Abstract
Fruit hardening, one of the physiological disorders influencing the quality of pear, is usually accompanied by fruit lignification. Nonetheless, the mechanism by which lignin accumulates in hardened pear fruit is still unknown. Here, Transcriptome analysis of normal fruits (NFs) and hardened fruits (HFs) of ‘Suli’ pear (Pyrus bretschneideri Rehd) was used to examine the molecular mechanism of fruit hardening. We found that hardened fruits (HFs), especially those in the top region within 35–180 days after flowering (DAF), possessed a prominently higher lignin level than normal fruits (NFs). Subsequently, transcriptome sequencing of fruit at 35 DAF identified 4391 [HF shoulder region (HF_S) vs. NF top region (NF_T)], 3849 (HF_T vs. NF_T) and 408 (HF_T vs. HF_S) differentially expressed genes. Among them, we clarified 26 unigenes that encode 10 enzymes as candidate genes participating in lignin biosynthesis. Thus, high expression of the lignin biosynthetic gene impels lignin to accumulate in HFs. Some transcription factors were evaluated to link with lignin formation and subsequently analysed. In addition, the genes participating in flavonoid/proanthocyanidin biosynthesis dropped from the HF_T to the HF_S, suggesting that the flavonoid metabolic pathway was reduced to promote intermediate metabolites to be converted to the lignin biosynthetic pathway. To conclude, this study lays a solid theoretical foundation and provides reference data for investigating the mechanism by which lignin accumulates in hardened pear fruit.
1. Introduction
‘Suli’ pear (Pyrus bretschneideri Rehd), has long been cultivated in a broad geographical area in China [1]. However, this variety in Shanxi Province of China has shown severe fruit hardening in recent years. Fruit hardening, known as hard-end, is principally manifested as an apparent protrusion in the top region of the diseased fruit with extremely hard texture [2]. Fruit hardening is an intractable disease hindering pear growth and production, and it appears in such European and Asian cultivars as ‘Anjou’, ‘Winter Nelis’, ‘Comice’, ‘Whangkeumbae’ and ‘Xueqing’ according to reports [3,4,5]. Currently, researchers speculate that texture hardening in pear fruit is attributable to scion-rootstock compatibility problems, which causes physiological imbalance, such as water stress and calcium deficiency [6,7,8]. Another study has confirmed that CaCl2 spray can alleviate the symptoms of hardening mainly by reducing the content of lignin in fruits [9]. Generally, there is close interrelationship between the changes of firmness in fruit and those of cell wall components [10]. Previous analysis of the variations in the composition of the cell wall has uncovered that only lignin content increased significantly in the hardened ‘Suli’ fruit [2]. Hence, screening and identification of lignin synthesis-associated key genes are pivotal for the determination of target genes useful for investigation on lignin metabolism regulation in the future.
Lignin is a phenylpropanoid pathway-derived secondary metabolite playing a role in plants [11]. Phenylalanine ammonia-lyase (PAL), 4-coumarate: coenzyme A ligase (4CL) and cinnamic acid 4-hydroxylase (C4H) catalyse phenylpropanoid biosynthesis. Caffeoyl shikimate esterase (CSE), Hydroxycinnamoyl-CoA: shikimate hydroxycinnamoyl transferase (HCT), p-coumaroyl shikimate 3′-hydroxylase (C3′H), caffeoyl CoA O-methyltransferase (CCoAMT), cinnamoyl-CoA reductase (CCR), caffeic acid 3-O-methyltransferase (COMT), ferulate 5-hydroxylase (F5H), cinnamyl alcohol dehydrogenase (CAD), peroxidase (PER) and laccase (LAC) are major enzymes controlling the lignin biosynthesis [12,13]. In pear fruit, stone cell growth relies in the biosynthesis of two types of lignin, namely, guaiacyl lignin (G-lignin) and syringyl lignin (S-lignin) [14,15]. For different pear varieties and developmental stages, structural genes were identified as hub genes involved in the lignin pathway [16,17,18,19,20], Table S1. Pears are rich in various lignin biosynthesis genes since the expansion of lignin synthesis gene family contributes to the large accumulation of lignin [21].
Largely deposited in the secondary cell wall of vascular plants, lignin enhances the mechanical strength to dredge the water transport of tissue and resist the invasion of the adverse external environment [12,22]. Fruits are susceptible to various biotic and abiotic stress, such as low-temperature stress in loquat fruit [23], physical impact in mangosteen [24,25] and injured or infected citrus fruit peel [26], which also speed up lignin accumulation. The synthesis of lignin in fruits is regulated by relevant genes and a variety of other genetic and physiological factors. For example, oleocellosis induces such lignin biosynthesis-associated genes, as PAL, CCR, F5H, C4H, UGT72E and POD, resulting in the accumulation of lignin in citrus fruit [27]. Nonetheless, the molecular mechanism by which lignin accumulates in hardened pear fruit remains largely unknown.
RNA-Seq, an efficacious technique for probation of plant functional genomics, has been applied in a wide range of studies of fruit trees. Based on this technology, large amounts of sequence information and expression data can be obtained [18,28]. In the present study, the molecular mechanism by which lignin forms in hardened pear fruit was probed using RNA-Seq. With the aim to clarify the genes involved in lignin, differentially expressed genes (DEGs) in the normal and hardened ‘Suli’ fruits were analysed, the findings of which elucidated the way fruit hardening forms in pear cultivars.
2. Materials and Methods
2.1. Fruit Materials
Fruits were gained from pear (Pyrus bretschneideri Rehd. cv. Suli) trees at commercial pear orchards of northern China, Qixian County in Shanxi Province. Hardened fruits (HFs) were collected from ‘Suli’ pear trees aged 10 years old in one orchard, while normal fruits (NFs) from healthy pear trees in another orchard. First, fruits were sampled at 15, 25, 35, 45, 55 and 180 DAF, and three biological replicates comprising 10–15 fruits each were used for each condition and at each sampling point. Next, the fleshy tissues harvested from the top region (the calyx end of the fruit) and the shoulder region (the pedicel of the fruit) were stochastically grouped accordingly. The samples in one group were utilised to measure the lignin level, and those in the other group were subjected to freezing in liquid nitrogen and reservation at −80 °C, followed by RNA isolation. As the lignin accumulation time uncovered that HFs stimulated the lignin accumulation after 25 DAP, the two regions presented no dramatic difference in the lignin level in NFs (shown in the ‘Results’). Therefore, fleshy tissues were chosen from the top region of NFs (NF_T), the shoulder region of HFs (HF_S) and the top region of HFs (HF_T) at 35 DAP for the transcriptomic analysis, so as to determine factors controlling lignin biosynthesis in the early phase.
