Stable Reference Gene Selection for qRT-PCR Normalization in Strawberry (Fragaria × ananassa) Leaves under Different Stress and Light-Quality Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions, Stress Treatments, and Material Collection
2.2. RNA Extraction and cDNA Synthesis
2.3. Candidate Reference Selection and Primer Design
2.4. qRT–PCR Analysis
2.5. Data Statistical Analysis
2.6. RNA-Seq Analysis
3. Results
3.1. Specific Analysis of Candidate Reference Genes by qRT-PCR
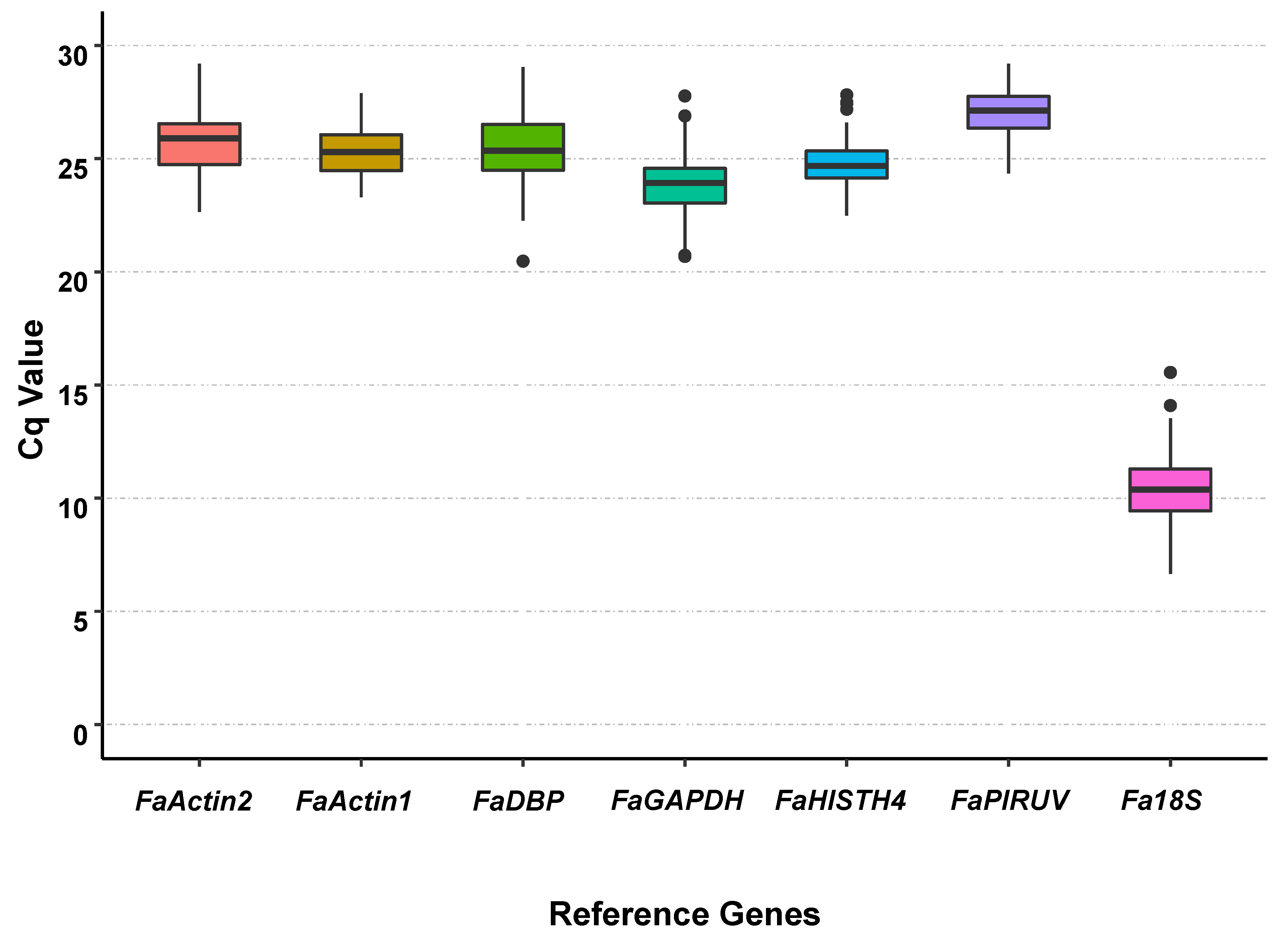
3.2. Cq Value Analysis of Candidate Reference Genes under All Treatments
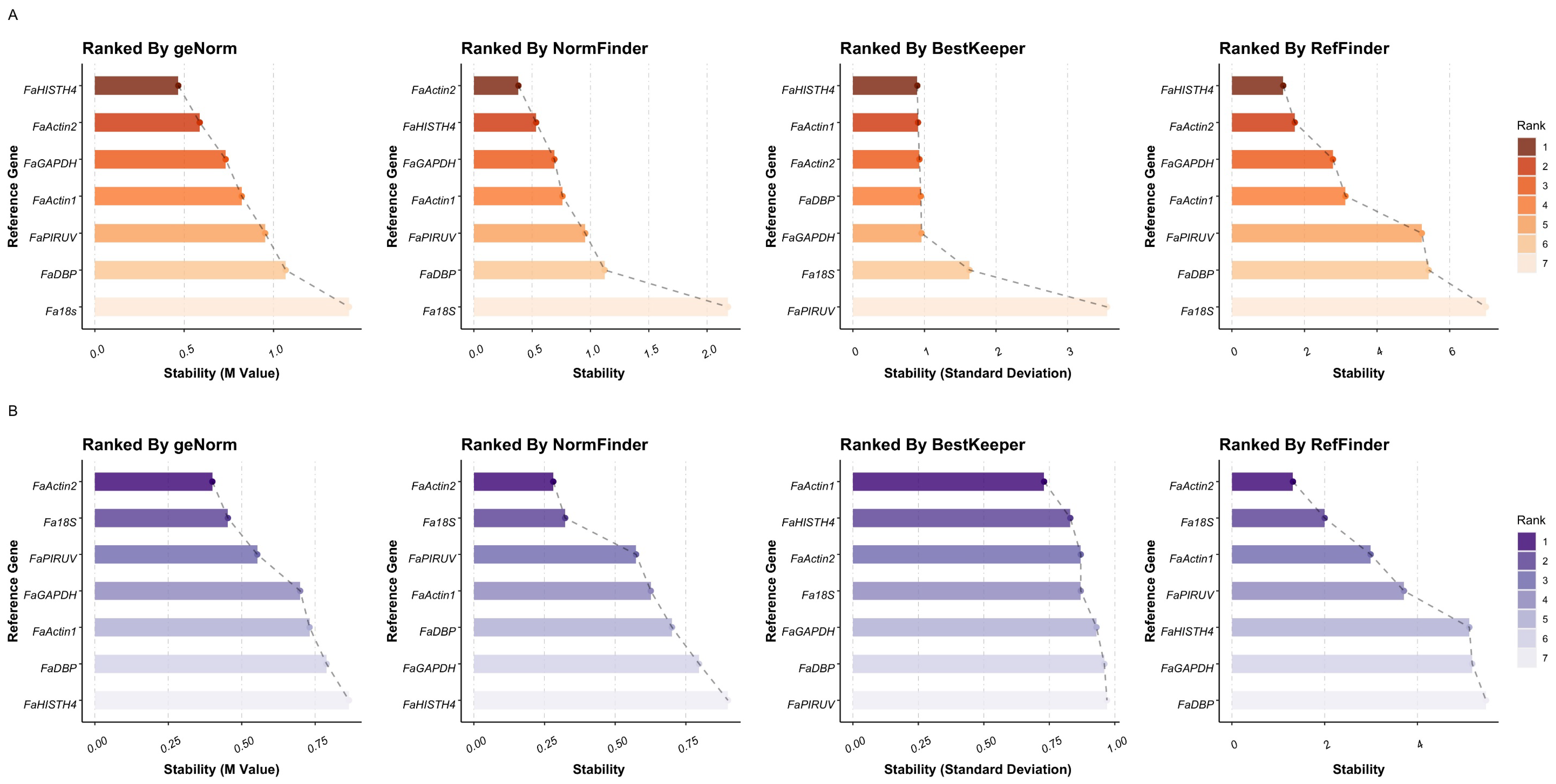
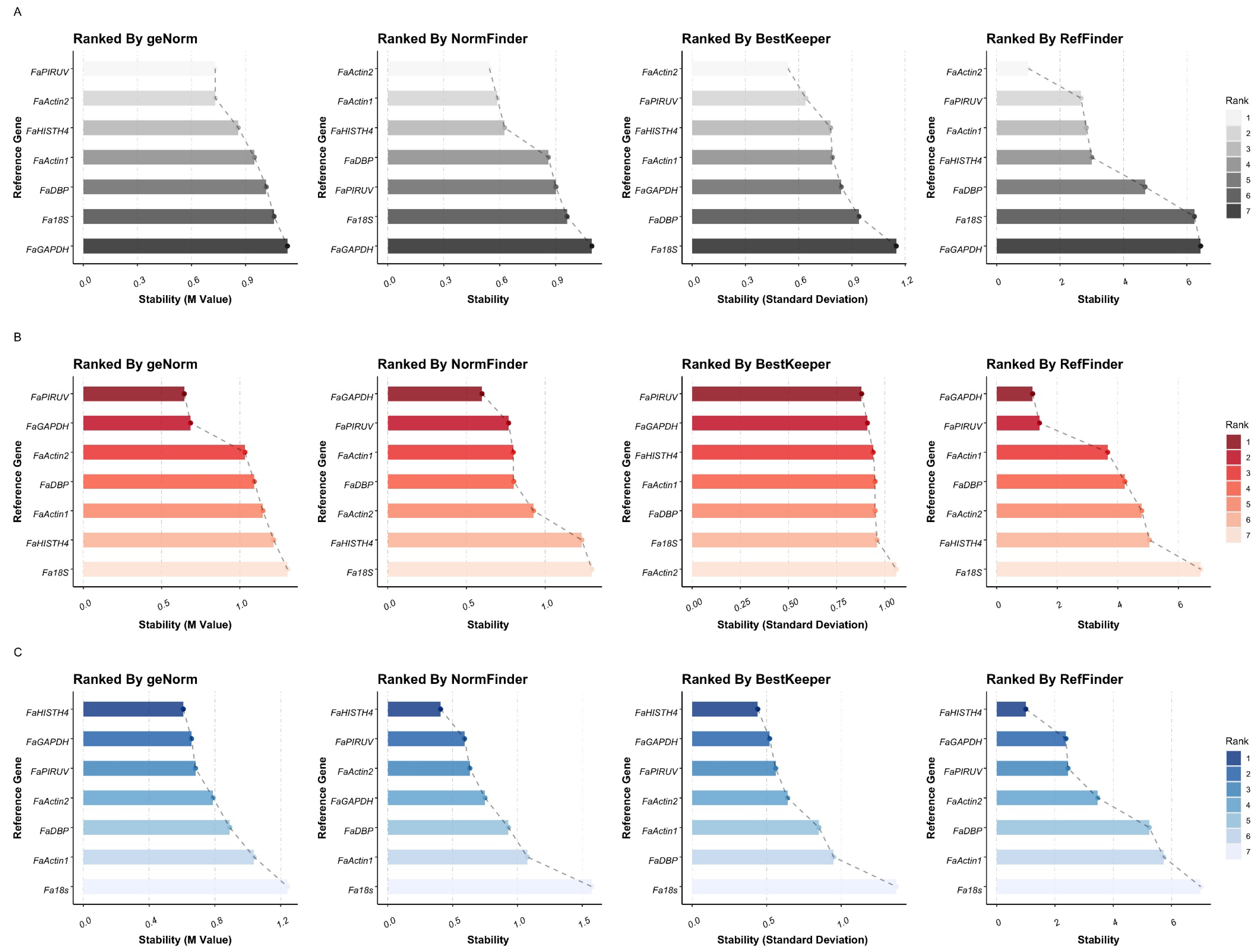
3.3. Reference Gene-Expression Stability Analysis under Treatments
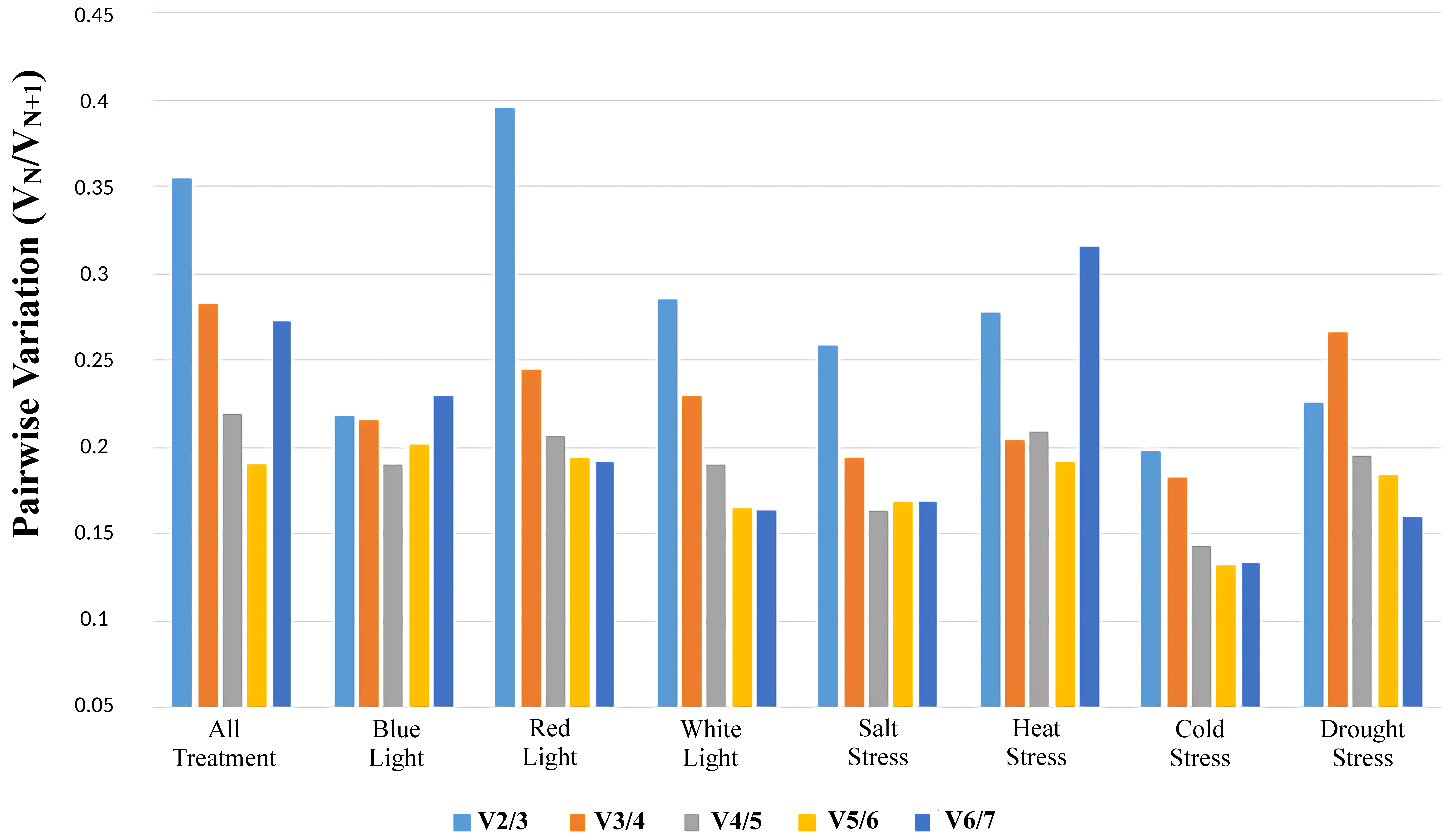
3.4. Identification of the Optimal Number of Reference Genes under Different Treatments
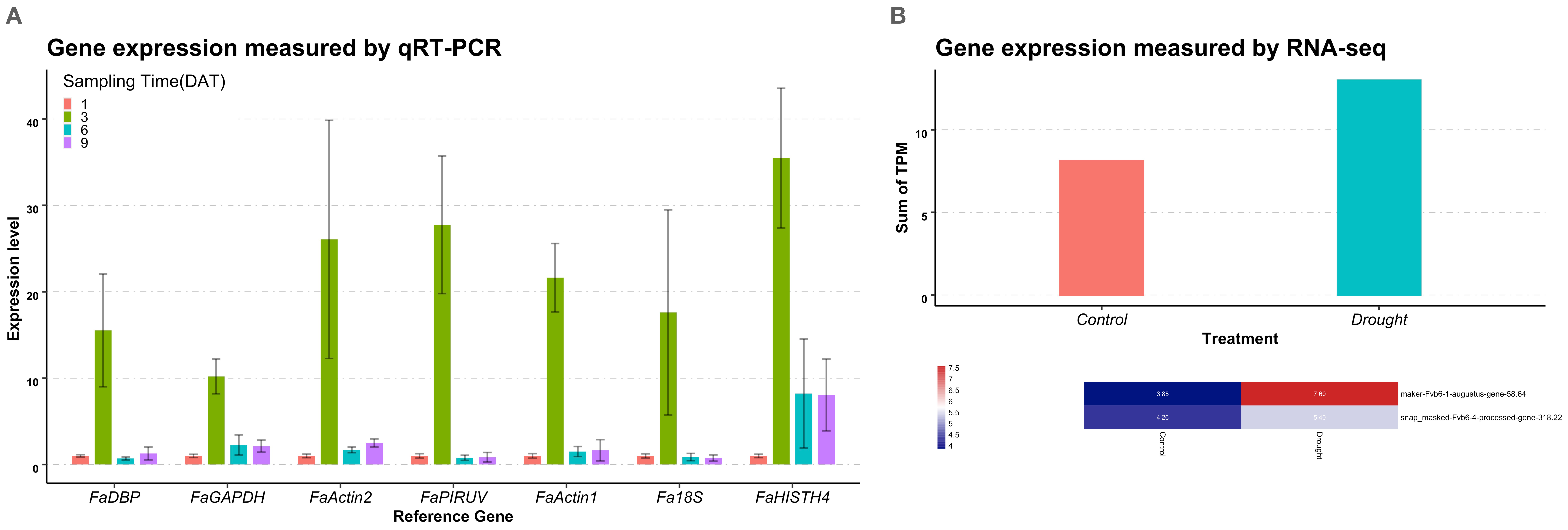
3.5. Validation of the Selected Reference Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Ali, A.; Yun, D.-J. Salt stress tolerance; what do we learn from halophytes? J. Plant Biol. 2017, 60, 431–439. [Google Scholar] [CrossRef]
