808–34,732,349 | 439 | 56.6 | ||
| CYP79B2 | BniCYP79B2-1 | BniB02g088940 | B2 (−) | 70,487,544–70,489,564 | 541 | 94.5 | AT4G39950 |
| BniCYP79B2-2 | BniB05g000130 | B5 (−) | 64,703–66,466 | 542 | 93.2 | ||
| BniCYP79B2-3 | BniB03g014770 | B3 (−) | 6,330,659–6,332,937 | 518 | 89.8 | ||
| CYP79B3 | BniCYP79B3 | BniB01g021760 | B1 (−) | 13,001,786–13,004,048 | 564 | 89.0 | AT2G22330 |
| CYP79C1 | BniCYP79C1-1 | BniB08g040770 | B8 (−) | 25,176,913–25,179,607 | 531 | 80.3 | AT1G79370 |
| BniCYP79C1-2 | BniB08g040860 | B8 (−) | 25,245,858–25,248,871 | 531 | 80 | ||
| BniCYP79C1-3 | BniB08g040830 | B8 (−) | 25,220,662–25,222,785 | 396 | 62.1 | ||
| BniCYP79C1-4 | BniB08g040890 | B8 (−) | 25,258,462–25,261,159 | 382 | 59.7 | ||
| CYP79C2 | BniCYP79C2-1 | BniB04g007960 | B4 (−) | 3,767,912–3,772,131 | 530 | 78.7 | AT1G58260 |
| BniCYP79C2-2 | BniB01g003800 | B1 (−) | 1,903,991–1,908,135 | 528 | 74.9 | ||
| BniCYP79C2-3 | BniB06g009140 | B6 (−) | 4,583,904–4,586,034 | 527 | 73.6 | ||
| CYP83A1 | BniCYP83A1-1 | BniB01g003820 | B1 (+) | 1,927,665–1,929,469 | 500 | 86.9 | AT4G13770 |
| BniCYP83A1-2 | BniB06g067060 | B6 (−) | 59,968,418–59,970,020 | 498 | 86.3 | ||
| CYP83B1 | BniCYP83B1 | BniB07g013920 | B7 (−) | 18,805,780–18,807,433 | 499 | 95.4 | AT4G31500 |
| CYTB5-C | BniCYTB5-C-1 | BniB01g000830 | B1 (−) | 449,656–450,237 | 135 | 80.1 | AT2G46650 |
| BniCYTB5-C-2 | BniS02554g140 | utg2554 + | 100,663–101,227 | 114 | 57.7 | ||
| GSTF9 | BniGSTF9-1 | BniB01g013320 | B1 (−) | 6,927,611–6,928,497 | 215 | 97.2 | AT2G30860 |
| BniGSTF9-2 | BniB08g019200 | B8 (+) | 9,243,485–9,244,436 | 215 | 96.7 | ||
| GSTF10 | BniGSTF10 | BniB08g019210 | B8 (+) | 9,246,072–9,247,215 | 215 | 94.4 | AT2G30870 |
| GSTF11 | BniGSTF11 | BniB01g061730 | B1 (+) | 57,396,702–57,397,557 | 214 | 82.2 | AT3G03190 |
| GSTU13 | BniGSTU13-1 | BniB04g039630 | B4 (−) | 21,519,677–21,520,647 | 227 | 81.1 | AT1G27130 |
| BniGSTU13-2 | BniB03g031270 | B3 (−) | 14,627,558–14,628,949 | 227 | 76.2 | ||
| GSTU20 | BniGSTU20 | BniB06g050340 | B6 (−) | 51,398,415–51,399,277 | 217 | 88.9 | AT1G78370 |
| GGP1 | BniGGP1-1 | BniB02g084810 | B2 (+) | 68,616,575–68,618,095 | 250 | 88.4 | AT4G30530 |
| BniGGP1-2 | BniB05g007160 | B5 (−) | 3,545,600–3,546,824 | 250 | 86.4 | ||
| BniGGP1-3 | BniB03g010950 | B3 (−) | 4,449,737–4,451,401 | 228 | 80 | ||
| BniGGP1-4 | BniB05g007170 | B5 (−) | 3,548,331–3,552,009 | 251 | 62.4 | ||
| GGP3 | BniGGP3 | BniB03g010940 | B3 (−) | 4,446,818–4,448,424 | 258 | 84.9 | AT4G30550 |
| SUR1 | BniSUR1-1 | BniB03g070290 | B3 (−) | 54,745,695–54,747,733 | 458 | 88.6 | AT2G20610 |
