Generation of a High-Density Genetic Map of Pepper (Capsicum annuum L.) by SLAF-seq and QTL Analysis of Phytophthora capsici Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Mapping Population
2.2. Pathogen Preparation and Plant Inoculation with P. capsici
2.3. Disease Evaluation
2.4. Statistical Analysis of Phenotypic Data
2.5. DNA Extraction, SLAF Library Construction, and High-Throughput Sequencing
2.6. SLAF-seq Data Grouping and Genotyping
2.7. High-Density Linkage Map Construction
2.8. QTL Mapping of P. capsici Resistance and Candidate Gene Prediction
2.9. qRT-PCR Analysis
3. Results
3.1. Sequencing and Genotyping Based on SLAF-seq
3.2. Genetic Map Construction
3.3. Quality and Accuracy of the Genetic Map
3.4. Phenotypic Analysis of P. capsici Resistance
3.5. QTL Mapping of P. capsici Resistance
3.6. Candidate Gene Prediction and qRT-PCR Analysis
4. Discussion
4.1. Genetic Map Constructed of Pepper
4.2. Identification QTL with the Resistance to P. capsici Traits
4.3. Candidate Gene Prediction
4.4. Candidate Gene qRT-PCR Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chaim, A.B.; Grube, R.C.; Lapidot, M.; Jahn, M.; Paran, I. Identification of quantitative trait loci associated with resistance to cucumber mosaic virus in Capsicum annuum. Theor. Appl. Genet. 2001, 102, 1213–1220. [Google Scholar] [CrossRef]
- Cai, W.-Q.; Fang, R.-X.; Shang, H.-S.; Wang, X.; Zhang, F.-L.; Li, Y.-R.; Zhang, J.-C.; Cheng, X.-Y.; Wang, G.-L.; Mang, K.-Q. Development of CMV-and TMV-resistant chili pepper: Field perfermance and biosafety assessment. Mol. Breed. 2003, 11, 25–35. [Google Scholar] [CrossRef]
- Hong, J.K.; Yang, H.J.; Jung, H.; Yoon, D.J.; Sang, M.K.; Jeun, Y.-C. Application of Volatile Antifungal Plant Essential Oils for Controlling Pepper Fruit Anthracnose by Colletotrichum gloeosporioides. Plant Pathol. J. 2015, 31, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Bosland, P.W. A Seedling Screen for Phytophthora Root Rot of Pepper, Capsicum annuum. Plant Dis. 1991, 75, 1048. [Google Scholar] [CrossRef]
- Parra, G.; Ristaino, J.B. Resistance to Mefenoxam and Metalaxyl Among Field Isolates of Phytophthora capsici Causing Phytophthora Blight of Bell Pepper. Plant Dis. 2001, 85, 1069–1075. [Google Scholar] [CrossRef]
- Oelke, L.M.; Bosland, P.W.; Steiner, R. Differentiation of Race Specific Resistance to Phytophthora Root Rot and Foliar Blight in Capsicum annuum. J. Am. Soc. Hortic. Sci. 2003, 128, 213–218. [Google Scholar] [CrossRef]
- Lefebvre, V.; Palloix, A. Both epistatic and additive effects of QTLs are involved in polygenic induced resistance to disease: A case study, the interaction pepper-Phytophthora capsici Leonian. Theor. Appl. Genet. 1996, 93, 503–511. [Google Scholar] [CrossRef]