- Wilkinson, S.; Davies, W.J. Drought, ozone, ABA and ethylene: New insights from cell to plant to community. Plant. Cell Environ. 2010, 33, 510–525. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Zhang, Q.; Jia, M.; Xing, Y.; Qin, L.; Li, B.; Jia, W. Genome-wide identification and expression analysis of MRLK family genes associated with strawberry (Fragaria vesca) fruit ripening and abiotic stress responses. PLoS ONE 2016, 11, e0163647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, X.; Khaliq, A.; Lu, S.; Xie, M.; Ma, Z.; Mao, J.; Chen, B. Genome-wide identification and characterization of the BES1 gene family in apple (Malus domestica). Plant Biol. 2020, 22, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Pocock, T. Light-emitting diodes and the modulation of specialty crops: Light sensing and signaling networks in plants. HortScience 2015, 50, 1281–1284. [Google Scholar] [CrossRef] [Green Version]
- Cao, K.; Yu, J.; Xu, D.; Ai, K.; Bao, E.; Zou, Z. Exposure to lower red to far-red light ratios improve tomato tolerance to salt stress. BMC Plant Biol. 2018, 18, 92. [Google Scholar] [CrossRef]
- Gachon, C.; Mingam, A.; Charrier, B. Real-time PCR: What relevance to plant studies? J. Exp. Bot. 2004, 55, 1445–1454. [Google Scholar] [CrossRef] [Green Version]
