Multi-Omics Analysis to Visualize Ecotype-Specific Heterogeneity of the Metabolites in the Mesocarp Tissue of Three Avocado (Persea Americana Mill.) Ecotypes
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
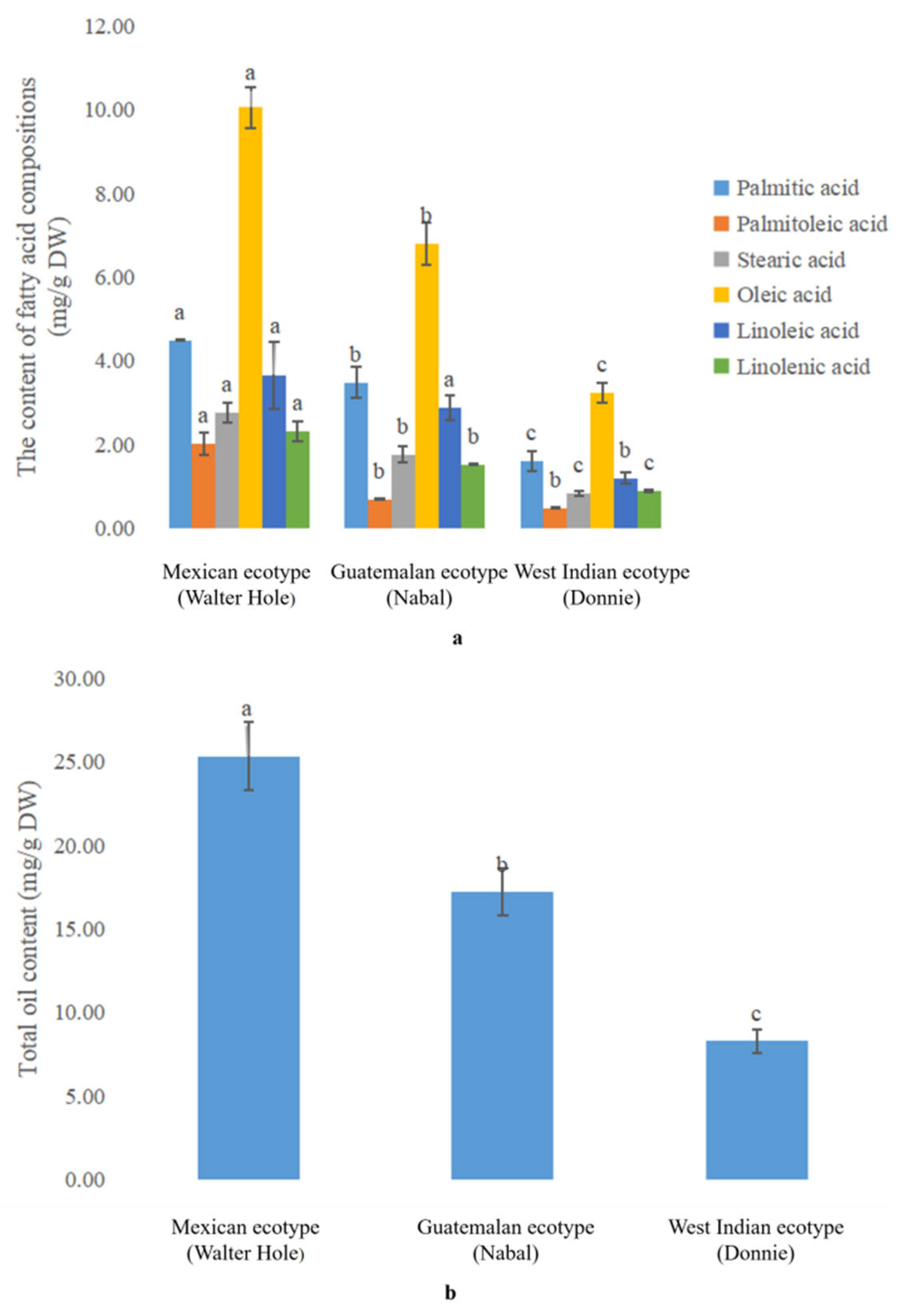
2.2. Analysis of the Fatty Acid Composition by Gas Chromatography-Mass Spectrometry
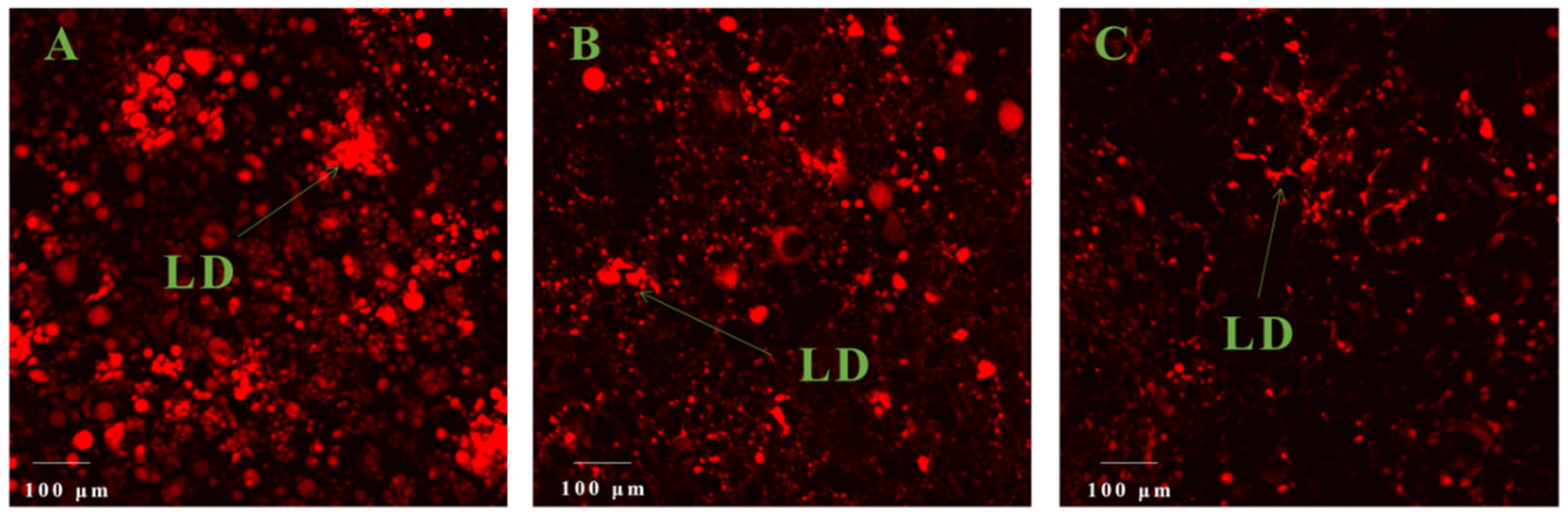
2.3. Histological Analyses
2.4. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
2.5. Analysis of the Lipids by Ultra-High-Performance Liquid Chromatography-Triple and Time-of-Flight Mass Spectrometry
2.6. Analysis of the Metabolites by Ultra-High Performance Liquid Chromatography-Q Exactive-Mass Spectrometry
3. Results
3.1. Comparative Analysis of the Oil Content in the Mature Avocado Mesocarps of Three Ecotypes
3.2. Expression-Level Changes to Lipid-Related Genes Involved in FA Synthesis and TAG Storage among the Mature Avocado Mesocarps of Three Ecotypes
3.3. Lipids Differences among the Mature Avocado Mesocarps of Three Ecotypes
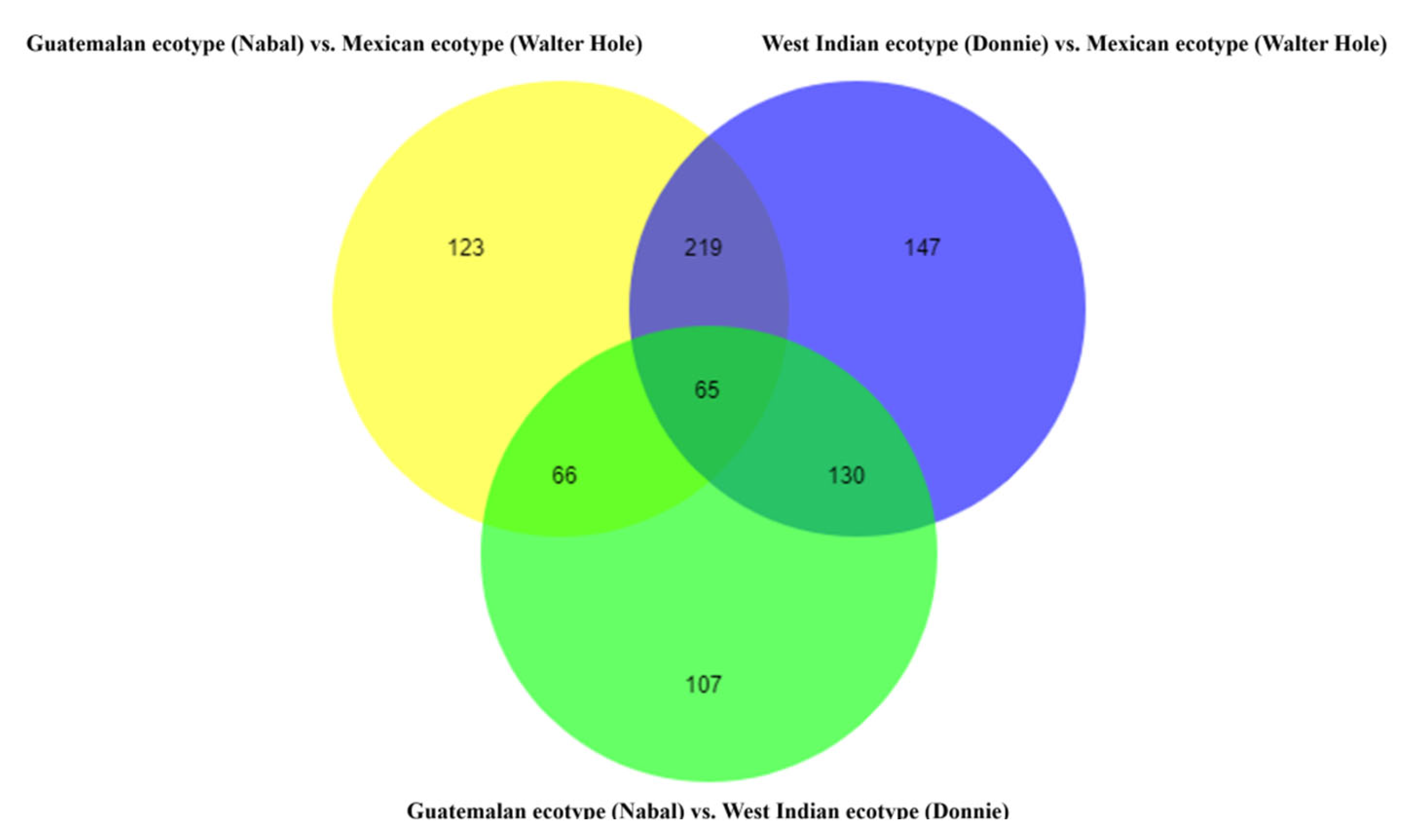
3.4. Metabolites Differences among the Mature Avocado Mesocarps of Three Ecotypes
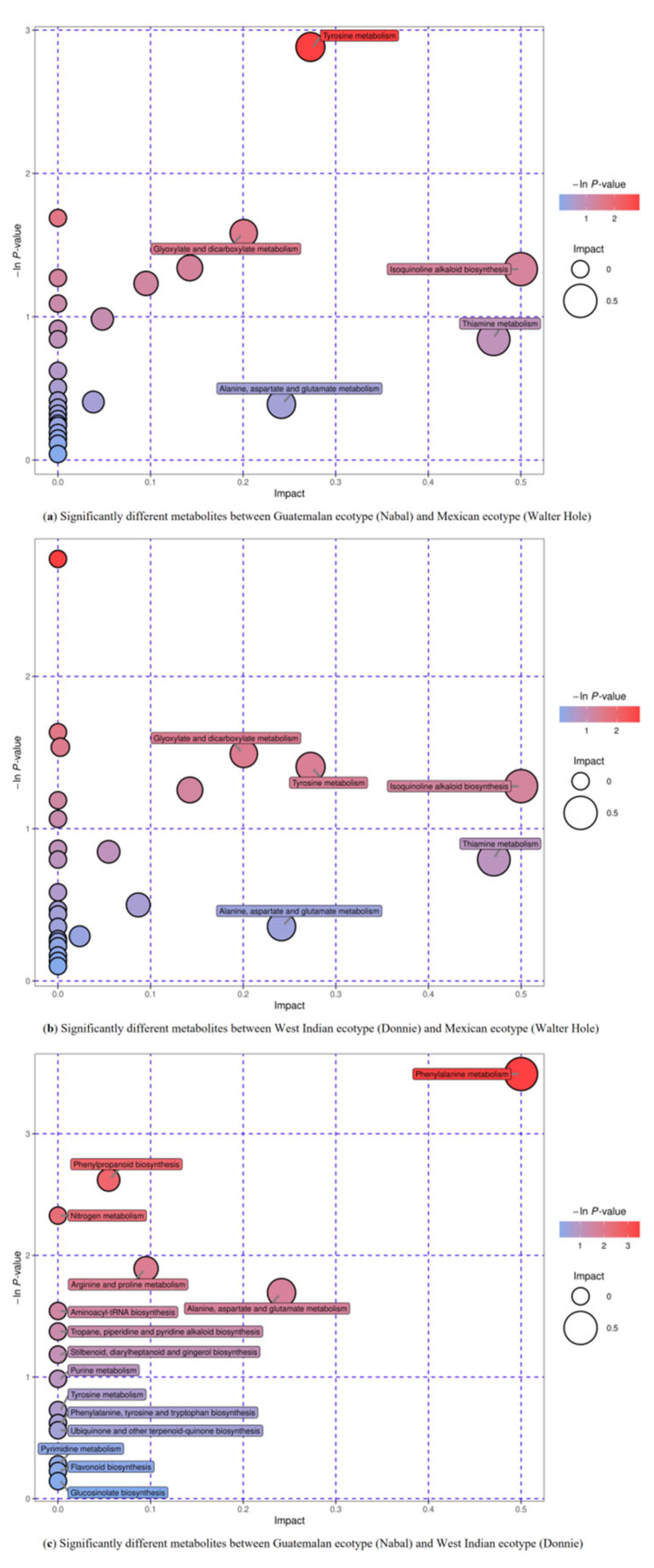
3.5. Metabolic Pathway Analysis of Differentially Accumulated Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ge, Y.; Cheng, Z.H.; Si, X.Y.; Ma, W.H.; Tan, L.; Zang, X.P.; Wu, B.; Xu, Z.N.; Wang, N.; Zhou, Z.X.; et al. Transcriptome profiling provides insight into the genes in carotenoid biosynthesis during the mesocarp and seed developmental stages of avocado (Persea Americana). Int. J. Mol. Sci. 2019, 20, 4117. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, D.N.; Livingstone, D.S., III; Richards, J.H.; Manosalva, P.; Van den Berg, P.; Chambers, A.H. Application of genomic tools to avocado (Persea americana) breeding: SNP discovery for genotyping and germplasm characterization. Sci. Hortic. 2019, 246, 1–11. [Google Scholar] [CrossRef]