| BniSUR1-2 | BniB02g015530 | B2 (−) | 9,570,907–9,578,729 | 445 | 83.3 | ||
| UGT74B1 | BniUGT74B1 | BniB04g039340 | B4 (−) | 21,237,953–21,239,435 | 465 | 85.2 | AT1G24100 |
| UGT74C1 | BniUGT74C1-1 | BniB06g020830 | B6 (+) | 11,872,313–11,874,700 | 456 | 85.1 | AT2G31790 |
| BniUGT74C1-2 | BniB01g012360 | B1 (−) | 6,333,513–6,335,348 | 456 | 83.6 | ||
| SOT16 | BniSOT16-1 | BniB03g052560 | B3 (−) | 28,106,607–28,107,620 | 337 | 92.9 | AT1G74100 |
| BniSOT16-2 | BniB05g070980 | B5 (−) | 64,926,130–64,927,143 | 337 | 92.9 | ||
| SOT17 | BniSOT17 | BniB04g033130 | B4 (+) | 17,622,762–17,623,781 | 339 | 86.5 | AT1G18590 |
| SOT18 | BniSOT18-1 | BniB03g052540 | B3 (+) | 28,101,782–28,102,844 | 334 | 80.9 | AT1G74090 |
| BniSOT18-2 | BniB04g019280 | B4 (−) | 9,875,796–9,876,833 | 345 | 78.6 | ||
| BniSOT18-3 | BniB03g038560-1 | B3 (−) | 19,264,549–19,265,610 | 353 | 73.7 | ||
| BniSOT18-4 | BniB03g052550 | B3 (+) | 28,105,157–28,106,167 | 319 | 72.9 | ||
| BniSOT18-5 | BniB06g047660 | B6 (+) | 49,729,557–49,730,615 | 352 | 72.7 | ||
| BniSOT18-6 | BniB06g047670 | B6 (+) | 49,740,347–49,741,381 | 344 | 72.6 | ||
| BniSOT18-7 | BniB06g047650 | B6 (+) | 49,723,604–49,724,665 | 353 | 72.2 | ||
| BniSOT18-8 | BniB03g038560-2 | B3 (−) | 19,273,089–19,274,147 | 352 | 71.3 | ||
| BniSOT18-9 | BniB04g020570 | B4 (−) | 10,687,342–10,688,406 | 354 | 70.4 | ||
| BniSOT18-10 | BniB02g035710 | B2 (+) | 43,650,924–43,651,937 | 337 | 70.1 | ||
| BniSOT18-11 | BniB04g020560 | B4 (+) | 10,684,415–10,685,521 | 368 | 69.4 | ||
| BniSOT18-12 | BniB06g047680 | B6 (+) | 49,744,098–49,745,159 | 333 | 68.3 | ||
| BniSOT18-13 | BniB03g038630 | B3 (−) | 19,304,714–19,306,727 | 367 | 49.6 | ||
| Secondary modification | |||||||
| FMOGS-OX1 | BniFMOGS-OX1 | BniB04g021440 | B4 (+) | 11,434,174–11,440,389 | 461 | 77.2 | AT1G65860 |
| FMOGS-OX2 | * | AT1G62540 | |||||
| FMOGS-OX3 | * | AT1G62560 | |||||
| FMOGS-OX4 | * | AT1G62570 | |||||
| FMOGS-OX5 | BniFMOGS-OX5-1 | BniB03g004970 | B3 (+) | 1,983,575–1,985,457 | 459 | 84.6 | AT1G12140 |
| BniFMOGS-OX5-2 | BniB02g006840 | B2 (+) | 3,630,178–3,632,014 | 459 | 82.8 | ||
| FMOGS-OX6 | * | AT1G12130 | |||||
| FMOGS-OX7 | * | AT1G12160 | |||||
| CYP81F1 | BniCYP81F1-1 | BniB05g001500 | B5 (−) | 845,091–846,941 | 499 | 86.2 | AT4G37430 |
| BniCYP81F1-2 | BniB03g041020 | B3 (+) | 20,720,008–20,721,740 | 496 | 80.4 | ||
| BniCYP81F1-3 | BniB05g001510 | B5 (−) | 849,140–850,741 | 504 | 79.5 | ||
| CYP81F2 | BniCYP81F2-1 | BniB02g036600 | B2 (+) | 44,402,518–44,404,789 | 491 | 90.7 | AT5G57220 |