2.2. Observation of Stone Cells
Stone cells were observed using the method specified by Liu et al. [2]. Specifically, fruits were fixed with FAA (formaldehyde, acetic acid, ethanol) for 3 h, soaked in glycerol and sectioned. Then the sections were observed under a Ni-U fluorescence microscope with C-HGF1 fluorescence emission source and Fi3 camera (Nikon, Tokoy, Japan). Finally, blue fluorescence of lignin in the stone cells was observed under the excitation wavelength of 380 nm of purple light.
2.3. Lignin Level Measurement
With the use of the modified version of method proposed by Syros et al., we tested the lignin level [29]. Subsequent to peeling, the pulp was dried, subjected to grinding into uniform powder and screened, and dry pulp powder was obtained. Then it was put into the 10 mL grinding glass test tube and added with 2 mL of 25% bromoacetyl-glacial acetic acid. Following this, the test tube mouth with a glass plug was sealed and the test tube oscillated every 10 min for half an hour in water bath at the constant temperature of 70 °C, which was then terminated by 3 mL of 2 mol L−1 NaOH. Next, the mixed solution was placed to a volumetric flask, whose volume was adjusted to 100 mL using glacial acetic acid. Finally, a spectrophotometer was employed to measure the absorbance at 280 nm.
2.4. Extraction of Total RNA and Analysis of RNA-Seq Data
Total RNA of NF and HF were extracted using CTAB-LiCl methods [30]. Electrophoresis was completed on 1% agarose gel to ascertain RNA integrity, and a Nanodrop 2000 spectrophotometer provided by Thermo Scientific (Waltham, MA, USA) was employed for RNA content measurement. Biomarker Technologies Corporation (Beijing, China) analysed RNA samples using the Illumina Hiseq4000 platform. In line with the manufacturer’s guidance, the cDNA library was established, and low-quality sequences were discarded by purification. Each sample had similar clean reads to the genome of Chinese white pear (http://peargenome.njau.edu.cn, accessed on 11 October 2020) [21]. Genes with the |log2| ≥ 1 and False Discovery Rate (FDR) < 0.05 obtained by DESeq2 were considered as DEGs. Using BMKCloud online tools (https://www.biocloud.net/, accessed on 14 August 2021), gene function assessment was implemented.
2.5. Quantitative Real-Time PCR Analysis (qRT-PCR)
As per the guidance of the manufacturer, 6 candidate DEGs participating in lignin biosynthesis were picked up for qRT-PCR using SYBR® Premix Ex TaqTM (TaKaRa Bio, Dalian, China) to testify the DEG results. Later, PrimeScript First-Stand cDNA Synthesis Kit (TaKaRa Bio, China) was utilised to synthesize cDNA. With gene-specific primers listed in Table S2, the 2−ΔΔCTmethod was adopted for the calculation of relative gene expressions [31], where PbrACTIN served as the housekeeping gene. Three independent replicates were set for data obtaining.
2.6. Processing of Data
SPSS19.0 (SPSS Inc. Chicago, IL, USA) was employed to statistically process data. The obtained data were expressed as means ± SE, and data comparison was implemented by t-test. Statistical significance could be confirmed at p < 0.05.
3. Results
3.1. Lignin Level in HFs at Varying Developmental Stages
To provide more detailed information on fruit lignification in hardened ‘Suli’ pear, the differences in stone cells content, fruit firmness and the lignin accumulation time between the shoulder and top regions of HFs and NFs were explored. The results revealed that there was an apparent protrusion in the HF_T (Figure 1A). Stone cells were observed using lignin autofluorescence method. We observed the largest range and brightness of lignin fluorescence in stone cells in the HF_T (Figure 1B). Besides, the HF_T had significantly greater firmness and higher stone cell content than the HF_S; however, there were no significant differences between the NF_T and NF_S (Figure 1C). Furthermore, lignin determination showed that the lignin level showed a step-wise rising trend at the early developmental stage of fruit, reached the peak at 45 DAF and then gradually decreased within 45–180 DAF (Figure 1D). Compared with NFs, HF presented no promotion on the lignin accumulation within 15–25 DAF but prominently elevated the lignin level at 35–180 DAF. This result indicated that the upstream regulators of lignin accumulation probably may play a role at 35 DAF. In addition, we have noticed that lignin content did not largely differ between the NF_T and NF_S at each stage of fruit development. However, the lignin levels in the top region were about 32.12%, 26.62%, 21.94% and 27.47%, respectively, which were higher than those in the HF_S at 35, 45, 55 and 180 DAF, suggesting that the HF_S and HF_T had different mechanisms for regulating lignin accumulation. Hence, the fleshy tissues from the NF_T, HF_S and the HF_T at 35 DAF were collected for the following transcriptomic analysis, so as to figure out the upstream regulators of lignin biosynthesis in hardened pear fruit at the early stage.
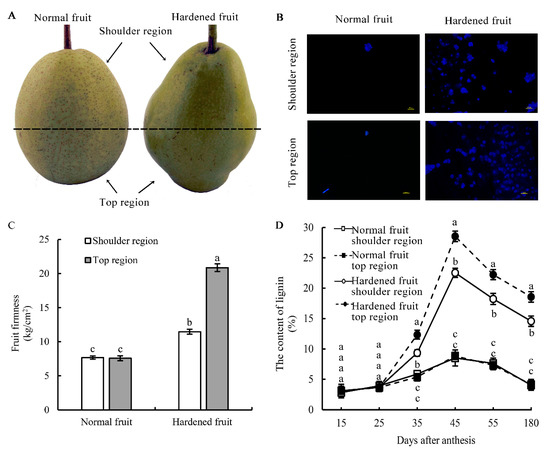
Figure 1.