- Schaffer, B.; Wolstenholme, B.N.; Whiley, A.W. The Avocado: Botany, Production and Uses, 2nd ed.; CPI Group (UK) Ltd.: Croydon, UK, 2012. [Google Scholar]
- Ge, Y.; Zhang, T.; Wu, B.; Tan, L.; Ma, F.N.; Zou, M.H.; Chen, H.H.; Pei, J.L.; Liu, Y.Z.; Chen, Z.H.; et al. Genome-wide assessment of avocado germplasm determined from specific length amplified fragment sequencing and transcriptomes: Population structure, genetic diversity, identification, and application of race-specific markers. Genes 2019, 10, 215. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Dong, X.S.; Wu, B.; Wang, N.; Chen, D.; Chen, H.H.; Zou, M.H.; Xu, Z.N.; Tan, L.; Zhan, R.L. Evolutionary analysis of six chloroplast genomes from three Persea americana ecological races: Insights into sequence divergences and phylogenetic relationships. PLoS ONE 2019, 14, e0221827. [Google Scholar] [CrossRef] [PubMed]
- Dreher, M.L.; Davenport, A.J. Hass avocado composition and potential health effects. Crit. Rev. Food Sci. 2013, 53, 738–750. [Google Scholar] [CrossRef]
- Ge, Y.; Zang, X.P.; Yang, Y.; Wang, T.; Ma, W.H. In-depth analysis of potential PaAP2/ERF transcription factor related to fatty acid accumulation in avocado (Persea americana Mill.) and functional characterization of two PaAP2/ERF genes in transgenic tomato. Plant. Physiol. Biochem. 2021, 158, 308–320. [Google Scholar] [CrossRef]
- Kilaru, A.; Cao, X.; Dabbs, P.B.; Sung, H.J.; Rahman, M.M.; Thrower, N.; Zynda, G.; Podicheti, R.; Ibarra-Laclette, E.; Herrera-Estrella, L.; et al. Oil biosynthesis in a basal angiosperm: Transcriptome analysis of Persea americana mesocarp. BMC Plant. Biol. 2015, 15, 203. [Google Scholar] [CrossRef]