| BniCYP81F2-2 | BniB05g036790 | B5 (+) | 19,374,525–19,376,334 | 493 | 88.4 | ||
| BniCYP81F2-3 | BniB08g013930 | B8 (−) | 6,333,770–6,33,6220 | 493 | 87.2 | ||
| CYP81F3 | BniCYP81F3-1 | BniB05g001540 | B5 (−) | 866,140–869,499 | 497 | 89.3 | AT4G37400 |
| BniCYP81F3-2 | BniB03g014280 | B3 (+) | 6,049,124–6,051,489 | 491 | 86.2 | ||
| CYP81F4 | BniCYP81F4-1 | BniB05g001530 | B5 (−) | 859,533–861,269 | 501 | 78.7 | AT4G37410 |
| BniCYP81F4-2 | BniB03g014290 | B3 (+) | 6,062,864–6,065,223 | 519 | 76.9 | ||
| AOP1 | BniAOP1-1 | BniB08g045800 | B8 (+) | 28,827,457–28,828,861 | 320 | 76.5 | AT4G03070 |
| BniAOP1-2 | BniB08g039120 | B8 (−) | 23,904,179–23,911,127 | 423 | 49.2 | ||
| AOP2 | BniAOP2-1 | BniB05g055540 | B5 (−) | 49,961,828–49,965,927 | 432 | 59.2 | AT4G03060 |
| BniAOP2-2 | BniB08g045740 | B8 (−) | 28,785,908–28,787,937 | 475 | 51.3 | ||
| AOP3 | * | AT4G03050 | |||||
| GSL-OH | * | AT2G25450 | |||||
| IGMT1 | BniIGMT1-1 | BniB04g035790 | B4 (−) | 19,012,288–19,013,601 | 372 | 90.4 | AT1G21100 |
| BniIGMT1-2 | BniB03g008950 | B3 (−) | 3,546,629–3,547,917 | 372 | 89.1 | ||
| IGMT2 | BniIGMT2 | BniB03g008960 | B3 (−) | 3,550,953–3,552,247 | 374 | 92.6 | AT1G21120 |
| IGMT5 | BniIGMT5 | BniB03g055850 | B3 (−) | 30,178,032–3,0179,487 | 370 | 79.4 | AT1G76790 |
| Co-substrate pathways | |||||||
| TSB1 | BniTSB1-1 | BniB02g033500 | B2 (−) | 42,216,546–42,218,333 | 472 | 92.6 | AT5G54810 |
| BniTSB1-2 | BniB05g034690 | B5 (−) | 17,928,767–17,930,499 | 309 | 55.2 | ||
| ASA1 | BniASA1-1 | BniB08g002350 | B8 (−) | 1,172,677–1,175,600 | 594 | 88.9 | AT5G05730 |
| BniASA1-2 | BniB02g054940 | B2 (+) | 53,796,911–53,799,668 | 604 | 88.2 | ||
| APK1 | BniAPK1-1 | BniB06g037540 | B6 (+) | 43,668,583–43,669,978 | 273 | 89.5 | AT2G14750 |
| BniAPK1-2 | BniB02g011750 | B2 (+) | 68,87,476–6,888,716 | 278 | 88.6 | ||
| APK2 | BniAPK2-1 | BniB02g088950 | B2 (−) | 70,492,417–70,493,784 | 293 | 90.1 | AT4G39940 |
| BniAPK2-2 | BniB05g000140 | B5 (−) | 71,545–73,229 | 293 | 89.8 | ||
| BniAPK2-3 | BniB03g014780 | B3 (−) | 6,338,724–6,339,871 | 227 | 63.9 | ||
| GSH1 | BniGSH1-1 | BniB05g014010 | B5 (+) | 6,646,285–6,649,224 | 514 | 93.1 | AT4G23100 |
| BniGSH1-2 | BniB02g080290-2 | B2 (−) | 66,603,961–66,606,919 | 520 | 92.2 | ||
| BniGSH1-3 | BniB02g080290-1 | B2 (−) | 66,598,228–66,600,769 | 469 | 62.6 | ||
| BniGSH1-4 | BniB03g021390 | B3 (+) | 9,550,645–9,553,182 | 359 | 61.7 | ||
| GSH2 | BniGSH2-1 | BniB07g038410 | B7 (−) | 45,708,701–45,711,311 | 531 | 79.8 | AT5G27380 |
| BniGSH2-2 | BniB02g079220 | B2 (+) | 66,040,689–66,048,921 | 478 | 75.1 | ||