- Foster, J.M.; Hausbeck, M.K. Resistance of Pepper to Phytophthora Crown, Root, and Fruit Rot Is Affected by Isolate Virulence. Plant Dis. 2010, 94, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Barksdale, T.H.; Papavizas, G.C.; Johnston, S.A. Resistance to foliar blight and crown rot of pepper caused by Phytophthora capsici. Plant Dis. 1984, 68, 506–509. [Google Scholar] [CrossRef]
- Flett, S.; Ashcroft, W.; Jerie, P.; Taylor, P. Control of Phytophthora root rot in processing tomatoes by metalaxyl and fosetyl-Al. Aust. J. Exp. Agric. 1991, 31, 279–283. [Google Scholar] [CrossRef]
- Polizzi, G.; Agosteo, G.E.; Cartia, G. Soil solarization for the control of Phytophthora capsici on pepper. Acta Hortic. 1994, 366, 331–338. [Google Scholar] [CrossRef]
- Lehmann, P. Structure and evolution of plant disease resistance genes. J. Appl. Genet. 2002, 43, 403–414. [Google Scholar]
- Stieg, J.R.; Walters, S.A.; Bond, J.P.; Babadoost, M. Effects of fungicides and cultivar resistance for Phytophthora capsici control in bell pepper production. HortScience 2006, 41, 1076. [Google Scholar] [CrossRef]
- Black, L. Studies on Phytophthora blight in pepper. In Talekar NS (ed) AVRDC Report 1998; Asian Vegetable Research and Development Center: Shanhua, Taiwan, 1999; pp. 25–27. [Google Scholar]
- Ortega, R.G.; Espanol, C.P.; Zueco, J.C. Genetics of Resistance to Phytophthora capsici in the Pepper Line ‘SCM-334’. Plant Breed. 1991, 107, 50–55. [Google Scholar] [CrossRef]
- Kim, B.S. Characteristics of bacterial spot resistant lines and Phytophthora Blight resistant lines of Capsicum pepper. Hortic. Environ. Biotechnol. 1988, 29, 247–252. [Google Scholar]
- Smith, P.G.; Kimble, K.A.; Grogan, R.G.; Millett, A.H. Inheritance of resistance in peppers to Phytophthora root rot. Phytopathology 1967, 57, 377–379. [Google Scholar]
- Sugita, T.; Yamaguchi, K.; Kinoshita, T.; Yuji, K.; Sugimura, Y.; Nagata, R.; Kawasaki, S.; Todoroki, A. QTL analysis for resistance to Phytophthora Blight (Phytophthora capsici Leon.) using an intraspecific Doubled-Haploid population of Capsicum annuum. Breed. Sci. 2006, 56, 137–145. [Google Scholar] [CrossRef]
- Kim, B.S.; Kwon, Y.S. Inheritance of resistance to Phytophthora Blight and to bacterial spot in pepper. J. Korean Soc. Hortic. Sci. 1990, 7, 17–24. [Google Scholar]
- Saini, S.S.; Sharma, P.P. Inheritance of resistance to fruit rot (Phytophthora capsici Leon.) and induction of resistance in bell pepper (Capsicum annuum L.). Euphytica 1978, 27, 721–723. [Google Scholar] [CrossRef]
- Wang, P.; Wang, L.; Guo, J.; Yang, W.; Shen, H. Molecular mapping of a gene conferring resistance to Phytophthora capsici Leonian race 2 in pepper line PI201234 (Capsicum annuum L.). Mol. Breed. 2016, 36, 1–11. [Google Scholar] [CrossRef]
- Reifschneider, F.J.B.; Boiteux, L.S.; Vecchia, P.T.D.; Poulos, J.M.; Kuroda, N. Inheritance of adult-plant resistance to Phytophthora capsici in pepper. Euphytica 1992, 62, 45–49. [Google Scholar] [CrossRef]
