Genome-Wide Identification and Characterization of DnaJ Gene Family in Grape (Vitis vinifera L.)
Abstract
:1. Introduction
2. Results
2.1. Identification and Analysis of VvDnaJs
2.2. Phylogenetic and Domain Organization Analysis of VvDnaJs
2.3. Analysis of Conserved Motif, Gene Structure, Protein Tertiary Structure, and Multiple Sequence Comparison in VvDnaJs
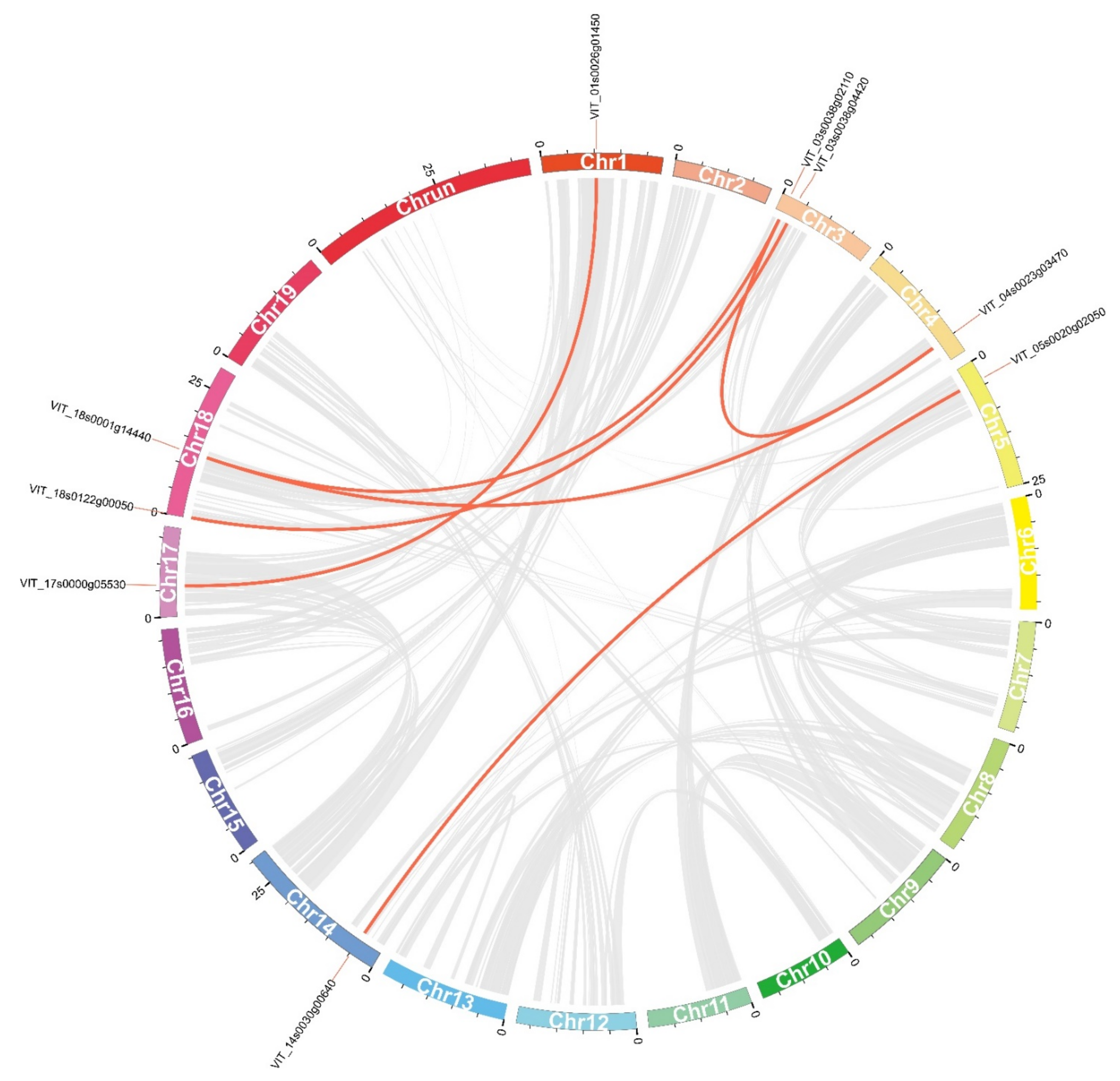
2.4. Chromosomal Distribution and Gene Duplication Analysis of VvDnaJs
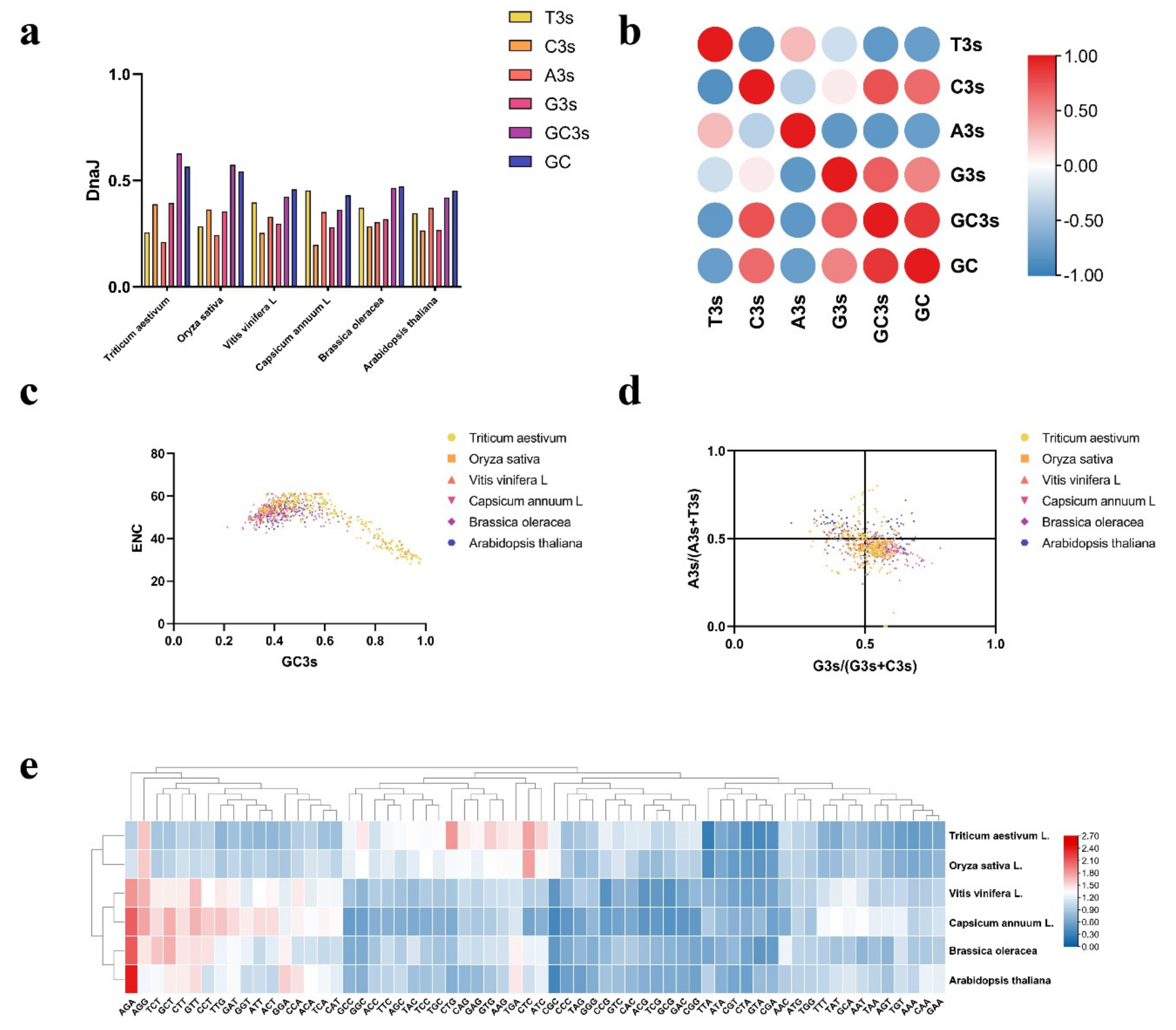
2.5. Analysis of Codon Usage Pattern in DnaJ Genes
2.6. Tissue-Specific Analysis of VvDnaJs
2.7. Prediction of VvDnaJs Cis-Acting Elements
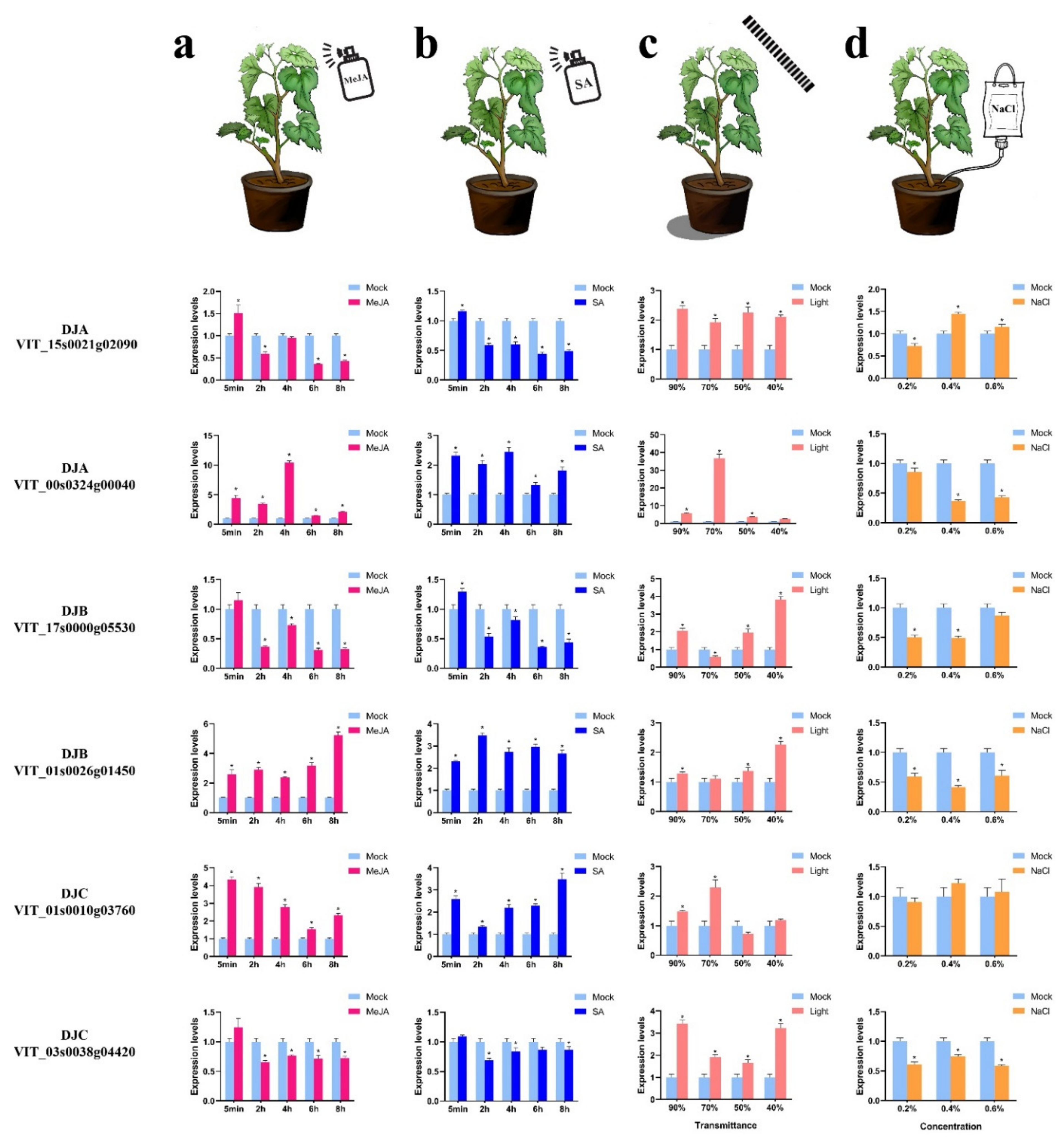
2.8. Analysis of VvDnaJ Expression under Hormone, Shade, Salt, and Heat Stress
3. Discussion
4. Materials and Methods
4.1. Identification of the DnaJ Family in Vitis vinifera L.
4.2. Chromosomal Localization, Phylogenetic, Collinearity, and Ks Analysis
4.3. Gene Structure, Conserved Motif, Protein Tertiary Structure, and Multiple Sequence Alignment Analysis
4.4. Codon Usage Pattern Analysis
4.5. cis-Acting Regulatory Element Analysis
4.6. Plant Materials and Hormone, Shade, Salt, and Heat Treatments
4.7. RNA Extraction and qRT-PCR Analysis
4.8. Subcellular Localization of VvDnaJs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Duan, S.; Wu, Y.; Fu, R.; Wang, L.; Chen, Y.; Xu, W.; Zhang, C.; Ma, C.; Shi, J.; Wang, S. Comparative Metabolic Profiling of Grape Skin Tissue along Grapevine Berry Developmental Stages Reveals Systematic Influences of Root Restriction on Skin Metabolome. Int. J. Mol. Sci. 2019, 20, 534. [Google Scholar] [CrossRef] [Green Version]
- Ramos, M.J.N.; Coito, J.L.; Silva, H.G.; Cunha, J.; Costa, M.M.R.; Rocheta, M. Flower development and sex specification in wild grapevine. BMC Genom. 2014, 15, 1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, J.; Luo, Y.; Sun, P.; Gao, J.; Zhao, D.; Yang, P.; Hu, T. Effects of shade stress on turfgrasses morphophysiology and rhizosphere soil bacterial communities. BMC Plant Biol. 2020, 20, 92. [Google Scholar] [CrossRef] [Green Version]
- Tanveer, M.; Yousaf, U. Plant single-cell biology and abiotic stress tolerance. In Plant Life under Changing Environment; Tripathi, D.K., Pratap Singh, V., Chauhan, D.K., Sharma, S., Prasad, S.M., Dubey, N.K., Ramawat, N., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 611–626. [Google Scholar]
- Zhao, Q.; Zhang, H.; Wang, T.; Chen, S.; Dai, S. Proteomics-based investigation of salt-responsive mechanisms in plant roots. J. Proteom. 2013, 82, 230–253. [Google Scholar] [CrossRef]
- De Leo, V.; Musacchio, M.C.; Cappelli, V.; Massaro, M.G.; Morgante, G.; Petraglia, F. Genetic, hormonal and metabolic aspects of PCOS: An update. Reprod. Biol. Endocrinol. 2016, 14, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.J.; Seo, Y.S. Heat Shock Proteins: A Review of the Molecular Chaperones for Plant Immunity. Plant Pathol. J. 2015, 31, 323–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martine, P.; Rébé, C. Heat Shock Proteins and Inflammasomes. Int. J. Mol. Sci. 2019, 20, 4508. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Kong, F.; Zhang, S.; Meng, X.; Wang, Y.; Meng, Q. A tomato chloroplast-targeted DnaJ protein protects Rubisco activity under heat stress. J. Exp. Bot. 2015, 66, 3027–3040. [Google Scholar] [CrossRef] [Green Version]
- Fan, C.-Y.; Lee, S.; Cyr, D.M. Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones 2003, 8, 309–316. [Google Scholar] [CrossRef]