| APS1 | BniAPS1-1 | BniB03g024880 | B3 (+) | 11,208,027–11,211,252 | 465 | 92.9 | AT3G22890 |
| BniAPS1-2 | BniB01g039550 | B1 (−) | 44,897,233–44,899,328 | 462 | 92.7 | ||
| APS3 | BniAPS3 | BniB05g021440 | B5 (+) | 10,445,377–10,447,307 | 470 | 89.9 | AT4G14680 |
| APR1 | BniAPR1-1 | BniB08g047710 | B8 (−) | 30,542,974–30,544,615 | 463 | 87 | AT4G04610 |
| BniAPR1-2 | BniB05g056120 | B5 (−) | 50,700,159–50,701,310 | 223 | 35.5 | ||
| APR2 | * | AT1G62180 | |||||
| APR3 | BniAPR3-1 | BniB02g079650 | B2 (−) | 66,283,619–66,285,188 | 464 | 92.5 | AT4G21990 |
| BniAPR3-2 | BniB05g015070 | B5 (+) | 7,144,150–7,145,729 | 464 | 92.2 | ||
| OASA1 | BniOASA1-1 | BniB07g020470 | B7 (−) | 33,626,673–33,628,609 | 322 | 97.5 | AT4G14880 |
| BniOASA1-2 | BniB05g021220 | B5 (+) | 10,294,312–10,296,240 | 324 | 94.8 | ||
| CHY1 | BniCHY1-1 | BniB04g000010 | B4 (−) | 7994–10,682 | 374 | 87 | AT5G65940 |
| BniCHY1-2 | BniB01g014580 | B1 (−) | 7,787,511–7,790,188 | 380 | 80.7 | ||
| BniCHY1-3 | BniB01g014490 | B1 (+) | 7,718,116–7,721,800 | 332 | 73.8 | ||
| BniCHY1-4 | BniB06g018370 | B6 (+) | 10,266,194–10,271,471 | 370 | 71.8 | ||
| BniCHY1-5 | BniB06g018590 | B6 (+) | 10,442,521–10,445,218 | 225 | 39.4 | ||
| AAO4 | BniAAO4 | BniB06g046290 | B6 (+) | 49,137,455–49,142,867 | 1337 | 86.8 | AT1G04580 |
| BZO1 | BniBZO1 | BniB03g043330 | B3 (−) | 22,256,842–22,258,819 | 566 | 76.4 | AT1G65880 |
| SCPL17 | BniSCPL17 | BniB07g056480 | B7 (+) | 55,233,985–55,236,333 | 436 | 65.3 | AT3G12203 |
| Name1 | Name2 | Gene ID | Chromosome Location | AA | Identity/% | AGI ID | |
|---|---|---|---|---|---|---|---|
| Activator | |||||||
| MYB28 | BniMYB28-1 | BniB02g075850 | B2 (+) | 64,495,904–64,497,224 | 352 | 78.6 | AT5G61420 |
| BniMYB28-2 | BniB07g041200 | B7 (−) | 47,224,440–47,225,564 | 329 | 68.3 | ||
| BniMYB28-3 | BniB04g002670 | B4 (−) | 1,321,731–1,323,290 | 329 | 60.2 | ||
| MYB29 | BniMYB29-1 | BniB08g003370 | B8 (+) | 1,668,386–1,669,814 | 339 | 74.3 | AT5G07690 |
| BniMYB29-2 | BniB02g053290 | B2 (−) | 53,047,760–53,050,344 | 298 | 46.2 | ||
| MYB76 | * | AT5G07700 | |||||
| MYB34 | BniMYB34-1 | BniB02g076220 | B2 (−) | 64,684,279–64,685,680 | 313 | 75.1 | AT5G60890 |
| BniMYB34-2 | BniB04g003010 | B4 (−) | 1,462,793–1,463,979 | 276 | 70.4 | ||
| BniMYB34-3 | BniB07g040970 | B7 (−) | 47,066,742–47,067,688 | 262 | 60.7 | ||
| MYB51 | BniMYB51-1 | BniB03g028100 | B3 (+) | 12,800,136–12,801,361 | 326 | 75.4 | AT1G18570 |
| BniMYB51-2 | BniB03g007660 | B3 (+) | 2,995,206–2,996,724 | 319 | 72.1 | ||
| BniMYB51-3 | BniB04g033080 | B4 (+) | 17,574,440–17,575,684 | 323 | 71.7 | ||