- Ge, Y.; Si, X.Y.; Cao, J.Q.; Zhou, Z.X.; Wang, W.L.; Ma, W.H. Morphological characteristics, nutritional quality, and bioactive constituents in fruits of two avocado (Persea americana) varieties from hainan province, China. J. Agric. Sci. 2017, 9, 8–17. [Google Scholar] [CrossRef]
- Ge, Y.; Ma, F.N.; Wu, B.; Tan, L. Morphological and chemical analysis of 16 avocado accessions (Persea americana) from China by principal component analysis and cluster analysis. J. Agri. Sci. 2018, 10, 80–89. [Google Scholar] [CrossRef]
- Chen, G.L.; Chen, S.G.; Zhao, Y.Y.; Luo, C.X.; Li, J.; Gao, Y.Q. Total phenolic contents of 33 fruits and their antioxidant capacities before and after in vitro digestion. Ind. Crop. Prod. 2014, 57, 150–157. [Google Scholar] [CrossRef]
- Kosinska, A.; Karamac, M.; Estrella, I.; Hernandez, T.; Bartolome, B.; Dykes, G.A. Phenolic compound profiles and antioxidant capacity of Persea americana Mill. peels and seeds of two varieties. J. Agric. Food Chem. 2012, 60, 4613–4619. [Google Scholar] [CrossRef]
- Vinha, A.F.; Moreira, J.; Barreira, S.V.P. Physicochemical parameters, phytochemical composition and antioxidant activity of the Algarvian avocado (Persea americana Mill.). J. Agric. Sci. 2013, 5, 100–109. [Google Scholar] [CrossRef]
- Weckwerth, W. Metabolomics in systems biology. Ann. Rev. Plant. Biol. 2003, 54, 669–689. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.L.; Gong, Z.; Guo, W.S.; Wang, H.Y.; Zhang, X.O.; Liu, S.B.; Yu, L.Z.; Xiong, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant. 2013, 6, 1769–1780. [Google Scholar] [CrossRef]