- Udvardi, M.K.; Czechowski, T.; Scheible, W.-R. Eleven golden rules of quantitative RT-PCR. Plant Cell 2008, 20, 1736–1737. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Peng, X.; Liu, Y.; Li, Y.; Luo, Y.; Wang, X.; Tang, H. Evaluation of suitable reference genes for qRT-PCR normalization in strawberry (Fragaria× ananassa) under different experimental conditions. BMC Mol. Biol. 2018, 19, 8. [Google Scholar] [CrossRef]
- Liu, Q.; Qi, X.; Yan, H.; Huang, L.; Nie, G.; Zhang, X. Reference gene selection for quantitative real-time reverse-transcriptase PCR in annual ryegrass (Lolium multiflorum) subjected to various abiotic stresses. Molecules 2018, 23, 172. [Google Scholar] [CrossRef]
- Jian, B.; Liu, B.; Bi, Y.; Hou, W.; Wu, C.; Han, T. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol. Biol. 2008, 9, 59. [Google Scholar] [CrossRef] [Green Version]
- Xiao, X.; Ma, J.; Wang, J.; Wu, X.; Li, P.; Yao, Y. Validation of suitable reference genes for gene expression analysis in the halophyte Salicornia europaea by real-time quantitative PCR. Front. Plant Sci. 2015, 5, 788. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.M.; Roche, J.R.; Donaghy, D.J.; Thrush, A.; Sathish, P. Validation of reference genes for quantitative RT-PCR studies of gene expression in perennial ryegrass (Lolium perenne L.). BMC Mol. Biol. 2010, 11, 8. [Google Scholar] [CrossRef] [Green Version]