- Caplan, A.J.; Cyr, D.M.; Douglas, M.G. Eukaryotic homologues of Escherichia coli dnaJ: A diverse protein family that functions with hsp70 stress proteins. Mol. Biol. Cell 1993, 4, 555–563. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Cai, G.; Kong, F.; Deng, Y.; Ma, N.; Meng, Q. Overexpression of tomato chloroplast-targeted DnaJ protein enhances tolerance to drought stress and resistance to Pseudomonas solanacearum in transgenic tobacco. Plant Physiol. Biochem. 2014, 82, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Rajan, V.B.; D’Silva, P. Arabidopsis thaliana J-class heat shock proteins: Cellular stress sensors. Funct. Integr. Genom. 2009, 9, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Pulido, P.; Leister, D. Novel DNAJ-related proteins in Arabidopsis thaliana. New Phytol. 2018, 217, 480–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.Z.; Whitham, S.A. Overexpression of a soybean nuclear localized type-III DnaJ domain-containing HSP40 reveals its roles in cell death and disease resistance. Plant J. 2013, 74, 110–121. [Google Scholar] [CrossRef]
- Walsh, P.; Bursać, D.; Law, Y.C.; Cyr, D.; Lithgow, T. The J-protein family: Modulating protein assembly, disassembly and translocation. EMBO Rep. 2004, 5, 567–571. [Google Scholar] [CrossRef] [Green Version]
- Kong, F.; Deng, Y.; Wang, G.; Wang, J.; Liang, X.; Meng, Q. LeCDJ1, a chloroplast DnaJ protein, facilitates heat tolerance in transgenic tomatoes. J. Integr. Plant Biol. 2014, 56, 63–74. [Google Scholar] [CrossRef]
- Bekh-Ochir, D.; Shimada, S.; Yamagami, A.; Kanda, S.; Ogawa, K.; Nakazawa, M.; Matsui, M.; Sakuta, M.; Osada, H.; Asami, T.; et al. A novel mitochondrial DnaJ/Hsp40 family protein BIL2 promotes plant growth and resistance against environmental stress in brassinosteroid signaling. Planta 2013, 237, 1509–1525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofius, D.; Maier, A.T.; Dietrich, C.; Jungkunz, I.; Börnke, F.; Maiss, E.; Sonnewald, U. Capsid protein-mediated recruitment of host DnaJ-like proteins is required for Potato virus Y infection in tobacco plants. J. Virol. 2007, 81, 11870–11880. [Google Scholar] [CrossRef] [Green Version]
- Tamadaddi, C.; Sagar, V.; Verma, A.K.; Afsal, F.; Sahi, C. Expansion of the evolutionarily conserved network of J-domain proteins in the Arabidopsis mitochondrial import complex. Plant Mol. Biol. 2021, 105, 385–403. [Google Scholar] [CrossRef]
- Park, M.; Kim, S. The Arabidopsis J Protein AtJ1 is Essential for Seedling Growth, Flowering Time Control and ABA Response. Plant Cell Physiol. 2014, 55, 2152–2163. [Google Scholar] [CrossRef] [Green Version]
- Verma, A.K.; Tamadaddi, C.; Tak, Y.; Lal, S.S.; Cole, S.J.; Hines, J.K.; Sahi, C. The expanding world of plant J-domain proteins. Crit. Rev. Plant Sci. 2019, 38, 382–400. [Google Scholar] [CrossRef]
- Yu, X.; Mo, Z.; Tang, X.; Gao, T.; Mao, Y. Genome-wide analysis of HSP70 gene superfamily in Pyropia yezoensis (Bangiales, Rhodophyta): Identification, characterization and expression profiles in response to dehydration stress. BMC Plant Biol. 2021, 21, 435. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Rinck, G.; Thiel, H.J.; Tautz, N. Cell-derived sequences in the N-terminal region of the polyprotein of a cytopathogenic pestivirus. J. Virol. 2003, 77, 10663–10669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ajit Tamadaddi, C.; Sahi, C. J domain independent functions of J proteins. Cell Stress Chaperones 2016, 21, 563–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeffreys, A.J.; Harris, S. Processes of gene duplication. Nature 1982, 296, 9–10. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Pei, J.; Li, Y.; Sun, H. Genome-wide identification and characterization of COMT gene family during the development of blueberry fruit. BMC Plant Biol. 2021, 21, 5. [Google Scholar] [CrossRef]