- Thabuis, A.; Palloix, A.; Pflieger, S.; Daubèze, A.-M.; Caranta, C.; Lefebvre, V. Comparative mapping of Phytophthora resistance loci in pepper germplasm: Evidence for conserved resistance loci across Solanaceae and for a large genetic diversity. Theor. Appl. Genet. 2003, 106, 1473–1485. [Google Scholar] [CrossRef] [PubMed]
- Pflieger, S.; Palloix, A.; Caranta, C.; Blattes, A.; Lefebvre, V. Defense response genes co-localize with quantitative disease resistance loci in pepper. Theor. Appl. Genet. 2001, 103, 920–929. [Google Scholar] [CrossRef]
- Minamiyama, Y.; Tsuro, M.; Kubo, T.; Hirai, M. QTL Analysis for Resistance to Phytophthora capsici in Pepper Using a High Density SSR-based Map. Breed. Sci. 2007, 57, 129–134. [Google Scholar] [CrossRef]
- Kim, H.-J.; Nahm, S.-H.; Lee, H.-R.; Yoon, G.-B.; Kim, K.-T.; Kang, B.-C.; Choi, D.; Kweon, O.Y.; Cho, M.-C.; Kwon, J.-K.; et al. BAC-derived markers converted from RFLP linked to Phytophthora capsici resistance in pepper (Capsicum annuum L.). Theor. Appl. Genet. 2008, 118, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Truong, H.T.H.; Kim, K.T.; Kim, D.W.; Kim, S.; Chae, Y.; Park, J.H.; Oh, D.G.; Cho, M.C. Identification of isolate-specific resistance QTLs to phytophthora root rot using an intraspecific recombinant inbred line population of pepper (Capsicum annuum). Plant Pathol. 2011, 61, 48–56. [Google Scholar] [CrossRef]
- Rehrig, W.Z.; Ashrafi, H.; Hill, T.; Prince, J.; Deynze, A.V. CaDMR1 Co-segregates with QTL Pc5.1 for resistance to Phytophthora capsici in pepper (Capsicum annuum). Plant Genome 2014, 7, 1–12. [Google Scholar] [CrossRef]
- Kim, N.; Kang, W.H.; Lee, J.; Yeom, S.I. Development of clustered resistance gene analogs-based markers of resistance to Phytophthora capsici in chili pepper. BioMed Res. Int. 2019, 2019, 1–12. [Google Scholar]
- Siddique, M.I.; Lee, H.Y.; Ro, N.Y.; Han, K.; Venkatesh, J.; Solomon, A.A.-O.; Patil, A.A.-O.; Changkwian, A.; Kwon, J.K.; Kang, B.C. Identifying candidate genes for Phytophthora capsici resistance in pepper (Capsicum annuum) via genotyping-by-sequencing-based QTL mapping and genome-wide association study. Sci. Rep. 2019, 9, 9962. [Google Scholar] [CrossRef]
- Mallard, S.; Cantet, M.; Massire, A.; Bachellez, A.; Ewert, S.; Lefebvre, V. A key QTL cluster is conserved among accessions and exhibits broad-spectrum resistance to Phytophthora capsici: A valuable locus for pepper breeding. Mol. Breed. 2013, 32, 349–364. [Google Scholar] [CrossRef]
- Bonnet, J.; Danan, S.; Boudet, C.; Barchi, L.; Sage-Palloix, A.-M.; Caromel, B.; Palloix, A.; Lefebvre, V. Are the polygenic architectures of resistance to Phytophthora capsici and P. parasitica independent in pepper? Theor. Appl. Genet. 2007, 115, 253–264. [Google Scholar] [CrossRef]