- Zheng, T.; Chen, Z.; Ju, Y.; Zhang, H.; Cai, M.; Pan, H.; Zhang, Q. Reference gene selection for qRT-PCR analysis of flower development in Lagerstroemia indica and L. speciose. PLoS ONE 2018, 13, e0195004. [Google Scholar]
- Wu, Z.-J.; Tian, C.; Jiang, Q.; Li, X.-H.; Zhuang, J. Selection of suitable reference genes for qRT-PCR normalization during leaf development and hormonal stimuli in tea plant (Camellia sinensis). Sci. Rep. 2016, 6, 19748. [Google Scholar] [CrossRef] [Green Version]
- Amil-Ruiz, F.; Garrido-Gala, J.; Blanco-Portales, R.; Folta, K.M.; Muñoz-Blanco, J.; Caballero, J.L. Identification and Validation of Reference Genes for Transcript Normalization in Strawberry (Fragaria × ananassa) Defense Responses. PLoS ONE 2013, 8, e70603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crystian, D.; Terto, J.; Silva, J.V.; Almeida, C. Validation of reference genes for accurate normalization by quantitative polymerase chain reaction in sugarcane drought stress studies using two cultivars. Ciência Rural. 2018, 48, 1–7. [Google Scholar] [CrossRef]
- Hancock, J.F. Strawberries; CABI: Oxon, UK, 2020. [Google Scholar]