- Uddin, D.A. Codon Usage Bias: A Tool for Understanding Molecular Evolution. J. Proteom. Bioinform. 2017, 10, e32. [Google Scholar] [CrossRef]
- Wu, H.; Bao, Z.; Mou, C.; Chen, Z.; Zhao, J. Comprehensive Analysis of Codon Usage on Porcine Astrovirus. Viruses 2020, 12, 991. [Google Scholar] [CrossRef]
- Hu, L.; Wang, P.; Hao, Z.; Lu, Y.; Xue, G.; Cao, Z.; Qu, H.; Cheng, T.; Shi, J.; Chen, J. Gibberellin Oxidase Gene Family in L. chinense: Genome-Wide Identification and Gene Expression Analysis. Int. J. Mol. Sci. 2021, 22, 7167. [Google Scholar] [CrossRef]
- Gräfe, K.; Shanmugarajah, K.; Zobel, T.; Weidtkamp-Peters, S.; Kleinschrodt, D.; Smits, S.H.J.; Schmitt, L. Cloning and expression of selected ABC transporters from the Arabidopsis thaliana ABCG family in Pichia pastoris. PLoS ONE 2019, 14, e0211156. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.; Kwon, S.I.; Choe, S. Antagonistic regulation of Arabidopsis growth by brassinosteroids and abiotic stresses. Mol. Cells 2014, 37, 795–803. [Google Scholar] [CrossRef] [Green Version]
- Müller, M.; Munné-Bosch, S. Hormone Profiling in Plant Tissues. In Plant Hormones: Methods and Protocols; Kleine-Vehn, J., Sauer, M., Eds.; Springer: New York, NY, USA, 2017; pp. 249–258. [Google Scholar]
- Zhang, Y.; Jiang, L.; Li, Y.; Chen, Q.; Ye, Y.; Zhang, Y.; Luo, Y.; Sun, B.; Wang, X.; Tang, H. Effect of Red and Blue Light on Anthocyanin Accumulation and Differential Gene Expression in Strawberry (Fragaria × ananassa). Molecules 2018, 23, 820. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Tang, J.; Yu, Z.; Khare, T.; Srivastav, A.; Datir, S.; Kumar, V. Light Stress Responses and Prospects for Engineering Light Stress Tolerance in Crop Plants. J. Plant Growth Regul. 2019, 38, 1489–1506. [Google Scholar] [CrossRef]
- Liu, J.; Shen, F.; Xiao, Y.; Fang, H.; Qiu, C.; Li, W.; Wu, T.; Xu, X.; Wang, Y.; Zhang, X.; et al. Genomics-assisted prediction of salt and alkali tolerances and functional marker development in apple rootstocks. BMC Genom. 2020, 21, 550. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhao, P.; Kong, N.; Lu, R.; Pei, Y.; Huang, C.; Ma, H.; Chen, Q. Genome-Wide Identification and Characterization of the Potato bHLH Transcription Factor Family. Genes 2018, 9, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Y.; Hu, Z.; Shang, L.; Chai, Z.; Tang, Y.Z. Transcriptional Responses of the Heat Shock Protein 20 (Hsp20) and 40 (Hsp40) Genes to Temperature Stress and Alteration of Life Cycle Stages in the Harmful Alga Scrippsiella trochoidea (Dinophyceae). Biology 2020, 9, 408. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.F.; Liu, F.; Yang, X.; Wan, H.; Kang, Y. Global analysis of expression profile of members of DnaJ gene families involved in capsaicinoids synthesis in pepper (Capsicum annuum L). BMC Plant Biol. 2020, 20, 326. [Google Scholar] [CrossRef]