Lignin accumulation in the ‘Suli’ pear fruit. (A) The phenotypes of ‘Suli’ pear NFs and HFs during the ripening period. (B) Observation of stone cells in NFs and HFs using fluorescence microscopy. Bar = 100 μm. (C) Fruit firmness in the top and shoulder regions of NFs and HFs. (D) Time courses of lignin accumulation at different fruit development stages. Three independent replicates consisting of 10–15 fruits each were collected for each sample. Small letters stand for significant differences (p = 0.05).
3.2. Library Establishement and Transcriptome Sequencing
Total RNA extraction from NFs and HFs was conducted, and nine cDNA libraries formed (each type of tissue had3 biological replicates; NF_T: NF_T 1, NF_T 2, NF_T 3; HF_S: HF_S 1, HF_S 2, HF_ S 3; HF_T: HF_T 1, HF_T 2, HF_T 3). After removing adapter sequences and reads with poor quality, 19–28 million clean reads were gained from each library (Table S3). In each sample, the GC content ranged from 47.17% to 47.72% and bases with a Q score of 30 or higher accounted for more than 92.66% of all the bases. Then the reads were subjected to quality control, and the majority of them from the NF_T (79.01–79.47%), HF_S (76.75–77.92%) and HF_T (76.36–79.21%) groups were mapped to the reference genome successfully. Around 70% of the mapped reads were in line with a single position. In each sample, the numbers of reads mapped to ‘+’ and ‘−’ strands were approximately equal. We also analysed the correlation, and it was uncovered that the 3 replicates for each sample had r2 > 0.95, demonstrating their the consistency. It can be concluded that the RNA-Seq results are highly valuable for subsequent research.
3.3. Identification of DEGs
The analysis of DEGs was implemented to reveal the potential mechanisms by which lignin differentially accumulates in pairs of samples (HF_S vs. NF_T, HF_T vs. NF_T, HF_T vs. HF_S). The expression levels of DEGs were measured as Fragments Per Kilobase of transcript per Million mapped (FPKM) reads values, followed by identification and filteration on the basis of criteria as follows: FDR < 0.05 and an |log2| ≥ 1. After screening, we identified 4391 (1870 upregulated and 2521 downregulated), 3849 (1410 rose and 2439 dropped) and 408 (24 rose and 384 dropped) DEGs for HF_S vs. NF_T, HF_T vs. NF_T and HF_T vs. HF_S, respectively (Figure 2A–C, Table S4). There were 93, 1203 and 836 DEGs detected independently in each compared group, denoting the involvement of these genes in the particular physiological process in HFs (Figure 2D–F, Table S4). 36 DEGs were common in all the compared samples, five of which participating in phenylpropanoid biosynthesis were determined.
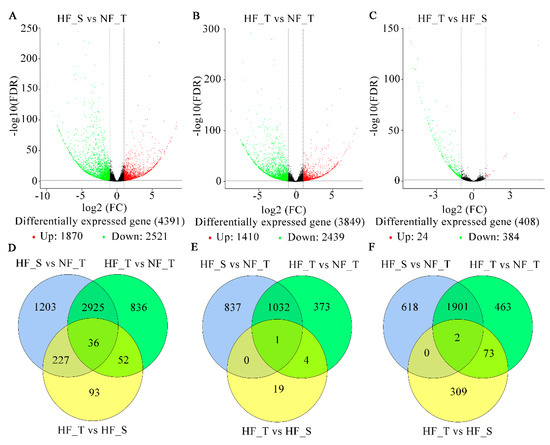
Figure 2.
RNA-Seq analysis was implemented to determine DEGs in NFs and HFs. (A–C) Volcano plot of the DEGs in HF_S vs. NF_T (A), HF_T vs. NF_T (B) and HF_T vs. HF_S (C), respectively. (D) Total number of DEGs. (E) Number of raised DEGs. (F) Number of decreased DEGs. Three independent replicates consisting of 10–15 fruits each were collected for each sample. (NF_T: normal fruit top region, HF_S: hardended fruit shoulder region, HF_T: hardended fruit top region).
3.4. Gene Ontology (GO) Enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis of the DEGs
GO enrichment analysis was implemented for DEGs, which were divided into ‘biological process’, ‘cellular component’ and ‘molecular function’ (Figure 3A–C, Table S5). The two most significant sub-categories in the ‘biological process’ category were ‘metabolic process’ and ‘cellular process’. The most abundant classes in the ‘cellular component’ category were ‘cell’ and ‘cell part’. ‘Catalytic activity’ and ‘binding’ were the two dominant sub-categories within the ‘molecular function’ category.
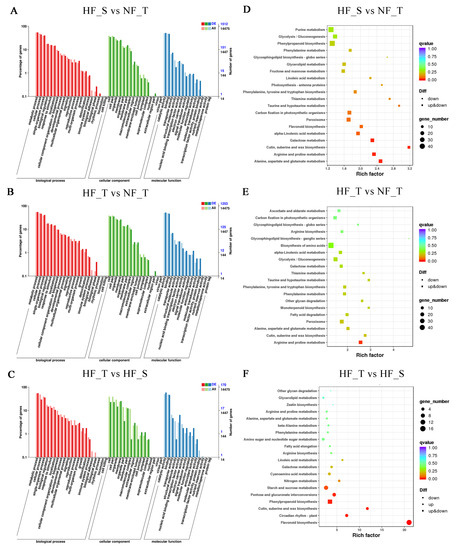
Figure 3.
Division of DEGs in light of function. (A–C) GO categories assigned to differentially expressed genes in HF_S vs. NF_T (A), HF_T vs. NF_T (B) and HF_T vs. HF_S (C), respectively. (D–F) KEGG pathways of DEGs in HF_S vs. NF_T (D), HF_T vs. NF_T (E) and HF_T vs. HF_S (F), respectively. Three independent replicates consisting of 10–15 fruits each were collected for each sample. (NF_T: normal fruit top region, HF_S: hardended fruit shoulder region, HF_T: hardended fruit top region).