- Sun, X.; Liu, D.; Zhang, X.; Li, W.; Liu, H.; Hong, W.; Jiang, C.; Guan, N.; Ma, C.; Zeng, H.; et al. SLAF-seq: An Efficient Method of Large-Scale De Novo SNP Discovery and Genotyping Using High-Throughput Sequencing. PLoS ONE 2013, 8, e58700. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Tian, L.; Zhang, J.; Huang, L.; Han, F.; Yan, S.; Wang, L.; Zheng, H.; Sun, J. Construction of a high-density genetic map based on large-scale markers developed by specific length amplified fragment sequencing (SLAF-seq) and its application to QTL analysis for isoflavone content in Glycine max. BMC Genom. 2014, 15, 1086. [Google Scholar] [CrossRef]
- Zhang, Z.; Shang, H.; Shi, Y.; Huang, L.; Li, J.; Ge, Q.; Gong, J.; Liu, A.; Chen, T.; Wang, D. Construction of a high-density genetic map by specific locus amplified fragment sequencing (SLAF-seq) and its application to Quantitative Trait Loci (QTL) analysis for boll weight in upland cotton (Gossypium hirsutum). BMC Plant Biol. 2016, 16, 79. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Huang, L.; Zhu, R.; Xin, D.; Liu, C.; Han, X.; Jiang, H.; Hong, W.; Hu, G.; Zheng, H.; et al. A High-Density Genetic Map for Soybean Based on Specific Length Amplified Fragment Sequencing. PLoS ONE 2014, 9, e104871. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, L.; Xin, H.; Li, D.; Ma, C.; Ding, X.; Hong, W.; Zhang, X. Construction of a high-density genetic map for sesame based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Plant Biol. 2013, 13, 141. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lu, L.; Zhu, B.; Xu, Q.; Qi, X.; Chen, X. QTL mapping of cucumber fruit flesh thickness by SLAF-seq. Sci. Rep. 2015, 5, 15829. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yin, Y.; Yang, K.; Li, J.; Sang, Y.; Huang, L.; Fan, S. Construction of a high-density genetic map using specific length amplified fragment markers and identification of a quantitative trait locus for anthracnose resistance in walnut (Juglans regia L.). BMC Genom. 2015, 16, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-F.; Wang, G.-Y.; Dong, T.-T.; Chen, B.; Du, H.-S.; Li, C.-B.; Zhang, F.-L.; Zhang, H.-Y.; Xu, Y.; Wang, Q.; et al. High-density genetic map construction and QTL mapping of first flower node in pepper (Capsicum annuum L.). BMC Plant Biol. 2019, 19, 167. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.H.; Zhang, S.Z.; Miao, H.R.; Cui, F.G.; Shen, Y.; Yang, W.Q.; Xu, T.T.; Chen, N.; Chi, X.Y.; Zhang, Z.M.; et al. High-Density Genetic Map Construction and Identification of QTLs Controlling Oleic and Linoleic Acid in Peanut using SLAF-seq and SSRs. Sci. Rep. 2018, 8, 5479. [Google Scholar] [CrossRef]
- Wang, L.; Yang, X.; Cui, S.; Zhao, N.; Li, L.; Hou, M.; Mu, G.; Liu, L.; Li, Z. High-density genetic map development and QTL mapping for concentration degree of floret flowering date in cultivated peanut (Arachis hypogaea L.). Mol. Breed. 2020, 40, 1–14. [Google Scholar] [CrossRef]
- Guo, G.; Wang, S.; Liu, J.; Pan, B.; Diao, W.; Ge, W.; Gao, C.; Snyder, J.C. Rapid identification of QTLs underlying resistance to Cucumber mosaic virus in pepper (Capsicum frutescens). Theor. Appl. Genet. 2016, 130, 41–52. [Google Scholar] [CrossRef]