- Nagaraju, M.; Kumar, A.; Rajasheker, G.; Manohar Rao, D.; Kavi Kishor, P.B. DnaJs, the critical drivers of Hsp70s: Genome-wide screening, characterization and expression of DnaJ family genes in Sorghum bicolor. Mol. Biol. Rep. 2020, 47, 7379–7390. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Fang, B.; Wang, W.; Yang, Y.; Rao, L.; Zhang, C. Genome-wide analysis of the rice J-protein family: Identification, genomic organization, and expression profiles under multiple stresses. 3 Biotech 2019, 9, 358. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Qiu, H.-L.; Qu, D.-H.; Ruan, Y.; Chen, D.-H. Phylogeny-dominant classification of J-proteins in Arabidopsis thaliana and Brassica oleracea. Genome 2018, 61, 405–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Liu, C.; Song, X.; Sun, F.; Xiao, D.; Wei, Y.; Hou, X.; Zhang, C. Genome-wide identification and analysis of the regulation wheat DnaJ family genes following wheat yellow mosaic virus infection. 3 Biotech 2020, 10, 363. [Google Scholar] [CrossRef]
- Yan, J.; Ma, Z.; Xu, X.; Guo, A.-Y. Evolution, functional divergence and conserved exon–intron structure of bHLH/PAS gene family. Mol. Genet. Genom. 2014, 289, 25–36. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly regulated genes are intron poor. Trends Genet. TIG 2008, 24, 375–378. [Google Scholar] [CrossRef]
- Cheng, C.; Wang, Y.; Chai, F.; Li, S.; Xin, H.; Liang, Z. Genome-wide identification and characterization of the 14–3-3 family in Vitis vinifera L. during berry development and cold and heat-stress response. BMC Genom. 2018, 19, 579. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Zhao, H.; Cheng, Z.; Ke, Y.; Liu, J.; Ma, H. Evolution Analysis of the Fasciclin-Like Arabinogalactan Proteins in Plants Shows Variable Fasciclin-AGP Domain Constitutions. Int. J. Mol. Sci. 2019, 20, 1945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Ortiz, C.; Peña-Garcia, Y.; Natarajan, P.; Bhandari, M.; Abburi, V.; Dutta, S.K.; Yadav, L.; Stommel, J.; Nimmakayala, P.; Reddy, U.K. The ankyrin repeat gene family in Capsicum spp: Genome-wide survey, characterization and gene expression profile. Sci. Rep. 2020, 10, 4044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, S.; Deb, B.; Barbhuiya, P.A.; Uddin, A. Analysis of codon usage patterns and influencing factors in Nipah virus. Virus. Res. 2019, 263, 129–138. [Google Scholar] [CrossRef]
- Carlini, D.B.; Stephan, W. In vivo introduction of unpreferred synonymous codons into the Drosophila Adh gene results in reduced levels of ADH protein. Genetics 2003, 163, 239–243. [Google Scholar] [CrossRef]
- Roberts, R.J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1981, 9, 213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, V.; Ravindran, P.; Kumar, P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Arfan, M.; Athar, H.R.; Ashraf, M. Does exogenous application of salicylic acid through the rooting medium modulate growth and photosynthetic capacity in two differently adapted spring wheat cultivars under salt stress? J. Plant Physiol. 2007, 164, 685–694. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, W.; Zhang, Y.; Zhang, X.; Lang, D.; Zhang, X. The roles of methyl jasmonate to stress in plants. Funct. Plant Biol. 2019, 46, 197–212. [Google Scholar] [CrossRef]
- Odabaşıoğlu, M.; Demirtaş, G.; Yildirim, K.; Gürsöz, S. Salt Stress on Grapes (Vitis spp). In Proceedings of the International Gap Agriculture & Livestock Congress, Şanlıurfa, Turkey, 25–27 April 2018. [Google Scholar]