Aiming at deeply understanding the biological pathways activated in HFs, the KEGG pathway enrichment was conducted, and the results showed that biosynthesis of ‘flavonoid biosynthesis’ (ko00941) and ‘phenylpropanoid biosynthesis’ (ko00940) represented the top 20 enriched KEGG pathways in all pair of samples (Figure 3D–F, Table S6).
3.5. DEGs Related to Lignin Biosynthesis between NFs and HFs
Generally, there is interrelationship between lignin biosynthesis and the phenylpropanoid metabolism pathway. It was revealed by the evaluation of RNA-seq data that some notable enzyme genes presented active transcription and expression in the phenylpropanoid metabolism pathway, and they were identify to regulate the entry of carbon sources for lignin biosynthesis, including PbrPAL (2), Pbr4CL (3), PbrHCT (1), PbrCOMT (2), PbrF5H (1), PbrCCR (6), PbrCAD (2), PbrC3H (1), PbrCES (2), PbrPER (3) and PbrLAC (3) (Figure 4A, Table S7). The expression levels of these notable enzyme genes in the lignin biosynthetic pathway were measured in view of the findings in transcriptome sequencing. In the NF_T group, the most important lignin biosynthesis enzyme genes wer elowly expressed; these notable enzyme genes in the lignin biosynthetic pathway showed an upregulation trend in at least one region of HFs.
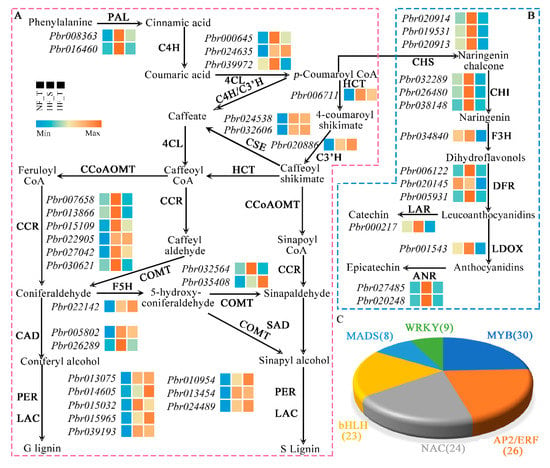
Figure 4.
Simplified lignin and flavonoid/proanthocyanidin biosynthetic pathways as well as lignin biosynthesis-related transcription factors. (A) The expression of structural genes associated with lignin biosynthetic pathway in hardened ‘Suli’ pear. (B) The expression of structural genes associated with flavonoid/proanthocyanidin biosynthetic pathway in hardened ‘Suli’ pear. Each procedure for lignin and flavonoid/proanthocyanidin biosynthesis presents the name of enzymes, gene IDs and expression patterns on the side. For each gene, the first square represents the NF_T, the first square represents the HF_S and the last square represents the HF_T. Different colours stand for varying expression levels. The colour gradient from blue to red harmonizes with transcript levels in an ascending order, whose values serve as the FPKM values. (C) Transcription factors of lignin biosynthesis in hardened ‘Suli’ pear.
To explain why the HF_T displays a level of lignin higher than that in the HF_S, we further analysed the difference in phenylpropane metabolism flavonoid metabolism between the two regions. Interestingly, in the flavonoid/proanthocyanidin biosynthetic pathway, the genes coding for Chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR), leucocyanidin dioxygenase (LDOX), leucoanthocyanidin reductase (LAR) and anthocyanidin reductase (ANR) were all downregulated (Figure 4B, Table S8). ANR and LAR are notable enzymes in the biosynthesis pathway of proanthocyanidins, responsible for the production of epicatechin and catechin, respectively. Biosynthesis of proanthocyanidins and lignin have common precursors. This suggests that the reduced flavonoid/proanthocyanidin biosynthesis probably impels the elevation of lignin.
Transcription factors control gene expression to participate in lignin biosynthesis in plants. In our data, we predicted transcription factors using BMKCloud online tools (https://www.biocloud.net/, accessed on 14 August 2021). One hundred and twenty-three differentially expressed transcription factors had relevance to lignin biosynthesis (Figure 4C, Table S9), of which MYBs (30) and AP2/ERFs (26) dominated, followed by NACs (24), bHLHs (23), MADSs (11), and WRKYs (9), and they were proven to impel lignin metabolism in the hardened ‘Suli’ pear.
3.6. qRT-PCR Validation
Six DEGs were chosen to confirm the veracity of transcriptome data. It was unveiled by the qRT-PCR results that, compared with NFs, all genes were highly expressed in two regions of HFs. However, the increasing trend of genes differed across the two regions of HFs, including four genes having higher expression in the top than in the shoulder region, one gene having no significant difference and one gene having lower expression in the top than in the shoulder region. As a result, the qRT-PCR results were in line with the RNA-Seq results (Figure 5).
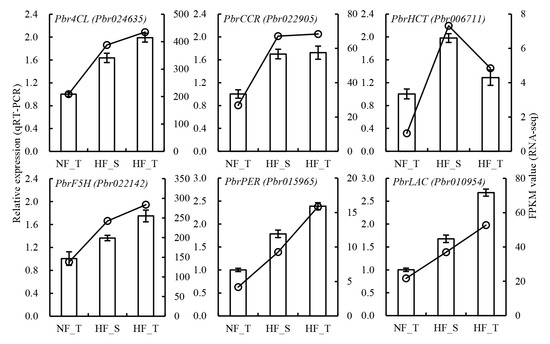
Figure 5.
Validation of DEG expression in the lignin biosynthesis pathway by qRT-PCR. Three independent replicates consisting of 10–15 fruits each were collected for each sample. (NF_T: normal fruit top region, HF_S: hardended fruit shoulder region, HF_T: hardended fruit top region).