| MYB115 | * | AT5G40360 | |||||
| MYB118 | BniMYB118-1 | BniB04g007120 | B4 (+) | 3,444,507–3,446,768 | 486 | 63 | AT3G27785 |
| BniMYB118-2 | BniB02g067910 | B2 (−) | 60,652,891–60,655,308 | 477 | 63 | ||
| MYB122 | BniMYB122-1 | BniB03g052520 | B3 (+) | 28,090,119–28,091,605 | 335 | 74.9 | AT1G74080 |
| BniMYB122-2 | BniB05g070970 | B5 (+) | 64,915,053–64,916,920 | 376 | 67.8 | ||
| MYC2 | BniMYC2-1 | BniB04g049150 | B4 (−) | 30,208,579–30,210,393 | 604 | 87.2 | AT1G32640 |
| BniMYC2-2 | BniB03g035750 | B3 (−) | 17,597,458–17,599,278 | 606 | 86.6 | ||
| BniMYC2-3 | BniB03g016310 | B3 (+) | 7,033,365–7,035,185 | 606 | 84.3 | ||
| MYC3 | BniMYC3-1 | BniB02g063410 | B2 (+) | 58,260,877–58,262,631 | 584 | 76.9 | AT5G46760 |
| BniMYC3-2 | BniB04g015560 | B4 (−) | 7,676,518–7,678,221 | 567 | 71.5 | ||
| MYC4 | BniMYC4 | BniB05g018490 | B5 (+) | 8,814,854–8,816,635 | 593 | 73.1 | AT4G17880 |
| IQD1 | BniIQD1-1 | BniB07g058100 | B7 (+) | 55,932,859–55,934,585 | 444 | 67.2 | AT3G09710 |
| BniIQD1-2 | BniB03g023450 | B3 (−) | 10,613,021–10,615,049 | 485 | 59.9 | ||
| SLIM1 | BniSLIM1-1 | BniB03g052120 | B3 (−) | 27,906,056–27,908,203 | 587 | 85.1 | AT1G73730 |
| BniSLIM1-2 | BniB03g038740 | B3 (+) | 19,356,898–19,358,816 | 584 | 82.1 | ||
| BniSLIM1-3 | BniB05g070840 | B5 (−) | 64,849,951–64,851,819 | 538 | 77.1 | ||
| OBP2 | BniOBP2-1 | BniB03g002340 | B3 (−) | 936,743–938,038 | 340 | 76.4 | AT1G07640 |
| BniOBP2-2 | BniB06g043020 | B6 (+) | 47,097,586–47,098,898 | 334 | 75.8 | ||
| BniOBP2-3 | BniB02g003700 | B2 (−) | 1,819,220–1,820,440 | 313 | 69.5 | ||
| CAMTA3 | BniCAMTA3-1 | BniB01g021790 | B1 (−) | 13,029,617–13,034,577 | 1034 | 86.2 | AT2G22300 |
| BniCAMTA3-2 | BniB03g062790 | B3 (−) | 45,548,997–45,550,544 | 429 | 34.9 | ||
| BniCAMTA3-3 | BniB02g018410 | B2 (+) | 11,860,712–11,862,740 | 358 | 27 | ||
| CCA1 | BniCCA1 | BniS02554g500 | utg2554 (−) | 328,706–331,283 | 572 | 74.4 | AT2G46830 |
| Activator/Repressor | |||||||
| HY5 | BniHY5-1 | BniB05g026170 | B5 (+) | 12,836,342–12,837,354 | 163 | 91.7 | AT5G11260 |
| BniHY5-2 | BniB02g050820 | B2 (+) | 51,949,284–51,950,286 | 168 | 91.1 | ||
| BniHY5-3 | BniB06g056490 | B6 (−) | 54,414,011–54,416,320 | 208 | 57.8 | ||
| Repressor | |||||||
| SD1 | BniSD1-1 | BniB02g069920 | B2 (−) | 61,609,407–61,611,242 | 306 | 92.5 | AT5G48850 |
| BniSD1-2 | BniB07g037550 | B7 (−) | 45,221,658–45,223,201 | 306 | 90.5 | ||
| BniSD1-3 | BniB04g005760 | B4 (+) | 2,848,170–2,850,064 | 299 | 88.9 | ||
| BniSD1-4 | BniB07g037520 | B7 (−) | 45,208,991–45,210,584 | 306 | 88.2 | ||