- Mimouni, H.; Wasti, S.; Manaa, A.; Gharbi, E.; Chalh, D.A.; Vandoorne, B.; Ben Ahmed, H. Does Salicylic Acid (SA) Improve Tolerance to Salt Stress in Plants? A Study of SA Effects On Tomato Plant Growth, Water Dynamics, Photosynthesis, and Biochemical Parameters. OMICS A J. Integr. Biol. 2016, 20, 180–190. [Google Scholar] [CrossRef]
- Lang, D.; Yu, X.; Jia, X.; Li, Z.; Zhang, X. Methyl jasmonate improves metabolism and growth of NaCl-stressed Glycyrrhiza uralensis seedlings. Sci. Hortic. 2020, 266, 109287. [Google Scholar] [CrossRef]
- Hatfield, J.L.; Prueger, J.H. Temperature extremes: Effect on plant growth and development. Weather. Clim. Extrem. 2015, 10, 4–10. [Google Scholar] [CrossRef] [Green Version]
- Li, K.-P.; Wong, C.-H.; Cheng, C.-C.; Cheng, S.-S.; Li, M.-W.; Mansveld, S.; Bergsma, A.; Huang, T.; Van Eijk, M.J.T.; Lam, H.-M. GmDNJ1, a type-I heat shock protein 40 (HSP40), is responsible for both Growth and heat tolerance in soybean. Plant Direct 2021, 5, e00298. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.W.; Rahman, M.A.; Kim, K.Y.; Choi, G.J.; Cha, J.Y.; Cheong, M.S.; Shohael, A.M.; Jones, C.; Lee, S.H. Overexpression of the alfalfa DnaJ-like protein (MsDJLP) gene enhances tolerance to chilling and heat stresses in transgenic tobacco plants. Turk. J. Biol. 2018, 42, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Chai, F.; Wang, Y.; Jiang, J.; Duan, W.; Wang, Y.; Wang, F.; Li, S.; Wang, L. Genome-wide Identification and Classification of HSF Family in Grape, and Their Transcriptional Analysis under Heat Acclimation and Heat Stress. Hortic. Plant J. 2018, 4, 133–143. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Li, J.; Zhao, X.-Q.; Wang, J.; Wong, G.K.-S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks Through Model Selection and Model Averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.; Duan, W.; Song, X.; Tang, J.; Wu, P.; Zhang, B.; Hou, X. Retention, Molecular Evolution, and Expression Divergence of the Auxin/Indole Acetic Acid and Auxin Response Factor Gene Families in Brassica Rapa Shed Light on Their Evolution Patterns in Plants. Genome Biol. Evol. 2016, 8, 302–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, M.A.; Haubold, B.; Mitchell-Olds, T. Comparative Evolutionary Analysis of Chalcone Synthase and Alcohol Dehydrogenase Loci in Arabidopsis, Arabis, and Related Genera (Brassicaceae). Mol. Biol. Evol. 2000, 17, 1483–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, P.; Zhong, K.; Zhang, F.; Xu, M.; He, L.; Jin, P.; Chen, J.; Yang, J. Wheat Yellow Mosaic Virus NIb Interacting with Host Light Induced Protein (LIP) Facilitates Its Infection through Perturbing the Abscisic Acid Pathway in Wheat. Biology 2019, 8, 80. [Google Scholar] [CrossRef] [Green Version]
- Wang, A. Dissecting the molecular network of virus-plant interactions: The complex roles of host factors. Annu. Rev. Phytopathol. 2015, 53, 45–66. [Google Scholar] [CrossRef]
- Bueso, E.; Rodriguez, L.; Lorenzo-Orts, L.; Gonzalez-Guzman, M.; Sayas, E.; Muñoz-Bertomeu, J.; Ibañez, C.; Serrano, R.; Rodriguez, P.L. The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J. 2014, 80, 1057–1071. [Google Scholar] [CrossRef]
- He, L.; Chen, X.; Yang, J.; Zhang, T.; Li, J.; Zhang, S.; Zhong, K.; Zhang, H.; Chen, J.; Yang, J. Rice black-streaked dwarf virus-encoded P5-1 regulates the ubiquitination activity of SCF E3 ligases and inhibits jasmonate signaling to benefit its infection in rice. New Phytol. 2020, 225, 896–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