4. Discussion
Fruit hardening is a new disorder that occurs in the Asian ‘Suli’ pear. The main symptom of the disorder is the accumulation of lignin, especially in the fruit top region, causing a rapid increase in fruit firmness [2]. However, the molecular mechanisms by which lignin forms in hardened ‘Suli’ pear remain elusive. In the present study, we observed that lignin accumulation began at 25–35 DAF in NFs and HFs, suggesting that 25–35 DAF is a decisive period for fruit hardening. Furthermore, within 35–180 DAF, the HF_T had a predominantly higher lignin level than the HF_S, but this level did not differ between the NF regions. This result indicated that there were regional differences in the lignin metabolism of HFs. Therefore, we selected fleshy tissues from the NF_T, HF_S and HF_T at 35 DAP for transcriptome sequencing to ascertain the molecular basis for lignin formation.
Generally, a higher lignin level reflects a higher expression level of lignin biosynthesis-associated genes. It has been reported before that of the highly expressed genes that encode PAL, 4CL, CAD and POD lead to the accumulation of more lignin in hard-end ‘Whangkeumbae’ pear [9]. In this study, transcriptome analysis identified 26 unigenes participating in the lignin biosynthesis, which manifested that most of the genes were significantly upregulated in at least one region of HFs compared with the NF_T (Figure 4A, Table S4). The above findings unveiled that lignin biosynthesis surged at 35 DAP, which resulted in higher lignin content in HFs.
In the phenylalanine metabolic pathway, phenylalanine, catalysed by multifold enzymes (PAL, C4H and 4CL), can yield p-coumaroyl-CoA [12,13]. The genes coding for PAL and 4CL and the function in producing lignin have been identified in Arabidopsis thaliana [32] and castor [33]. In our study, two genes coding for PbrPAL (Pbr008363 and Pbr016460) and three genes coding for Pbr4CL (Pbr000645, Pbr024635 and Pbr039972) were upregulated in at least one region of HFs compared with NFs, suggesting that excess of p-coumaroyl-CoA stimulates the up-regulation of downstream genes associated with monolignol biosynthesis, which eventually drives monolignol biosynthesis in the HF fruit. Furthermore, several key enzyme genes needed by lignin biosynthesis were identified in a study on the early development of lignin in ‘Dangshan Su’ pear, including PbrPAL (Pbr008363 and Pbr016460) and Pbr4CL (Pbr024635) [16].
p-coumaroyl shikimate is synthesised by HCT, followed by hydroxylation of the hydroxycinnamoyl moiety at position 3 via C3′H to yield caffeoyl shikimate [12,13]. In Arabidopsis thaliana, deficiency in HCT or C3′H gives rise to lignin reduction and growth impediment [34]. This result indicates that both HCT and C3′H belong to key genes in lignin biosynthesis. In two regions of HFs, two upregulated genes, PbrHCT (Pbr006711) and PbrC3′H (Pbr020886) are likely to participate in the lignin biosynthetic pathway.
CSE has been recently characterized as an enzyme central to the lignin biosynthetic pathway in Arabidopsis thaliana [35]. In the top and shoulder regions of HFs, two upregulated genes coding PbrCSE (Pbr024538 and Pbr032606) are likely to participate in the lignin biosynthetic pathway.
Feruloyl-CoA is processed to coniferaldehyde by CCR and converted to coniferyl alcohol through CAD [12,13]. The downregulation of CCR or CAD has been demonstrated to cause lignin level reduction and monolignol precursor accumulation [36,37,38]. Here, six CCRs with differential expressions. Among them, Pbr015109, Pbr022905 and Pbr027042 were upregulated in both shoulder and top in HFs, suggesting that lignin accumulation is associated with their expression. The other PbrCCR genes (Pbr007658, Pbr013866 and Pbr030621) showed upregulation in the HF_S but downregulation in the HF_T, indicating that their function could be redundant, which requires additional verification. Two genes coding for PbrCAD (Pbr005802 and Pbr026289) were upregulated in HFs. Notably, PbrCAD (Pbr026289) is involved in lignin biosynthesis in ‘Dangshan Su’ and ‘Lianglizaosu’ pear fruits [17].
Both COMT and F5H perform functions in exact of monolignol lignin biosynthesis [12,13]. F5H is capable of converting guaiacyl (G) monolignol to syringyl (S) monolignol and utilising such substrates as coniferaldehyde, ferulic acid and coniferyl alcohol; COMT catalyses O-methylation to multifold substrates, including 5-hydroxy-coniferaldehyde, caffeic acid, caffeoyl alcohol, 5-hydroxyl-feruloyl-CoA and 5-hydroxy-coniferyl alcohol. It has been unveiled that variations in lignin syringyl monomer content can be attributed to the controlling of COMT or F5H expression [39]. Consistently, the levels of PbrF5H (Pbr022142) and PbrCOMT (Pbr032564 and Pbr035408) transcripts were increased in HFs, unveiling that impelling the conversion of G-lignin to S-lignin monomer is beneficial for lignin accumulation. Besides, PbrF5H (Pbr022142) was reported for lignin biosynthesis in ‘Dangshan Su’ pear fruit [16].
LAC and POD perform functions in the final procedure of lignin biosynthesis. In Arabidopsis thaliana, interference with AtLAC4 and AtLAC17 or over-expression of PRX gene AtPrx17 reduced or increased lignin content, respectively [40,41]. Therefore, the increased expression levels of PbrLACs and PbrPERs, especially PbrPRX64 (Pbr039193) screened in previous studies [42], may play crucial roles in lignin accumulation in hardened ‘Suli’ pear.
The activity of transcription factors determines a multitude of physiological activities of plants, including lignin biosynthesis [43]. So far, most lignin activators characterized are members of the MYB family and have been identified in Arabidopsis [44,45,46], Pinus taeda [47,48], Eucalyptus gunnii [49] and Populus trichocarpa [50]. Meanwhile, the identification and characterisation of some lignin biosynthesis-associated transcription factors, such as NAC, bHLH and AP2/EFRs, have been achieved in Arabidopsis thaliana, pear, and loquat fruit [51,52,53,54]. Upstream transcription factors control the lignin. For example, the MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) is responsible for the formation of lignified tissues by binding to the AtPRX17 promoter and regulating the AtPRX17 expression level [41]. Here, transcriptome data evaluation presented 86 differentially expressed key transcription factors, including AP2/ERFs, MYB, WRKY, MADS, bHLH, WD40 and NACs (Table S8), and they are likely to be candidates for controlling lignin biosynthesis in the hardened ‘Suli’ pear fruit.