- Sawada, Y.; Akiyama, K.; Sakata, A.; Kuwahara, A.; Otsuki, H.; Sakurai, T.; Saito, K.; Hirai, M.Y. Widely targeted metabolomics based on large-scale MS/MS data for elucidating metabolite accumulation patterns in plants. Plant. Cell Physiol. 2009, 50, 37–47. [Google Scholar] [CrossRef]
- Tan, H.; Zhang, J.; Qi, X.; Shi, X.L.; Zhou, J.G.; Wang, X.C.; Xiang, X.E. Correlation analysis of the transcriptome and metabolome reveals the regulatory network for lipid synthesis in developing Brassica napus embryos. Plant. Mol. Biol. 2019, 99, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Khan, W.A.; Hou, X.; Han, K.; Khan, N.; Dong, H.; Saqib, M.; Zhang, Z.; Naseri, E.; Hu, C. Lipidomic study reveals the effect of morphological variation and other metabolite interactions on the lipid composition in various cultivars of Bok choy. Biochem. Biophys. Res. Commun. 2018, 506, 755–764. [Google Scholar] [CrossRef]
- Zhi, Y.; Taylor, M.C.; Campbell, P.M.; Warden, A.C.; Shrestha, P.; El Tahchy, A.; Rolland, V.; Vanhercke, T.; Petrie, J.R.; White, R.G.; et al. Comparative lipidomics and proteomics of lipid droplets in the mesocarp and seed tissues of Chinese tallow (Triadica sebifera). Front. Plant. Sci. 2017, 8, 1339. [Google Scholar] [CrossRef]
- Ge, Y.; Dong, X.S.; Liu, Y.Z.; Yang, Y.; Zhan, R.L. Molecular and biochemical analyses of avocado (Persea americana) reveal differences in the oil accumulation pattern between the mesocarp and seed during the fruit developmental period. Sci. Hortic. 2021, 276, 109717. [Google Scholar] [CrossRef]
- Rendón-Anaya, M.; Ibarra-Laclette, E.; Méndez-Bravo, A.; Lan, T.Y.; Zheng, C.F.; Carretero-Paulet, L.; Perez-Torres, C.A.; Chacón-López, A.; Hernandez-Guzmán, G.; Chang, T.H. The avocado genome informs deep angiosperm phylogeny, highlights introgressive hybridization, and reveals pathogen-influenced gene space adaptation. Proc. Natl. Acad. Sci. USA 2019, 116, 17081–17089. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Walther, T.C.; Chung, J.Y.; Farese, R.V., Jr. Lipid droplet biogenesis. Annu. Rev. Cell Dev. Bi. 2017, 33, 491–510. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.L.; Ji, H.Y.; Liu, D.T. Oil biosynthesis in underground oil-rich storage vegetative tissue: Comparison of Cyperus esculentus ruber with oil seeds and fruits. Plant. Cell Physiol. 2016, 57, 2519–2540. [Google Scholar] [CrossRef]
- Gidda, S.K.; Watt, S.; Collins-Silva, J.; Kilaru, A.; Arondel, V.; Yurchenko, O.; Horn, P.J.; James, C.N.; Shintani, D.; Ohlrogge, J.B. Lipid droplet-associated proteins (LDAPs) are involved in the compartmentalization of lipophilic compounds in plant cells. Plant. Signal. Behav. 2013, 8, e27141. [Google Scholar] [CrossRef] [PubMed]
- Horn, P.J.; James, C.N.; Gidda, S.K.; Kilaru, A.; Dyer, J.M.; Mullen, R.T.; Ohlrogge, J.B.; Chapman, K.D. Identification of a new class of lipid droplet-associated proteins in plants. Plant. Physiol. 2013, 162, 1926–1936. [Google Scholar] [CrossRef] [PubMed]
- Gross-German, E.; Viruel, M.A. Molecular characterization of avocado germplasm with a new set of SSR and EST-SSR markers: Genetic diversity, population structure, and identification of race-specific markers in a group of cultivated genotypes. Tree Genet. Genomes 2013, 9, 539–555. [Google Scholar] [CrossRef]
- Ashworth, V.E.T.M.; Clegg, M.T. Microsatellite markers in avocado (Persea americana Mill.). genealogical relationships among cultivated avocado genotypes. J. Hered. 2003, 94, 407–415. [Google Scholar] [CrossRef]
- Schnell, R.J.; Brown, J.S.; Olano, C.T.; Power, E.J.; Krol, C.A.; Kuhn, D.N.; Motamayor, J.C. Evaluation of avocado germplasm using microsatellite markers. J. Amer. Soc. Hort. Sci. 2003, 128, 881–889. [Google Scholar] [CrossRef]