- Zhu, Z.; Sun, B.; Wei, J.; Cai, W.; Huang, Z.; Chen, C.; Cao, B.; Chen, G.; Lei, J. Construction of a high density genetic map of an interspecific cross of Capsicum chinense and Capsicum annuum and QTL analysis of floral traits. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, C.L.; Wendel, J.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef] [PubMed]
- Peichel, C.L.; Nereng, K.S.; Ohgi, K.A.; Cole, B.L.E.; Colosimo, P.F.; Buerkle, C.A.; Schluter, D.; Kingsley, D.M. The genetic architecture of divergence between threespine stickleback species. Nat. Cell Biol. 2001, 414, 901–905. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Huang, S.; Liu, Y.; Fang, Z.; Yang, L.; Hua, W.; Yuan, S.; Liu, S.; Sun, J.; Zhuang, M.; et al. Construction and analysis of a high-density genetic linkage map in cabbage (Brassica oleracea L. var. capitata). BMC Genom. 2012, 13, 523. [Google Scholar] [CrossRef]
- Burdon, J.J.; Thrall, P.H. Coevolution of Plants and Their Pathogens in Natural Habitats. Science 2009, 324, 755–756. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Voorrips, R.E.; Westende, W.V.; van Kaauwen, M.; Visser, R.G.F.; Vosman, B. Aphid resistance in Capsicum maps to a locus containing LRR-RLK gene analogues. Theor. Appl. Genet. 2020, 133, 227–237. [Google Scholar] [CrossRef]
- Ogundiwin, E.A.; Berke, T.F.; Massoudi, M.; Black, L.L.; Huestis, G.; Choi, D.; Lee, S.; Prince, J.P. Construction of 2 intra-specific linkage maps and identification of resistance QTLs for Phytophthora capsici root-rot and foliar-blight diseases of pepper (Capsicum annuum L.). Genome 2005, 48, 698–711. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Jahufer, M.Z.Z.; Brouwer, J.B.; Pang, E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Rose, L.E.; Bittner-Eddy, P.D.; Langley, C.H.; Holub, E.B.; Michelmore, R.W.; Beynon, J.L. The Maintenance of Extreme Amino Acid Diversity at the Disease Resistance Gene, RPP13, in Arabidopsis thaliana. Genetics 2004, 166, 1517–1527. [Google Scholar] [CrossRef] [PubMed]
- Serra, H.; Choi, K.; Zhao, X.; Blackwell, A.R.; Kim, J.; Henderson, I.R. Interhomolog polymorphism shapes meiotic crossover within the Arabidopsis RAC1 and RPP13 disease resistance genes. PLoS Genet. 2018, 14, e1007843. [Google Scholar] [CrossRef]
- Alder, M.N.; Rogozin, I.B.; Iyer, L.M.; Glazko, G.V.; Cooper, M.D.; Pancer, Z. Diversity and Function of Adaptive Immune Receptors in a Jawless Vertebrate. Science 2005, 310, 1970–1973. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, V. Role of extracytoplasmic leucine rich repeat proteins in plant defence mechanisms. Microbiol. Res. 2005, 160, 83–94. [Google Scholar] [CrossRef]
- Zhang, X.S.; Choi, J.H.; Heinz, J.; Chetty, C.S. Domain-Specific Positive Selection Contributes to the Evolution of Arabidopsis Leucine-Rich Repeat Receptor-Like Kinase (LRR RLK) Genes. J. Mol. Evol. 2006, 63, 612–621. [Google Scholar] [CrossRef]