Lignin and flavonoids/PAs are synthesised via the phenylpropanoid pathway, in which p-coumaroyl CoA is turned into different metabolites via two different steps catalysed by HCT or CHS [11]. HCT can condense p-coumaroyl CoA with 3 malonyl CoA molecules to catalyse the conversion of p-coumaroyl CoA to p-coumaroyl shikimate [30]. In plants, the disequilibrium in HCT and CHS genes expression modulated the accumulation of lignin and flavonoids. In a report of Besseau et al., impediment of lignin synthesis brinds about the redirection of the metabolic flux into flavonoids through CHS activity in Arabidopsis plants silenced by HCT [55]. A similar result was also verified by Li et al., in which transgenes inhibiting the expression of HCT promoted flavonoids accumulation [56]. The HF_T showed a predominantly higher lignin level relative to the HF_S. Correspondingly, PbrCHS, PbrCHI, PbrDFR, PbrLAR and PbrANR were considered as enzymes impeding the rate in the flavonoids/PAs biosynthetic pathway, and their expressions were all downregulated in the HF_T compared with the HF_S (Figure 4). Therefore, we speculate that the impediment of the flavonoid metabolic pathway impels intermediate metabolites to be converted to the lignin biosynthetic pathway.
5. Conclusions
Fruit hardening is a new disorder in ‘Suli’ pear, accompanied by the accumulation of lignin, especially in the top region. This study investigated the transcriptome profiles of the NFs and HFs of ‘Suli’ pear using Illumina RNA-seq technology. The transcriptome analysis identified thousands of DEGs between NFs and HFs. In addition, gene clustering, GO and KEGG pathway enrichment analyses were adopted to describe the transcriptional patterns of genes involved in lignin biosynthesis and accumulation. These data provide valuable information about the candidate genes participating in the lignin biosynthesis and accumulation in hardened ‘Suli’ pear.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/horticulturae7110467/s1. Table S1: Reported genes related to lignin biosynthesis in pear. Table S2: Primer sequences for qRT-PCR. Table S3: Descriptive statistics of sequencing data. Table S4: DEGs in the normal and the hardened fruit. Table S5: GO enrichment of DEGs in the normal and the hardened fruit. Table S6: KEGG enrichment of DEGs in the normal and the hardened fruit. Table S7: DEGs related to lignin biosynthesis. Table S8: DEGs related to flavonoid/proanthocyanidin biosynthesis. Table S9: Differentially expressed transcription factors associated with lignin metabolism in hardened ‘Suli’ pear.
Author Contributions
Conceptualisation, X.F. and X.L.; Data Curation, X.F. and Y.J.; Formal Analysis, X.F. and H.L.; Writing—Original Draft Preparation, X.F.; Writing—Review and Editing, L.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Scientific and Technological Innovation Programs of Higher Education Institutions in Shanxi (Grant No. 2021L101) and the Technology Innovation Fund of Shanxi Agricultural University (Grant No. 2017YJ45).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found below: https://www.ncbi.nlm.nih.gov/, accessed on 14 August 2021, PRJNA763913.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Liu, G.Q.; Li, W.S.; Zheng, P.H.; Xu, T.; Chen, L.J.; Liu, D.F.; Hussain, S.; Teng, Y.W. Transcriptomic analysis of ‘Suli’ pear (Pyrus pyrifolia white pear group) buds during the dormancy by RNA-Seq. BMC Genom. 2012, 13, 700. [Google Scholar] [CrossRef]
- Liu, X.F.; Li, S.R.; Feng, X.X.; Li, L.L. Study on cell wall composition, fruit quality and tissue structure of hardened ‘Suli’ pears (Pyrus bretschneideri Rehd). J. Plant Growth Regul. 2020, 40, 2007–2016. [Google Scholar] [CrossRef]
- Yamamoto, T.; Watanabe, S. Initial time of development of hard end disorder in ‘Bartlett’ pear. J. Jpn. Soc. Hort. Sci. 1982, 51, 42–151. [Google Scholar] [CrossRef][Green Version]
- Pierson, C.F.; Ceponis, M.J.; Mccolloch, L.P. Market Diseases of Apples, Pears, and Quinces; Agriculture Handbook; USDA: Washington, DC, USA, 1971; No. 376. [Google Scholar]
- Raese, J.T.; Drake, S.R. Calcium foliar sprays for control of alfalfa greening, cork spot, and hard end in ‘Anjou’ pears. J. Plant Nutr. 2006, 29, 543–552. [Google Scholar] [CrossRef]
- Fumio, T. Recent advances in research on Japanese pear rootstocks. J. Jpn. Soc. Hort. Sci. 2012, 81, 1–10. [Google Scholar]
- Raese, J.T. Fruit disorders, mineral composition and tree performance influenced by rootstocks of ‘Anjou’ pears. Acta Hortic. 1994, 367, 372–379. [Google Scholar] [CrossRef]
- Wang, Y.L.; Zhang, X.F.; Wang, Y.Z.; Yang, S.L.; Qu, H.Y. The changes of intracellular calcium concentration and distribution in the hard end pear (Pyrus pyrifolia cv. ‘Whangkeumbae’) fruit. Cell Calcium 2018, 71, 15–23. [Google Scholar] [CrossRef]
- Lu, G.L.; Li, Z.J.; Zhang, X.F.; Wang, R.; Yang, S.L. Expression analysis of lignin-associated genes in hard end pear (Pyrus pyrifolia Whangkeumbae) and its response to calcium chloride treatment conditions. J. Plant Growth Regul. 2015, 34, 251–262. [Google Scholar] [CrossRef]
- Chen, H.J.; Cao, S.F.; Fang, X.J.; Mu, H.L.; Yang, H.L.; Wang, X.; Xu, Q.Q.; Gao, H.Y. Changes in fruit firmness, cell wall composition and cell wall degrading enzymes in postharvest blueberries during storage. Sci. Hortic. 2015, 188, 44–48. [Google Scholar] [CrossRef]