| SD2 | BniSD2-1 | BniB06g046090 | B6 (+) | 49,043,247–49,044,406 | 303 | 88.2 | AT1G04770 |
| BniSD2-2 | BniB03g001070 | B3 (−) | 446,167–447,323 | 299 | 83.8 | ||
| FRS7 | BniFRS7 | BniB01g057400 | B1 (+) | 55,333,501–55,335,611 | 534 | 51.1 | AT3G06250 |
| FRS12 | BniFRS12 | BniB08g009620 | B8 (+) | 4,493,856–4,496,198 | 780 | 86.4 | AT5G18960 |
| Mediator | |||||||
| MED5 | BniMED5-1 | BniB01g038210 | B1 (−) | 43,391,779–43,397,254 | 1308 | 89.5 | AT3G23590 |
| BniMED5-2 | BniB07g048400 | B7 (−) | 51,150,764–51,156,016 | 1297 | 87.3 | ||
| MED25 | BniMED25-1 | BniB03g018400 | B3 (−) | 8,065,052–8,070,109 | 830 | 87.9 | AT1G25540 |
| BniMED25-2 | BniB04g040670 | B4 (+) | 22,229,344–22,231,613 | 396 | 23.7 | ||
| Transporter | |||||||
| BAT5 | BniBAT5-1 | BniB08g037040 | B8 (+) | 22,004,327–22,006,322 | 408 | 90 | AT4G12030 |
| BniBAT5-2 | BniB08g049400 | B8 (−) | 32,467,906–32,474,047 | 393 | 79.1 | ||
| SULTR1;1 | BniSULTR1;1 | BniB05g033180 | B5 (−) | 16,848,645–16,851,311 | 519 | 69.8 | AT4G08620 |
| SULTR1;2 | BniSULTR1;2-1 | BniB05g074030 | B5 (+) | 67,138,702–67,142,049 | 652 | 93.7 | AT1G78000 |
| BniSULTR1;2-2 | BniB03g057280 | B3 (+) | 31,113,533–31,116,846 | 655 | 92.7 | ||
| BniSULTR1;2-3 | BniB03g057270 | B3 (+) | 31,102,957–31,106,559 | 671 | 91.8 | ||
| GTR1 | BniGTR1-1 | BniB06g024230 | B6 (+) | 14,337,898–14,340,309 | 634 | 83.4 | AT3G47960 |
| BniGTR1-2 | BniB08g061180 | B8 (−) | 57,886,043–57,888,835 | 615 | 80.7 | ||
| BniGTR1-3 | BniB05g052360 | B5 (−) | 44,394,313–44,396,700 | 617 | 78 | ||
| GTR2 | BniGTR2-1 | BniB04g002130 | B4 (+) | 1,022,088–1,024,566 | 612 | 92 | AT5G62680 |
| BniGTR2-2 | BniB06g010940 | B6 (−) | 5,574,061–5,576,460 | 612 | 91.6 | ||
| BniGTR2-3 | BniB07g041760 | B7 (−) | 47,577,010–47,579,359 | 606 | 85.2 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Yu, Y.; Xu, L.; Guo, E.; Zang, Y.; He, Y.; Zhu, Z. Transcriptome Profiling Reveals Candidate Key Genes Involved in Sinigrin Biosynthesis in Brassica nigra. Horticulturae 2021, 7, 173. https://doi.org/10.3390/horticulturae7070173
Li Y, Yu Y, Xu L, Guo E, Zang Y, He Y, Zhu Z. Transcriptome Profiling Reveals Candidate Key Genes Involved in Sinigrin Biosynthesis in Brassica nigra. Horticulturae. 2021; 7(7):173. https://doi.org/10.3390/horticulturae7070173
Chicago/Turabian StyleLi, Yang, Youjian Yu, Liai Xu, Erbiao Guo, Yunxiang Zang, Yong He, and Zhujun Zhu. 2021. "Transcriptome Profiling Reveals Candidate Key Genes Involved in Sinigrin Biosynthesis in Brassica nigra" Horticulturae 7, no. 7: 173. https://doi.org/10.3390/horticulturae7070173