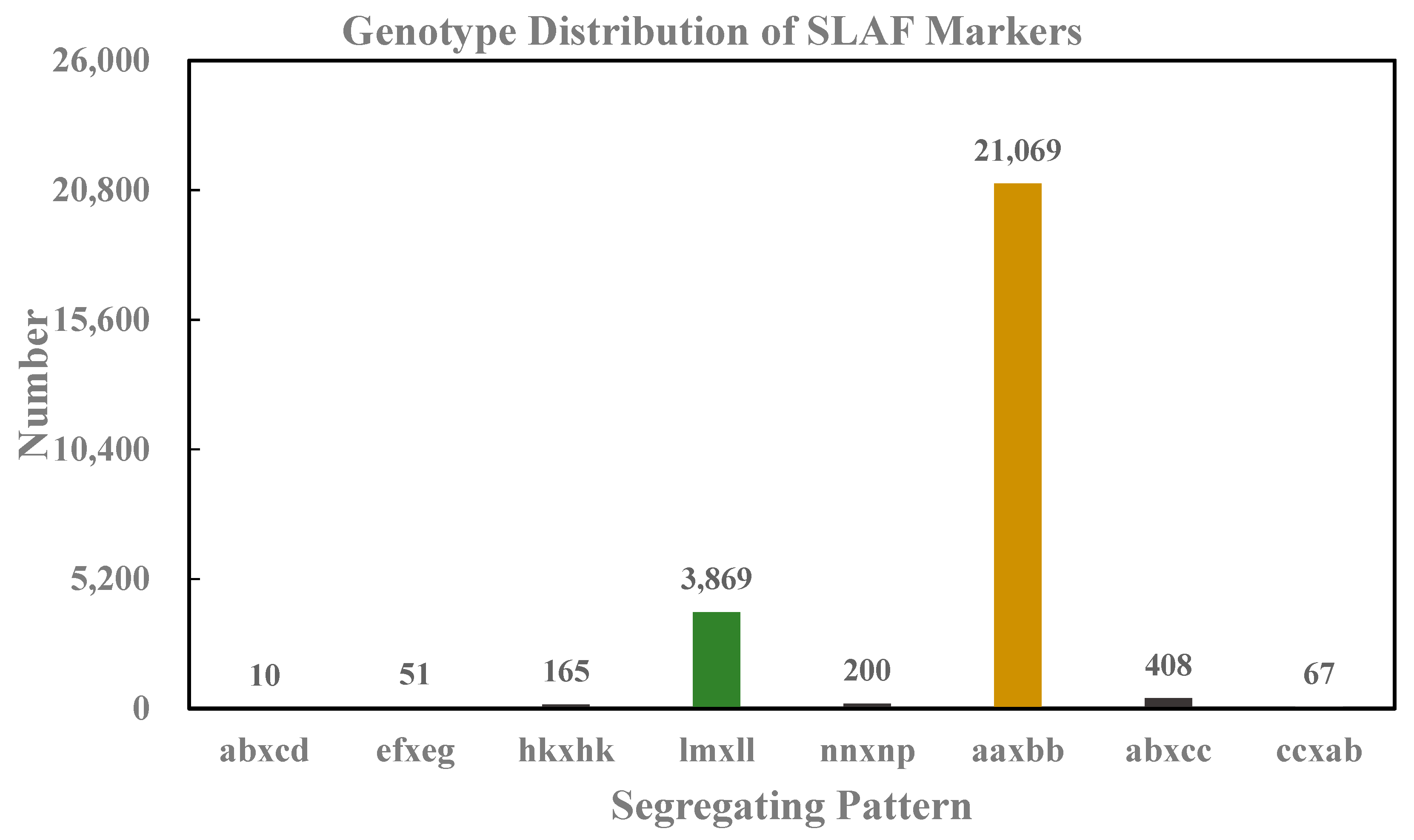

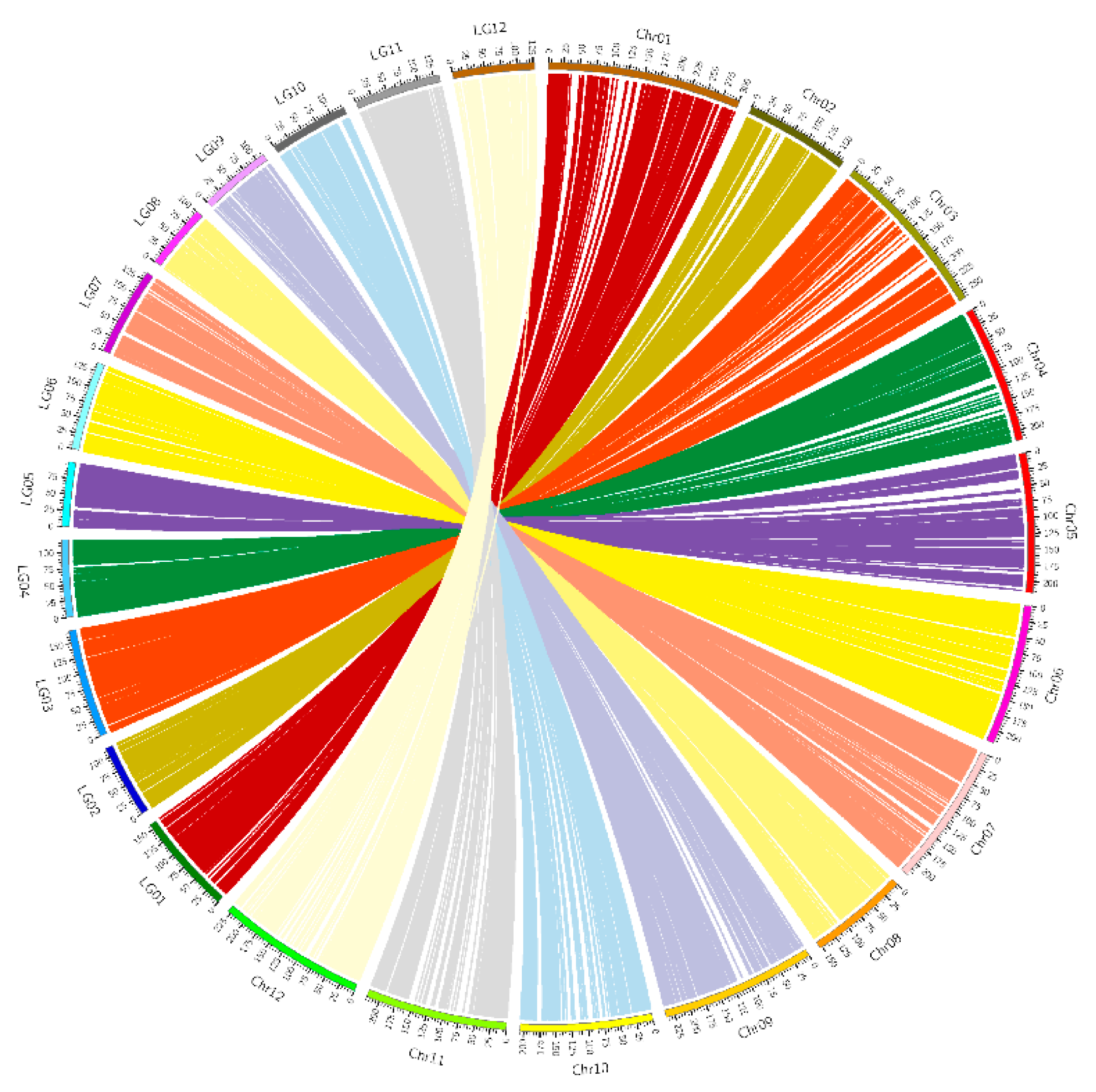
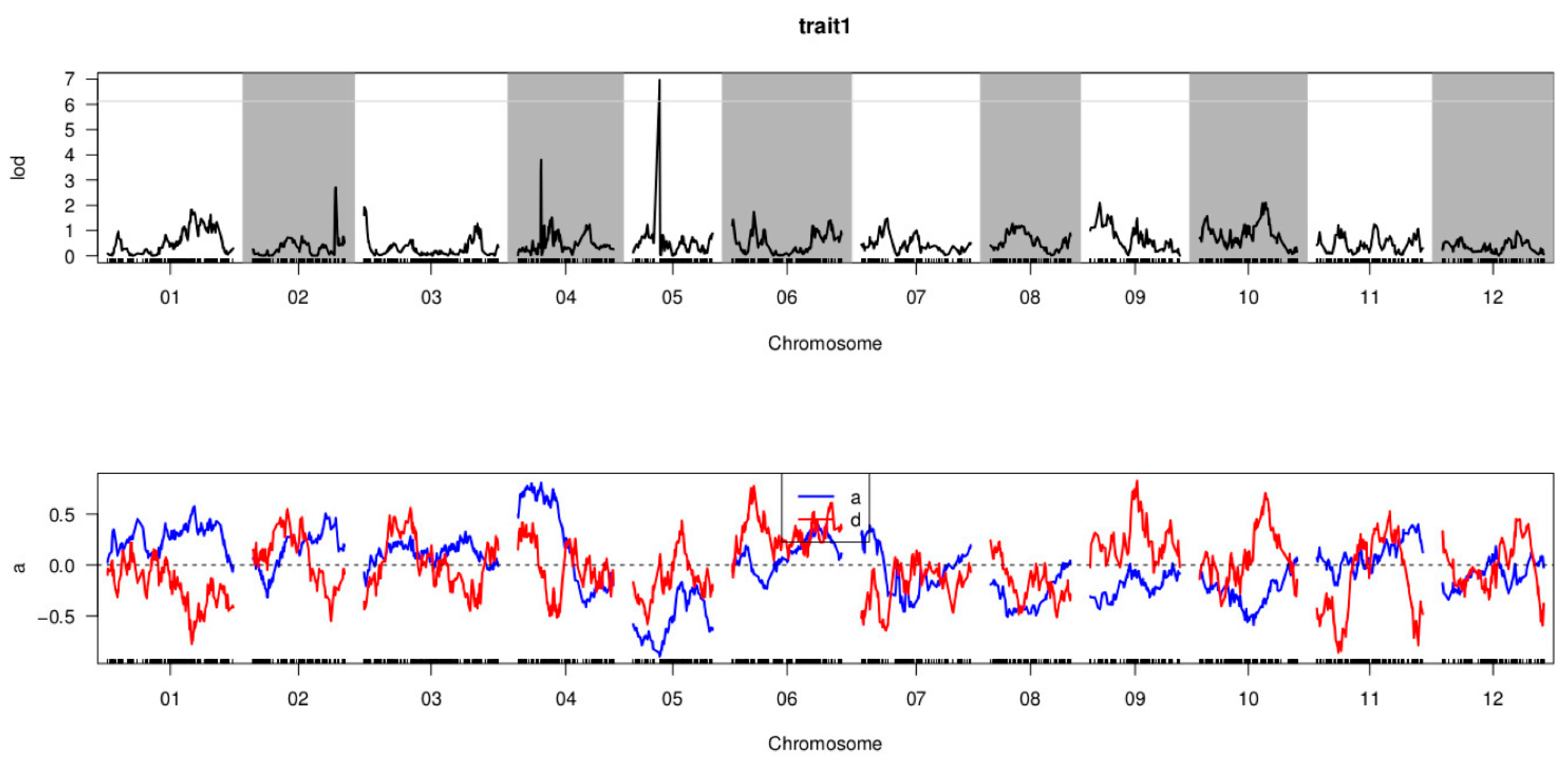

| Samples | Total Read | Total Bases | Q30 Percentage (%) | GC Content (%) | SLAF Number | Total Depth | Average Depth(X) |
|---|---|---|---|---|---|---|---|
| PI201234 | 12,250,440 | 2,449,757,552 | 93.98 | 38.42 | 139,046 | 8,875,578 | 63.83 |
| 1287 | 13,232,257 | 2,646,335,364 | 94.71 | 38.15 | 141,584 | 10,239,208 | 72.32 |
| Offspring | 2,371,153 | 474,202,396 | 94.37 | 38.36 | 124,582 | 1,825,928 | 14.66 |
| Total | 381,155,587 | 76,226,452,308 | 94.37 | 38.86 | 405,212 | / | / |
| Linkage Group | SLAF Number | Polymorphic |
|---|---|---|
| Chr01 | 16,109 | 3221 |
| Chr02 | 9259 | 1626 |
| Chr03 | 15,231 | 3159 |
| Chr04 | 12,696 | 1569 |
| Chr05 | 13,024 | 2986 |
| Chr06 | 12,887 | 2640 |
| Chr07 | 11,667 | 1907 |
| Chr08 | 9426 | 1263 |
| Chr09 | 14,507 | 3250 |
| Chr10 | 11,356 | 1687 |
| Chr11 | 11,890 | 3937 |
| Chr12 | 13,089 | 2343 |
| Other | 23,052 | 4844 |
| Total | 174,193 | 34,432 |
| Linkage Group | Marker Number | Average Distance between Markers (cM) | Size (cM) | Gaps ≤ 5 | Max Gap (cM) | Correlation Coefficient |
|---|---|---|---|---|---|---|
| Chr01 | 437 | 0.36 | 158.32 | 99.08% | 6.77 | 0.9968 |
| Chr02 | 415 | 0.28 | 114.98 | 100.00% | 4.57 | 0.9273 |
| Chr03 | 444 | 0.38 | 169.18 | 100.00% | 4.63 | 0.9979 |
| Chr04 | 336 | 0.36 | 120.20 | 99.70% | 6.38 | 0.9948 |
| Chr05 | 460 | 0.22 | 99.98 | 99.78% | 7.29 | 0.9054 |
| Chr06 | 722 | 0.19 | 137.92 | 99.58% | 6.31 | 0.9977 |
| Chr07 | 517 | 0.27 | 137.66 | 99.22% | 8.71 | 0.9712 |
| Chr08 | 373 | 0.27 | 100.81 | 100.00% | 3.94 | 0.9568 |
| Chr09 | 414 | 0.27 | 112.88 | 99.52% | 5.82 | 0.9875 |
| Chr10 | 532 | 0.23 | 122.63 | 99.81% | 9.99 | 0.9963 |
| Chr11 | 458 | 0.29 | 133.80 | 100.00% | 4.70 | 0.9974 |
| Chr12 | 457 | 0.28 | 127.33 | 99.56% | 6.38 | 0.9803 |
| Maximum | 722 | 0.38 | 169.18 | 100.00% | 9.99 | 0.9979 |
| Minimum | 336 | 0.19 | 99.98 | 99.08% | 3.94 | 0.9054 |
| Total | 5565 | 0.28 | 1535.69 | / | / | / |
| Average | 463.75 | / | 127.97 | 99.70% | / | 0.9758 |
| Samples | Marker Numbers | Total Depth(X) | Average Depth(X) |
|---|---|---|---|
| PI201234 | 5565 | 605,153 | 108.74 |
| 1287 | 5565 | 702,568 | 126.25 |
| Offspring | 5513 | 125,293 | 22.73 |
| Traits | F2 Population | Parent | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Range | Average | Standard Error | Var | Skew | Kurt | PI201234 | 1287 | Midparent | |
| Disease index (100%) | 0 | 1 | 0–1 | 0.4772 | 0.029 | 0.75 | −0.17 | −1.43 | 7.9 | 84.3 | 46.1 |
| QTL | LOD Threshold | Chr ID | Physical Distance Interval (bp) | Genetic Distance Interval (cM) | Max LOD | ADD | DOM | PVE (%) |
|---|---|---|---|---|---|---|---|---|
| CQPc5.1 | 6.125 | 05 | 17,967,630–19,446,349 | 33.103–33.448 | 6.972 | −0.897 | 0.079 | 11.758 |
| Gene | Start | Stop | COG | KEGG | Swiss-Prot | Nr |
|---|---|---|---|---|---|---|