- Singh, R.; Rastogi, S.; Dwivedi, U.N. Phenylpropanoid metabolism in ripening fruits. Compr. Rev. Food Sci. Food Saf. 2010, 9, 398–416. [Google Scholar] [CrossRef]
- Dixon, R.A.; Barros, J. Lignin biosynthesis: Old roads revisited and new roads explored. Open Biol. 2019, 9, 190215. [Google Scholar] [CrossRef]
- Yoon, J.; Choi, H.; An, G. Roles of lignin biosynthesis and regulatory genes in plant development. J. Integr. Plant Biol. 2015, 57, 902–912. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.P.; Li, G.Q.; Nie, J.Q.; Lin, Y.; Nie, F.; Zhang, J.Y.; Xu, Y.L. Study of the structure and biosynthetic pathway of lignin in stone cells of pear. Sci. Hortic. 2010, 125, 374–379. [Google Scholar] [CrossRef]
- Yan, C.C.; Yin, M.; Zhang, N.; Jin, Q.; Fang, Z.; Lin, Y.; Cai, Y.P. Stone cell distribution and lignin structure in various pear varieties. Sci. Hortic. 2014, 174, 142–150. [Google Scholar] [CrossRef]
- Su, X.Q.; Zhao, Y.; Wang, H.; Li, G.H.; Cheng, X.; Jin, Q.; Cai, Y.P. Transcriptomic analysis of early fruit development in Chinese white pear (Pyrus bretschneideri Rehd.) and functional identification of PbCCR1 in lignin biosynthesis. BMC Plant Biol. 2019, 19, 417. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Cheng, X.; Jin, Q.; Su, X.Q.; Li, M.L.; Yan, C.C.; Jiao, X.Y.; Li, D.H.; Lin, Y.; Cai, Y.P. Comparison of the transcriptomic analysis between two Chinese white pear (Pyrus bretschneideri Rehd.) genotypes of different stone cells contents. PLoS ONE 2017, 12, e0187114. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.P.; Liu, T.T.; Hu, C.H.; Song, Y.Q.; Hao, R.J.; Feng, X.X.; Cui, T.T.; Han, Y.Z.; Li, L.L. Comparative analysis of transcriptomic profiling to identify genes involved in the bulged surface of pear fruit (Pyrus bretschneideri Rehd. cv. Yuluxiangli). Physiol. Mol. Biol. Plants 2021, 27, 69–80. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, X.Y.; Liu, L.; Yao, D.Z.; Li, J.C.; Fang, J.C.; Chen, X.N.; Zhu, L.W.; Liu, P.; Ye, Z.F.; et al. Transcriptome and metabolomic analysis to reveal the browning spot formation of ‘Huangguan’ pear. BMC Plant Biol. 2021, 21, 321. [Google Scholar]
- Mamat, A.; Tusong, K.; Xu, J.; Yan, P.; Mei, C.; Wang, J.X. Integrated transcriptomic and proteomic analysis reveals the complex molecular mechanisms underlying stone cell formation in Korla pear. Sci. Rep. 2021, 11, 7688. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, Z.W.; Shi, Z.B.; Zhang, S.; Ming, R.; Zhu, S.L.; Khan, M.A.; Tao, S.T.; Korban, S.S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri Rehd). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Boudet, A.M.; Kajita, S.; Grima-Pettenati, J.; Goffner, D. Lignins and lignocellulosics: A better control of synthesis for new and improved uses. Trends Plant Sci. 2003, 8, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.K.; Li, X.; Zhang, J.; Ge, H.; Yin, X.R.; Chen, K.S. Regulation of loquat fruit low temperature response and lignification involves interaction of heat shock factors and genes associated with lignin biosynthesis. Plant Cell Environ. 2016, 39, 1780–1789. [Google Scholar] [CrossRef] [PubMed]
- Ketsa, S.; Koolpluksee, M. Some physical and biochemical characteristics of damaged pericarp of mangosteen fruit after impact. Postharvest Biol. Technol. 1993, 2, 209–215. [Google Scholar] [CrossRef]
- Bunsiri, A.; Ketsa, S.; Paull, R.E. Phenolic metabolism and lignin synthesis in damaged pericarp of mangosteen fruit after impact. Postharvest Biol. Technol. 2003, 29, 61–71. [Google Scholar] [CrossRef]
- Deng, L.L.; Cui, W.J.; Yao, S.X.; Zeng, K.F. Effect of mechanical damage on organizational structure of postharvest navel orange rind. Food Sci. 2019, 40, 118–126. [Google Scholar]
- Zhou, X.Y.; Yue, J.Q.; Yang, H.B.; Zhu, C.H.; Zhu, F.; Li, J.X.; Xu, R.W.; Gao, J.Y.; Zhou, D.G.; Deng, X.X.; et al. Integration of metabolome, histochemistry and transcriptome analysis provides insights into lignin accumulation in oleocellosis-damaged flavedo of citrus fruit. Postharvest Biol. Technol. 2021, 172, 111362. [Google Scholar] [CrossRef]
- Zheng, J.; Liu, L.B.; Tao, H.H.; An, Y.Y.; Wang, L.J. Transcriptomic profiling of apple calli with a focus on the key genes for ALA-induced anthocyanin accumulation. Front. Plant Sci. 2021, 12, 640606. [Google Scholar] [CrossRef] [PubMed]
- Syros, T.; Yupsanis, T.; Zafiriadis, H.; Economou, A. Activity and isoforms of peroxidases, lignin and anatomy, during adventitious rooting in cuttings of Ebenus cretica L. J. Plant Physiol. 2004, 161, 69–77. [Google Scholar] [CrossRef]
- Jaakola, L.; Pirttila, A.M.; Halonen, M.; Hohtola, A. Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol. Biotechnol. 2001, 19, 201–203. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Goujon, T.; Sibout, R.; Eudes, A.; Mackay, J.; Jouanin, L. Genes involved in the biosynthesis of lignin precursors in Arabidopsis thaliana. Plant Physiol. Biochem. 2003, 41, 677–687. [Google Scholar] [CrossRef]
- Lu, J.Y.; Shi, Y.Z.; Li, W.J.; Chen, S.; Wang, Y.F.; He, X.L.; Yin, X.G. RcPAL, a key gene in lignin biosynthesis in Ricinus communis L. BMC Plant Biol. 2019, 19, 181. [Google Scholar]