- Zhang, J.; Wang, X.; Yu, O.; Tang, J.; Gu, X.; Wan, X.; Fang, C. Metabolic profiling of strawberry (Fragaria× ananassa Duch.) during fruit development and maturation. J. Exp. Bot. 2011, 62, 1103–1118. [Google Scholar] [CrossRef] [PubMed]
- Keutgen, A.J.; Pawelzik, E. Contribution of amino acids to strawberry fruit quality and their relevance as stress indicators under NaCl salinity. Food Chem. 2008, 111, 642–647. [Google Scholar] [CrossRef]
- Ferreira, J.F.S.; Liu, X.; Suarez, D.L. Fruit yield and survival of five commercial strawberry cultivars under field cultivation and salinity stress. Sci. Hortic. 2019, 243, 401–410. [Google Scholar] [CrossRef]
- Yu, D.; Tang, H.; Zhang, Y.; Du, Z.; Yu, H.; Chen, Q. Comparison and improvement of different methods of RNA isolation from strawberry (Fragria× ananassa). J. Agric. Sci. 2012, 4, 51. [Google Scholar] [CrossRef]
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Galli, V.; Messias, R.S.; Guzman, F.; Perin, E.C.; Margis, R.; Rombaldi, C.V. Transcriptome analysis of strawberry (Fragaria× ananassa) fruits under osmotic stresses and identification of genes related to ascorbic acid pathway. Physiol. Plant. 2019, 166, 979–995. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.L.; Wai, C.M.; et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Villalba, A.; Ruijter, J.M.; van den Hoff, M.J.B. Use and misuse of Cq in qPCR data analysis and reporting. Life 2021, 11, 496. [Google Scholar] [CrossRef]
- de Spiegelaere, W.; Dern-Wieloch, J.; Weigel, R.; Schumacher, V.; Schorle, H.; Nettersheim, D.; Bergmann, M.; Brehm, R.; Kliesch, S.; Vandekerckhove, L.; et al. Reference Gene Validation for RT-qPCR, a Note on Different Available Software Packages. PLoS ONE 2015, 10, e0122515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Xie, F.; He, Q.; Li, J.; Liu, J.; Sun, B.; Luo, Y.; Zhang, Y.; Chen, Q.; Zhang, F.; et al. Expression Analysis of XTH in Stem Swelling of Stem Mustard and Selection of Reference Genes, (n.d.). Genes 2020, 11, 113. [Google Scholar] [CrossRef] [Green Version]
- Hamalainen, H.K.; Tubman, J.C.; Vikman, S.; Kyrölä, T.; Ylikoski, E.; Warrington, J.A.; Lahesmaa, R. Identification and Vali-dation of Endogenous Reference Genes for Expression Profiling of T Helper Cell Differentiation by Quantitative Real-Time RT-PCR. Anal. Biochem. 