| Capana05g000592 | 18,024,840 | 18,035,001 | – | K17550 (protein phosphatase 1 regulatory subunit 7) | Protein phosphatase 1 regulatory inhibitor subunit PPP1R7 homolog | PREDICTED: protein phosphatase 1 regulatory subunit pprA-like |
| Capana05g000594 | 18,317,198 | 18,327,621 | – | K13099 (CD2 antigen cytoplasmic tail-binding protein 2) | – | PREDICTED: CD2 antigen cytoplasmic tail-binding protein 2 |
| Capana05g000595 | 18,357,315 | 18,357,857 | – | – | Disease resistance RPP8-like protein | Hypothetical protein T459_14155 |
| Capana05g000596 | 18,358,568 | 18,359,167 | General function prediction only | – | Putative disease resistance RPP13-like protein | Hypothetical protein T459_14156 |
| Capana05g000597 | 18,359,457 | 18,359,987 | – | – | Disease resistance protein RPP13 | Hypothetical protein BC332_12877 |
| Capana05g000598 | 18,387,075 | 18,390,244 | Transcription | – | Probable LRR receptor-like serine/threonine-protein kinase At3g47570 | PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 |
| Capana05g000599 | 18,390,817 | 18,396,058 | Carbohydrate transport and metabolism | K05298 (glyceraldehyde-3-phosphate dehydrogenase (NADP+)) | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic (Fragment) | PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic |
| Capana05g000600 | 18,401,537 | 18,405,948 | General function prediction only | – | Protein high chlorophyll fluorescent 107 | PREDICTED: protein high chlorophyll fluorescent 107 |
| Capana05g000601 | 18,410,078 | 18,412,165 | Post-translational modification, protein turnover, chaperones | K00587 (protein-S-isoprenylcysteine O-methyltransferase) | Protein-S-isoprenylcysteine O-methyltransferase B | PREDICTED: protein-S-isoprenylcysteine O-methyltransferase A-like isoform X1 |
| Capana05g000602 | 18,413,946 | 18,415,745 | – | – | Pentatricopeptide repeat-containing protein At2g13600 OS = Arabidopsis thaliana | PREDICTED: pentatricopeptide repeat-containing protein At2g13600 |
| Capana05g000603 | 18,424,439 | 18,427,748 | Coenzyme transport and metabolism | – | FAD synthetase 1, chloroplastic | PREDICTED: FAD synthetase 1, chloroplastic-like |
| Capana05g000604 | 18,449,805 | 18,454,056 | Transcription | K10268 (F-box and leucine-rich repeat protein) | F-box/LRR-repeat protein 4 | PREDICTED: F-box/LRR-repeat protein 20 |
| Capana05g000605 | 18,709,353 | 18,710,879 | – | – | UPF0481 protein At3g47200 | PREDICTED: putative UPF0481 protein At3g02645 isoform X1 |
| Capana05g000607 | 18,749,830 | 18,750,435 | – | – | – | PREDICTED: uncharacterized protein LOC107870380 isoform X1 |
| Capana05g000608 | 18,775,198 | 18,776,266 | Cell wall/membrane/envelope biogenesis | K18819 (inositol 3-alpha-galactosyltransferase) | Galactinol synthase 2 | Galactinol synthase 2 |
| Capana05g000609 | 18,823,996 | 18,827,956 | – | – | – | Hypothetical protein CQW23_12126 |