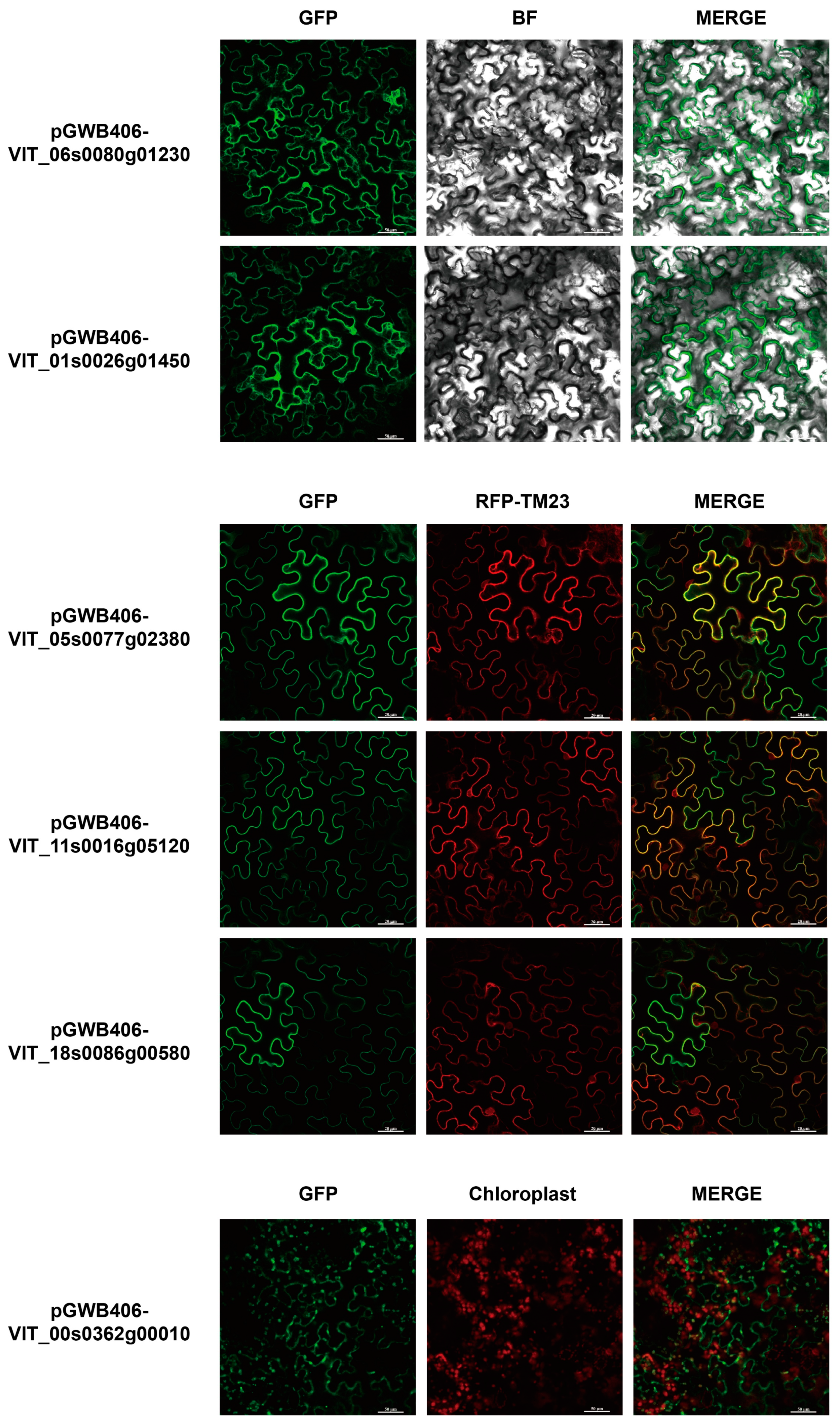
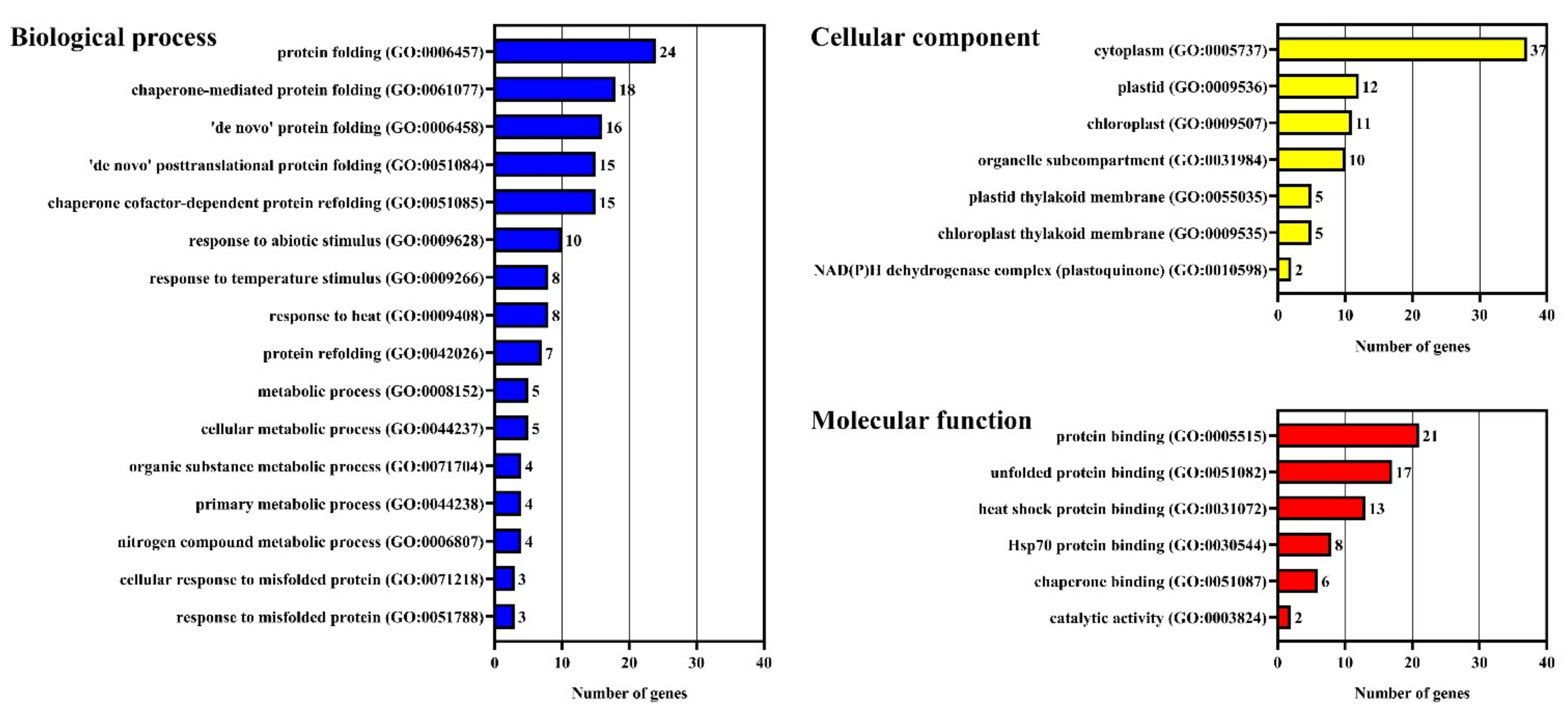
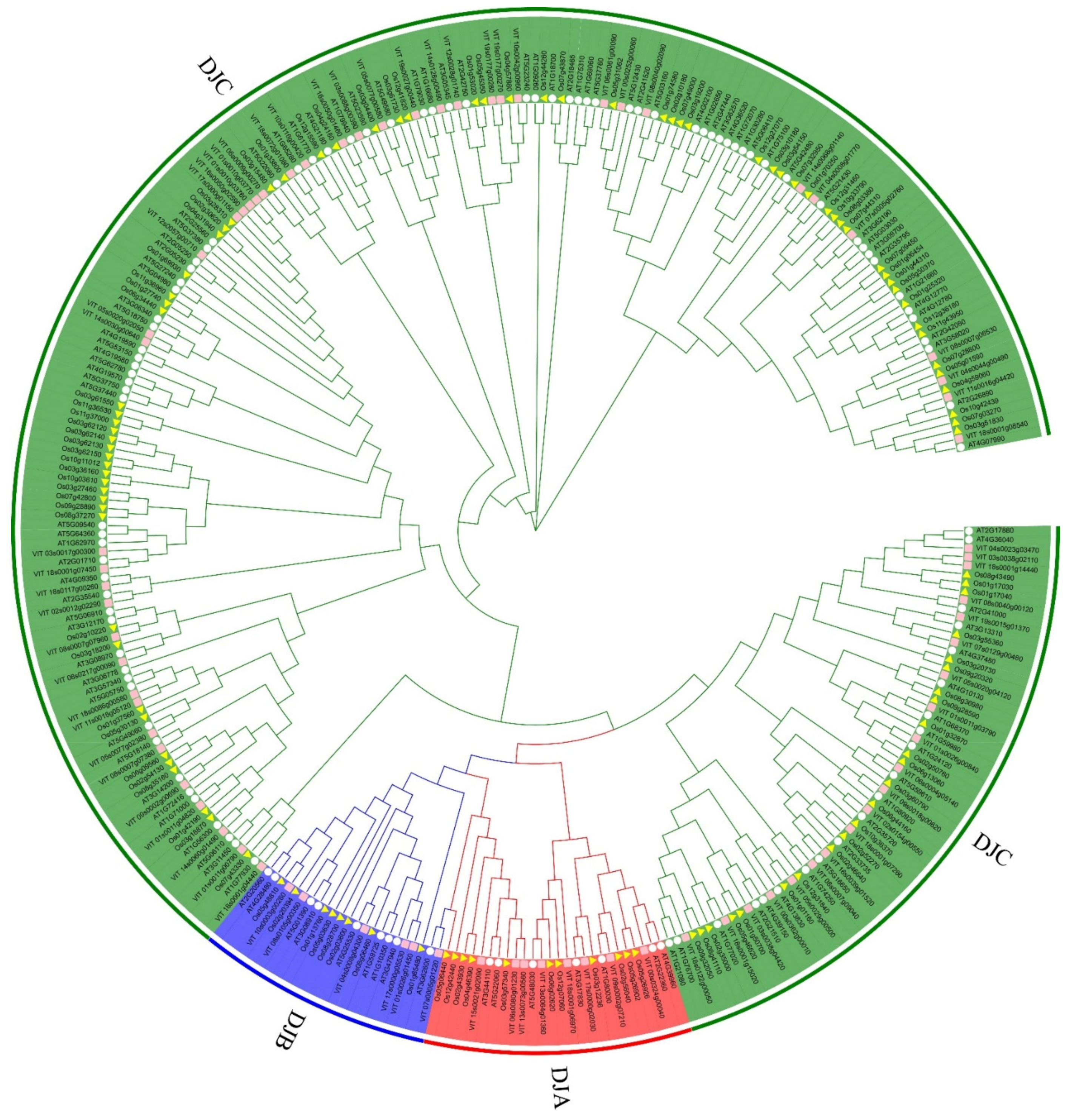

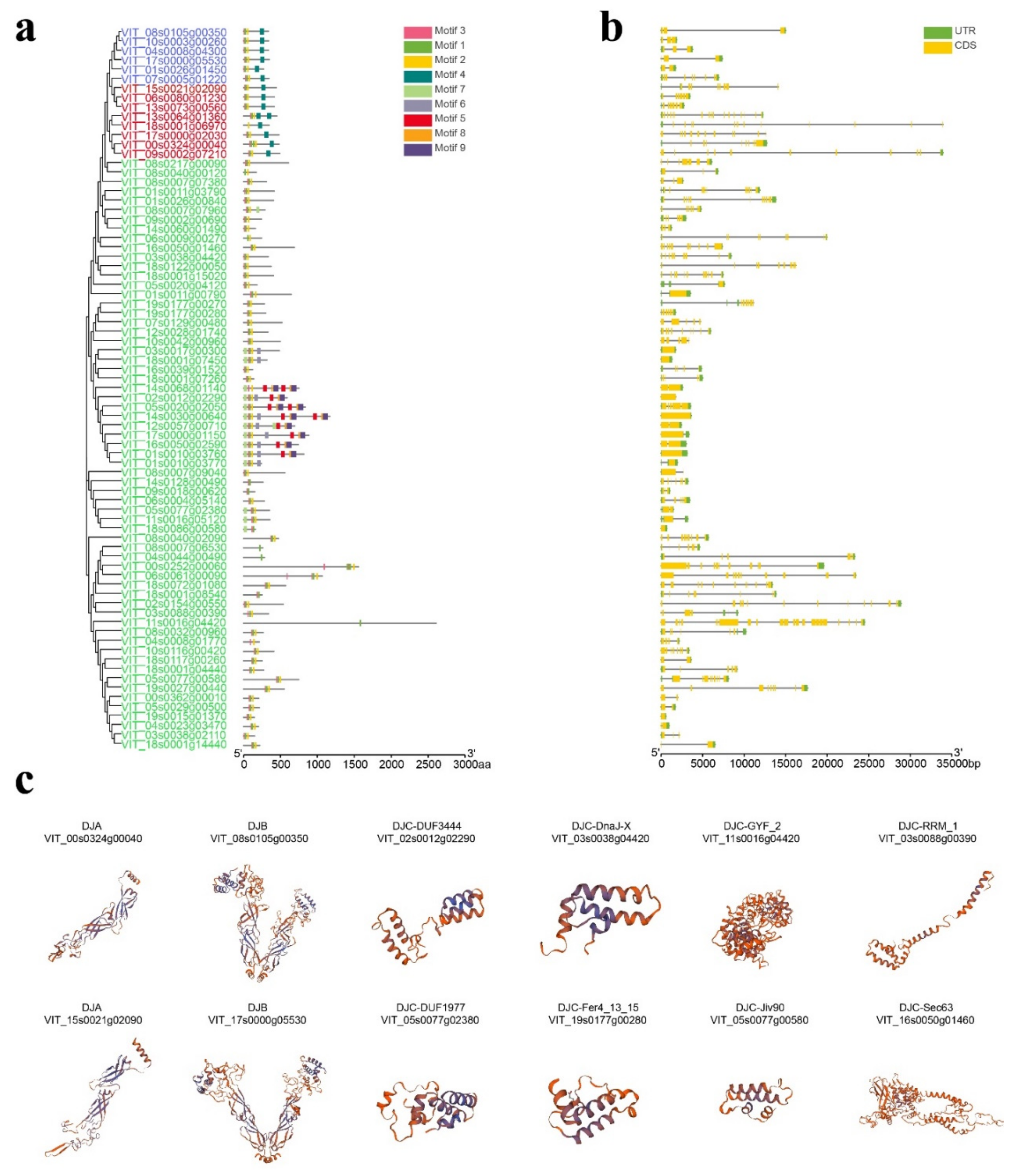



| Gene Name | Type | Chromosomal Distribution | CDS Length | Protein Size | Theoretical pI | Molecular Weight | Subcellular Localization | Signal Peptides |
|---|---|---|---|---|---|---|---|---|
| VIT_01s0011g00790 | DJC | Chr1: 710539-714104 | 1950 | 649 | 8.15 | 74,483 | Extracellular | None |
| VIT_01s0011g03790 | DJC | Chr1: 3429615-3441498 | 1248 | 415 | 5.92 | 45,912 | Plasma membrane | None |
| VIT_01s0011g04820 | DJC | Chr1: 4436645-4438196 | 507 | 168 | 6.18 | 19,610 | Extracellular | None |
| VIT_01s0026g00840 | DJC | Chr1: 9697764-9711600 | 1227 | 408 | 8.53 | 46,044 | Plasma membrane | None |
| VIT_01s0026g01450 | DJB | Chr1: 10418267-10420055 | 822 | 273 | 9.52 | 31,045 | Cytoplasmic | None |
| VIT_01s0010g03760 | DJC | Chr1: 21117742-21120902 | 2454 | 817 | 8.59 | 90,798 | Extracellular | None |
| VIT_01s0010g03770 | DJC | Chr1: 21126528-21128523 | 729 | 242 | 9.55 | 26,551 | Extracellular | None |
| VIT_02s0154g00550 | DJC | Chr2: 5296962-5325864 | 1614 | 537 | 8.98 | 59,496 | Plasma membrane | None |
| VIT_02s0012g02290 | DJC | Chr2: 9220337-9222116 | 1776 | 591 | 9.26 | 68,065 | Extracellular | None |
| VIT_03s0038g02110 | DJC | Chr3: 1449330-1451521 | 456 | 151 | 9.68 | 17,257 | Extracellular | None |
| VIT_03s0038g04420 | DJC | Chr3: 3205478-3213940 | 1011 | 336 | 6.64 | 37,724 | Extracellular | None |
| VIT_03s0088g00390 | DJC | Chr3: 8414516-8423780 | 1026 | 341 | 9.29 | 38,310 | Extracellular | None |
| VIT_03s0017g00300 | DJC | Chr3: 14583926-14585707 | 1461 | 486 | 5.15 | 53,581 | Extracellular | None |
| VIT_04s0008g01770 | DJC | Chr4: 1382423-1384601 | 645 | 214 | 6.77 | 24,104 | Extracellular | None |
| VIT_04s0008g04300 | DJB | Chr4: 3680859-3684659 | 1020 | 339 | 8.86 | 37,058 | Cytoplasmic | None |
| VIT_04s0023g03470 | DJC | Chr4: 20028906-20029894 | 606 | 201 | 9.02 | 22,502 | Extracellular | None |
| VIT_04s0044g00490 | DJC | Chr4: 21276992-21300325 | 858 | 285 | 9.77 | 32,810 | Extracellular | None |
| VIT_05s0077g00580 | DJC | Chr5: 388198-396327 | 2244 | 747 | 9.1 | 82,639 | Extracellular | None |
| VIT_05s0077g02380 | DJC | Chr5: 1910184-1911679 | 1062 | 353 | 7.63 | 41,094 | Plasma membrane | None |
| VIT_05s0020g02050 | DJC | Chr5: 3805550-3809148 | 2508 | 835 | 5.34 | 94,619 | Extracellular | None |
| VIT_05s0020g04120 | DJC | Chr5: 5772852-5780510 | 549 | 182 | 4.43 | 20,345 | Extracellular | None |
| VIT_05s0029g00500 | DJC | Chr5: 15301515-15303244 | 651 | 216 | 9.69 | 24,466 | Membrane bound chloroplast | None |
| VIT_06s0004g05140 | DJC | Chr6: 6080876-6084357 | 849 | 282 | 6.06 | 31,904 | Extracellular | Yes |
| VIT_06s0009g00270 | DJC | Chr6: 10340297-10360270 | 741 | 246 | 9.29 | 29,363 | Extracellular | None |
| VIT_06s0061g00090 | DJC | Chr6: 17293997-17317439 | 3210 | 1069 | 5.66 | 118,065 | Extracellular | None |
| VIT_06s0080g01230 | DJA | Chr6: 21412098-21415608 | 1254 | 417 | 5.78 | 46,358 | Cytoplasmic | None |
| VIT_07s0005g01220 | DJB | Chr7: 3750414-3757370 | 1038 | 345 | 5.99 | 38,873 | Endoplasmic reticulum | Yes |
| VIT_07s0005g02760 | DJC | Chr7: 5033390-5035659 | 219 | 72 | 9.99 | 8036 | Extracellular | None |
| VIT_07s0129g00480 | DJC | Chr7: 15715343-15720091 | 1569 | 522 | 6.71 | 59,464 | Extracellular | None |