- Lu, S.P.; Sturtevant, D.; Aziz, M.; Jin, C.; Li, Q.; Chapman, K.D.; Guo, L. Spatial analysis of lipid metabolites and expressed genes reveals tissue-specific heterogeneity of lipid metabolism in high- and low-oil Brassica napus L. seeds. Plant. J. 2018, 94, 915–932. [Google Scholar] [CrossRef]




| Lipid Classes | Relative Total Amount | ||
|---|---|---|---|
| Mexican Ecotype (Walter Hole) | Guatemalan Ecotype (Nabal) | West Indian Ecotype (Donnie) | |
| DAG | 2.98 × 10−2 ± 1.41 × 10−4 a | 1.93 × 10−0.2 ± 4.06 × 10−4 b | 1.55 × 10−0.2 ± 6.27 × 10−4 b |
| TAG | 2.47 × 10−2 ± 2.65 × 10−4 a | 2.13 × 10−2 ± 2.67 × 10−4 b | 2.03 × 10−2 ± 8.61 × 10−4 b |
| MGDG | 6.74 × 10−2 ± 5.58 × 10−4 a | 9.03 × 10−3 ± 9.23 × 10−4 b | 5.51 × 10−3 ± 4.06 × 10−4 a |
| DGDG | 3.83 × 10−3 ± 2.15 × 10−4 a | 6.20 × 10−3 ± 1.92 × 10−4 b | 6.60 × 10−3 ± 2.94 × 10−4 b |
| PA | 3.54 × 10−4 ± 3.07 × 10−5 a | 8.42 × 10−4 ± 3.68 × 10−5 b | 6.37 × 10−4 ± 7.34 × 10−5 c |
| PC | 5.24 × 10−3 ± 1.05 × 10−3 a | 3.77 × 10−2 ± 5.12 × 10−3 b | 2.71 × 10−2 ± 3.87 × 10−3 b |
| PE | 1.47 × 10−2 ± 1.49 × 10−3 a | 3.64 × 10−2 ± 1.90 × 10−3 b | 3.12 × 10−2 ± 1.90 × 10−4 b |
| PG | 2.51 × 10−3 ± 3.97 × 10−4 a | 2.75 × 10−3 ± 1.35 × 10−4 a | 2.13 × 10−3 ± 1.33 × 10−4 a |
| PI | 8.09 × 10−4 ± 3.95 × 10−5 a | 6.15 × 10−4 ± 3.45 × 10−5 b | 4.59 × 10−4 ± 2.33 × 10−5 c |
| PS | 1.68 × 10−4 ± 1.00 × 10−5 a | 3.59 × 10−4 ± 4.18 × 10−5 b | 2.82 × 10−4 ± 2.65×10−5 c |
| Metabolite Super-Classes | Relative Total Amount | ||
|---|---|---|---|
| Mexican Ecotype (Walter Hole) | Guatemalan Ecotype (Nabal) | West Indian Ecotype (Donnie) | |
| Benzenoids | 15.06 ± 0.51 a | 22.01 ± 1.35 b | 19.52 ± 1.93 b |