| Capana05g000611 | 18,835,570 | 18,835,953 | – | K13495 (cis-zeatin O-glucosyltransferase) | Zeatin O-xylosyltransferase | PREDICTED: zeatin O-glucosyltransferase |
| Capana05g000612 | 18,851,478 | 18,852,197 | – | K13495 (cis-zeatin O-glucosyltransferase) | Zeatin O-glucosyltransferase | PREDICTED: zeatin O-xylosyltransferase-like |
| Capana05g000613 | 18,857,046 | 18,857,423 | – | K13495 (cis-zeatin O-glucosyltransferase) | Zeatin O-glucosyltransferase | Hypothetical protein T459_14175 |
| Capana05g000614 | 18,879,064 | 18,879,435 | – | – | Putative cis-zeatin O-glucosyltransferase | PREDICTED: zeatin O-xylosyltransferase-like |
| Capana05g000615 | 18,879,694 | 18,880,182 | – | K13495 (cis-zeatin O-glucosyltransferase) | Zeatin O-xylosyltransferase | Hypothetical protein T459_14174 |
| Capana05g000617 | 19,193,439 | 19,194,854 | – | K13495 (cis-zeatin O-glucosyltransferase) | Zeatin O-glucosyltransferase | PREDICTED: zeatin O-glucosyltransferase-like |
| Capana05g000618 | 19,354,157 | 19,355,500 | Transcription | – | Receptor-like protein Cf-9 | Hypothetical protein T459_14173 |
| Total | 9 | 11 | 20 | 23 |
| Gene Name | Primer Sequence (5′-3′) | PCR Product Size (bp) | TM (°C) |
|---|---|---|---|
| Capana05g000595 | F:AAGGAGGCATTTAGCCGCAA | 115 | 59.0 |
| R:TGTCTCAAGGCGAGCAACAT | |||
| Capana05g000596 | F:CTGCAAGAAAGCGTGTCAGG | 98 | 59.0 |
| R:AGCCTCCACATCTTTCCACC | |||
| Capana05g000597 | F:CAATCCCTCAAGCGACGAGT | 121 | 55.0 |
| R:CCAGGTCGGACCGATTGTTA | |||
| Capana05g000598 | F:ACCTTCCGTGGTGAAATCCC | 190 | 55.0 |
| R:CGATCCGCGTAACAGGTTTG | |||
| Capana05g000604 | F:TTAGCTGTTGCTGAGGGGTG | 163 | 59.0 |
| R:GCTTGCGTCCAGAGAGACAAA | |||
| Actin (AY572427) | F:AGCAACTGGGACGATATGGAGAAG | 198 | 50.0 |
| R:AAGAGACAACACCGCCTGAATAGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.-F.; Zhang, S.-C.; Yang, X.-M.; Wang, C.-P.; Huang, Q.-Z.; Huang, R.-Z. Generation of a High-Density Genetic Map of Pepper (Capsicum annuum L.) by SLAF-seq and QTL Analysis of Phytophthora capsici Resistance. Horticulturae 2021, 7, 92. https://doi.org/10.3390/horticulturae7050092
Li Y-F, Zhang S-C, Yang X-M, Wang C-P, Huang Q-Z, Huang R-Z. Generation of a High-Density Genetic Map of Pepper (Capsicum annuum L.) by SLAF-seq and QTL Analysis of Phytophthora capsici Resistance. Horticulturae. 2021; 7(5):92. https://doi.org/10.3390/horticulturae7050092
Chicago/Turabian StyleLi, Yi-Fei, Shi-Cai Zhang, Xiao-Miao Yang, Chun-Ping Wang, Qi-Zhong Huang, and Ren-Zhong Huang. 2021. "Generation of a High-Density Genetic Map of Pepper (Capsicum annuum L.) by SLAF-seq and QTL Analysis of Phytophthora capsici Resistance" Horticulturae 7, no. 5: 92. https://doi.org/10.3390/horticulturae7050092
APA StyleLi, Y.-F., Zhang, S.-C., Yang, X.-M., Wang, C.-P., Huang, Q.-Z., & Huang, R.-Z. (2021). Generation of a High-Density Genetic Map of Pepper (Capsicum annuum L.) by SLAF-seq and QTL Analysis of Phytophthora capsici Resistance. Horticulturae, 7(5), 92. https://doi.org/10.3390/horticulturae7050092