| VIT_08s0105g00350 | DJB | Chr8: 7542314-7557322 | 1017 | 338 | 9.24 | 37,358 | Cytoplasmic | None |
| VIT_08s0217g00090 | DJC | Chr8: 8204077-8210204 | 1833 | 610 | 9.41 | 66,957 | Extracellular | Yes |
| VIT_08s0040g00120 | DJC | Chr8: 11031671-11038538 | 519 | 172 | 9.57 | 19,237 | Plasma membrane | None |
| VIT_08s0040g02090 | DJC | Chr8: 13199639-13205358 | 1425 | 474 | 5.92 | 52,783 | Extracellular | Yes |
| VIT_08s0007g06530 | DJC | Chr8: 20258210-20262858 | 786 | 261 | 9.13 | 30,015 | Extracellular | None |
| VIT_08s0007g07380 | DJC | Chr8: 20936611-20939266 | 930 | 309 | 9.41 | 34,898 | Extracellular | None |
| VIT_08s0007g07960 | DJC | Chr8: 21361191-21366038 | 876 | 291 | 8.11 | 33,289 | Nuclear | None |
| VIT_08s0007g09040 | DJC | Chr8: 22381356-22383986 | 1686 | 561 | 5.28 | 64,094 | Extracellular | None |
| VIT_09s0002g00690 | DJC | Chr9: 460481-463477 | 732 | 243 | 6.1 | 26,948 | Extracellular | None |
| VIT_09s0002g07210 | DJA | Chr9: 7150525-7184468 | 1470 | 489 | 8.96 | 52,719 | Extracellular | None |
| VIT_09s0018g00620 | DJC | Chr9: 16881863-16882927 | 459 | 152 | 5.25 | 17,357 | Chloroplast | None |
| VIT_10s0116g00420 | DJC | Chr10: 204741-208123 | 1236 | 411 | 9.31 | 47,334 | Plasma membrane | None |
| VIT_10s0003g00260 | DJB | Chr10: 1559664-1561563 | 1029 | 342 | 9.12 | 37,846 | Cytoplasmic | None |
| VIT_10s0042g00960 | DJC | Chr10: 14458409-14461750 | 1497 | 498 | 9.22 | 54,930 | Extracellular | None |
| VIT_11s0016g04420 | DJC | Chr11: 3708675-3733206 | 7830 | 2,609 | 5.84 | 284,387 | Extracellular | None |
| VIT_11s0016g05120 | DJC | Chr11: 4399836-4403087 | 1074 | 357 | 8.13 | 40,166 | Plasma membrane | None |
| VIT_12s0028g01740 | DJC | Chr12: 2373445-2379438 | 996 | 331 | 8.43 | 37,242 | Extracellular | None |
| VIT_12s0057g00710 | DJC | Chr12: 9329408-9331895 | 2085 | 694 | 7.85 | 76,528 | Extracellular | None |
| VIT_13s0073g00560 | DJA | Chr13: 14441307-14444078 | 1251 | 416 | 6.11 | 46,240 | Cytoplasmic | None |
| VIT_13s0064g01360 | DJA | Chr13: 23242305-23254584 | 1356 | 451 | 8.93 | 49,119 | Mitochondrial | None |
| VIT_14s0060g01490 | DJC | Chr14: 1175091-1176374 | 486 | 161 | 5.05 | 17,980 | Extracellular | None |
| VIT_14s0128g00490 | DJC | Chr14: 3110690-3113953 | 798 | 265 | 5.44 | 29,331 | Extracellular | None |
| VIT_14s0030g00640 | DJC | Chr14: 4739769-4743418 | 3510 | 1169 | 8.23 | 131,034 | Extracellular | None |
| VIT_14s0068g01140 | DJC | Chr14: 24933132-24935740 | 2259 | 752 | 5.78 | 85,287 | Extracellular | None |
| VIT_15s0021g02090 | DJA | Chr15: 12895594-12909716 | 1332 | 443 | 6.42 | 49,332 | Cytoplasmic | None |
| VIT_16s0039g01520 | DJC | Chr16: 1113429-1118305 | 372 | 123 | 6.19 | 14,482 | Plasma membrane | None |
| VIT_16s0050g01460 | DJC | Chr16: 18328905-18336293 | 2067 | 688 | 5.72 | 76,840 | Extracellular | Yes |
| VIT_16s0050g02590 | DJC | Chr16: 19570187-19573227 | 2232 | 743 | 9 | 82,581 | Extracellular | None |
| VIT_17s0000g01150 | DJC | Chr17: 818277-821660 | 2661 | 886 | 8.99 | 97,930 | Extracellular | None |
| VIT_17s0000g02030 | DJA | Chr17: 1644223-1656845 | 1446 | 481 | 9.08 | 53,139 | Extracellular | None |
| VIT_17s0000g05530 | DJB | Chr17: 6022005-6029405 | 1050 | 349 | 8.8 | 39,030 | Cytoplasmic | None |
| VIT_18s0122g00050 | DJC | Chr18: 70299-86511 | 1125 | 374 | 5.45 | 41,925 | Extracellular | None |
| VIT_18s0001g04440 | DJC | Chr18: 3852065-3861217 | 819 | 272 | 9.63 | 32,183 | Extracellular | Yes |
| VIT_18s0001g06970 | DJA | Chr18: 5184425-5218375 | 1044 | 347 | 8.54 | 38,433 | Mitochondrial | Yes |