| Hydrocarbons | 0.79 ± 0.06 a | 3.15 ± 0.13 b | 3.55 ± 0.29 b |
| Lipids and lipid-like molecules | 135.18 ± 3.75 a | 93.86 ± 4.88 b | 84.99 ± 5.53 b |
| Organic acids and derivatives | 40.14 ± 4.43 a | 26.82 ± 2.05 b | 45.99 ± 2.30 a |
| Organic oxygen compounds | 199.28 ± 10.39 a | 523.53 ± 37.08 b | 567.93 ± 34.98 b |
| Organic heterocyclic compounds | 22.57 ± 2.58 a | 22.64 ± 1.67 a | 23.63 ± 0.63 a |
| Phenylpropanoids and polyketides | 0.78 ± 0.08 a | 5.07 ± 0.67 b | 1.06 ± 0.23 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, Y.; Zang, X.; Liu, Y.; Wang, L.; Wang, J.; Li, Y.; Ma, W. Multi-Omics Analysis to Visualize Ecotype-Specific Heterogeneity of the Metabolites in the Mesocarp Tissue of Three Avocado (Persea Americana Mill.) Ecotypes. Horticulturae 2021, 7, 94. https://doi.org/10.3390/horticulturae7050094
Ge Y, Zang X, Liu Y, Wang L, Wang J, Li Y, Ma W. Multi-Omics Analysis to Visualize Ecotype-Specific Heterogeneity of the Metabolites in the Mesocarp Tissue of Three Avocado (Persea Americana Mill.) Ecotypes. Horticulturae. 2021; 7(5):94. https://doi.org/10.3390/horticulturae7050094
Chicago/Turabian StyleGe, Yu, Xiaoping Zang, Yuanzheng Liu, Lixia Wang, Jiashui Wang, Yanxia Li, and Weihong Ma. 2021. "Multi-Omics Analysis to Visualize Ecotype-Specific Heterogeneity of the Metabolites in the Mesocarp Tissue of Three Avocado (Persea Americana Mill.) Ecotypes" Horticulturae 7, no. 5: 94. https://doi.org/10.3390/horticulturae7050094
APA StyleGe, Y., Zang, X., Liu, Y., Wang, L., Wang, J., Li, Y., & Ma, W. (2021). Multi-Omics Analysis to Visualize Ecotype-Specific Heterogeneity of the Metabolites in the Mesocarp Tissue of Three Avocado (Persea Americana Mill.) Ecotypes. Horticulturae, 7(5), 94. https://doi.org/10.3390/horticulturae7050094