- Li, X.; Bonawitz, N.D.; Weng, J.K.; Chapple, C. The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 2010, 22, 1620–1632. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; Cesarino, I.; Rataj, K.; Xiao, Y.G.; Sundin, L.; Goeminne, G.; Kim, H.; Cross, J.; Morreel, K.; Araujo, P.; et al. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 2013, 341, 1103–1106. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Li, M.L.; Li, D.H.; Zhang, J.Y.; Jin, Q.; Sheng, L.L.; Cai, Y.P.; Lin, Y. Characterization and analysis of CCR and CAD gene families at the whole-genome level for lignin synthesis of stone cells in pear (Pyrus bretschneideri) fruit. Biol. Open 2017, 6, 1602–1613. [Google Scholar] [CrossRef]
- Rahantamalala, A.; Rech, P.; Martinez, Y.; Chaubet-Gigot, N.; Grima-Pettenati, J.; Pacquit, V. Coordinated transcriptional regulation of two key genes in the lignin branch pathway—CAD and CCR—is mediated through MYB-binding sites. BMC Plant Biol. 2010, 10, 130. [Google Scholar] [CrossRef] [PubMed]
- Sirisha, V.L.; Prashant, S.; Kumar, D.R.; Pramod, S.; Jalaja, N.; Kumari, P.H.; Rao, P.M.; Rao, S.N.; Mishra, P.; Karumanchi, S.R.; et al. Cloning, characterization and impact of up- and down-regulating subabul cinnamyl alcohol dehydrogenase (CAD) gene on plant growth and lignin profiles in transgenic tobacco. Plant Growth Regul. 2012, 66, 239–253. [Google Scholar] [CrossRef]
- Weng, J.K.; Mo, H.; Chapple, C. Over-expression of F5H in COMT-deficient Arabidopsis leads to enrichment of an unusual lignin and disruption of pollen wall formation. Plant J. 2010, 64, 898–911. [Google Scholar] [CrossRef] [PubMed]
- Berthet, S.; Demont-Caulet, N.; Pollet, B.; Bidzinski, P.; Cézard, L.; Le Bris, P.; Borrega, N.; Hervé, J.; Blondet, E.; Balzergue, S.; et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 2011, 23, 1124–1137. [Google Scholar] [CrossRef]
- Cosio, C.; Ranocha, P.; Francoz, E.; Burlat, V.; Zheng, Y.; Perry, S.E.; Ripoll, J.J.; Yanofsky, M.; Dunand, C. The class III peroxidase PRX17 is a direct target of the MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) and participates in lignified tissue formation. New Phytol. 2017, 213, 250–263. [Google Scholar] [CrossRef]
- Cao, Y.P.; Han, Y.H.; Meng, D.D.; Li, D.H.; Jin, Q.; Lin, Y.; Cai, Y.P. Structural, Evolutionary, and Functional Analysis of the Class III Peroxidase Gene Family in Chinese Pear (Pyrus bretschneideri). Front. Plant Sci. 2016, 7, 1874. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Dixon, R.A. Transcriptional networks for lignin biosynthesis: More complex than we thought? Trends Plant Sci. 2011, 16, 227–233. [Google Scholar] [CrossRef]
- Zhou, J.L.; Lee, C.H.; Zhong, R.Q.; Ye, Z.H. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 2009, 21, 248–266. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, R.L.; Zhong, R.Q.; Ye, Z.H. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 2009, 50, 1950–1964. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.Q.; Richardson, E.A.; Ye, Z.H. The MYB46 Transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2776–2792. [Google Scholar] [CrossRef] [PubMed]
- Bomal, C.; Bedon, F.; Caron, S.; Mansfield, S.D.; Levasseur, C.; Cooke, J.E.; Blais, S.; Tremblay, L.; Morency, M.J.; Pavy, N.; et al. Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wall biogenesis: A comparative in planta analysis. J. Exp. Bot. 2008, 59, 3925–3939. [Google Scholar] [CrossRef] [PubMed]
- Patzlaff, A.; Newman, L.J.; Dubos, C.; Whetten, R.W.; Smith, C.; McInnis, S.; Bevan, M.W.; Sederoff, R.R.; Campbell, M.M. Characterisation of PtMYB1, an R2R3-MYB from pine xylem. Plant Mol. Biol. 2003, 53, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Goicoechea, M.; Lacombe, E.; Legay, S.; Mihaljevic, S.; Grima-Pettenati, J. EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J. 2010, 43, 553–567. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, R.L.; Zhong, R.; Fowler, S.; Lyskowski, D.; Piyasena, H.; Carleton, K.; Spicer, C.; Ye, Z.H. The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol. 2010, 51, 1084–1090. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Lee, C.; Zhou, J.; McCarthy, R.L.; Ye, Z.H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef]
- Li, M.T.; Cheng, C.X.; Zhang, X.F.; Zhou, S.P.; Wang, C.H.; Ma, C.H.; Yang, S.L. PpNAC187 enhances lignin synthesis in ‘Whangkeumbae’ pear (Pyrus pyrifolia) ‘hard-end’ fruit. Molecules 2019, 24, 4338. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Li, S.J.; Liu, X.F.; Yin, X.R.; Grierson, D.; Chen, K.S. Ternary complex EjbHLH1-EjMYB2-EjAP2-1 retards low temperature-induced flesh lignification in loquat fruit. Plant Physiol. Biochem. 2019, 139, 731–737. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.K.; Li, X.; Xu, Q.; Chen, J.Y.; Yin, X.R.; Ferguson, I.B.; Chen, K.S. EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol. J. 2015, 13, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Besseau, S.; Hoffmann, L.; Geoffroy, P.; Lapierre, C.; Legrand, M. Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 2007, 19, 148–162. [Google Scholar] [CrossRef]
- Hoffmann, L.; Besseau, S.; Geoffroy, P.; Ritzenthaler, C.; Meyer, D.; Lapierre, C.; Pollet, B.; Legrand, M. Silencing of hydroxycinnamoyl-coenzyme a shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 2004, 16, 1446–1465. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).