| VIT_18s0001g07260 | DJC | Chr18: 5484997-5490030 | 417 | 138 | 4.71 | 16,458 | Extracellular | None |
| VIT_18s0001g07450 | DJC | Chr18: 5687109-5688423 | 957 | 318 | 8.8 | 34,752 | Extracellular | None |
| VIT_18s0001g08540 | DJC | Chr18: 6978788-6992668 | 750 | 249 | 9.28 | 29,677 | Extracellular | None |
| VIT_18s0001g14440 | DJC | Chr18: 12432955-12439459 | 660 | 219 | 10 | 24,129 | Extracellular | None |
| VIT_18s0001g15020 | DJC | Chr18: 13043063-13050564 | 1218 | 405 | 5.88 | 45,726 | Extracellular | None |
| VIT_18s0086g00580 | DJC | Chr18: 17892148-17892855 | 486 | 161 | 8.34 | 17,904 | Plasma membrane | None |
| VIT_18s0072g01080 | DJC | Chr18: 20620084-20633488 | 1716 | 571 | 8.86 | 64,865 | Extracellular | None |
| VIT_18s0117g00260 | DJC | Chr18: 23472866-23476496 | 762 | 253 | 5.42 | 28,812 | Extracellular | None |
| VIT_19s0177g00270 | DJC | Chr19: 6120151-6131282 | 855 | 284 | 9.24 | 32,355 | Extracellular | None |
| VIT_19s0177g00280 | DJC | Chr19: 6133013-6134771 | 909 | 302 | 8.85 | 34,404 | Extracellular | None |
| VIT_19s0015g01370 | DJC | Chr19: 9657864-9658472 | 447 | 148 | 9.35 | 16,343 | Extracellular | None |
| VIT_19s0027g00440 | DJC | Chr19: 19247402-19265051 | 1662 | 553 | 8.75 | 63,182 | Extracellular | None |
| VIT_00s0252g00060 | DJC | Chrun: 17649954-17669572 | 4677 | 1558 | 6.23 | 169,621 | Extracellular | None |
| VIT_00s0324g00040 | DJA | Chrun: 23409521-23422273 | 1446 | 481 | 9.32 | 52,150 | Extracellular | None |
| VIT_00s0362g00010 | DJC | Chrun: 25688160-25690181 | 627 | 208 | 9.35 | 23,579 | Chloroplast | None |
| Duplicated Gene Pairs | Ka | Ks | Ka/Ks | Duplicated Type | Time (Mya) |
|---|---|---|---|---|---|
| VIT_17s0000g05530/VIT_01s0026g01450 | 0.23 | 1.23 | 0.19 | Segmental | 7.59 |
| VIT_18s0122g00050/VIT_03s0038g04420 | 0.34 | 3.25 | 0.11 | Segmental | 11.40 |
| VIT_04s0023g03470/VIT_18s0001g14440 | 0.50 | 1.70 | 0.29 | Segmental | 16.59 |
| VIT_03s0038g02110/VIT_18s0001g14440 | 0.23 | 1.57 | 0.15 | Segmental | 7.68 |
| VIT_03s0038g02110/VIT_04s0023g03470 | 0.27 | 1.19 | 0.22 | Segmental | 8.90 |
| VIT_14s0030g00640/VIT_05s0020g02050 | 0.35 | 1.32 | 0.26 | Segmental | 11.56 |
| VIT_01s0010g03760/VIT_01s0010g03770 | 0.02 | 0.05 | 0.32 | Tandem | 0.54 |
| VIT_19s0177g00270/VIT_19s0177g00280 | 0.15 | 0.24 | 0.63 | Tandem | 5.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, T.; Xu, T.; Zhang, T.; Liu, T.; Shen, L.; Chen, Z.; Wu, Y.; Yang, J. Genome-Wide Identification and Characterization of DnaJ Gene Family in Grape (Vitis vinifera L.). Horticulturae 2021, 7, 589. https://doi.org/10.3390/horticulturae7120589
Chen T, Xu T, Zhang T, Liu T, Shen L, Chen Z, Wu Y, Yang J. Genome-Wide Identification and Characterization of DnaJ Gene Family in Grape (Vitis vinifera L.). Horticulturae. 2021; 7(12):589. https://doi.org/10.3390/horticulturae7120589
Chicago/Turabian StyleChen, Tianchi, Tao Xu, Tianye Zhang, Tingting Liu, Leyi Shen, Zhihui Chen, Yueyan Wu, and Jian Yang. 2021. "Genome-Wide Identification and Characterization of DnaJ Gene Family in Grape (Vitis vinifera L.)" Horticulturae 7, no. 12: 589. https://doi.org/10.3390/horticulturae7120589
APA StyleChen, T., Xu, T., Zhang, T., Liu, T., Shen, L., Chen, Z., Wu, Y., & Yang, J. (2021). Genome-Wide Identification and Characterization of DnaJ Gene Family in Grape (Vitis vinifera L.). Horticulturae, 7(12), 589. https://doi.org/10.3390/horticulturae7120589