2001, 299, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Huang, X.; Lin, Y.; Wang, X.; Yan, Z.; Wang, Q.; Ding, J.; Gu, T.; Li, Y. Identification of reference genes for transcript normalization in various tissue types and seedlings subjected to different abiotic stresses of woodland strawberry Fragaria vesca. Sci. Hortic. 2020, 261, 108840. [Google Scholar] [CrossRef]
- Galli, V.; Borowski, J.M.; Perin, E.C.; Messias, R.d.; Labonde, J.; Pereira, I.d.S.; Silva, S.D.d.A.; Rombaldi, C.V. Validation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in strawberry fruits using different cultivars and osmotic stresses. Gene 2015, 554, 205–214. [Google Scholar] [CrossRef]
- Benítez-Burraco, A.; Blanco-Portales, R.; Redondo-Nevado, J.; Bellido, M.L.; Moyano, E.; Caballero, J.; Muñoz-Blanco, J. Clon-ing and characterization of two ripening-related strawberry (Fragaria × ananassa cv. Chandler) pectate lyase genes. J. Exp. Bot. 2003, 54, 633–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Hu, L.; Wang, X.; Liu, H.; Tian, H.; Wang, J. Selection of reliable reference genes for gene expression analysis in seeds at different developmental stages and across various tissues in Paeonia ostia. Mol. Biol. Rep. 2019, 46, 6003–6011. [Google Scholar] [CrossRef]
- Hu, Y.; Han, Y.; Wei, W.; Li, Y.; Zhang, K.; Gao, Y.; Zhao, F.; Feng, J. Identification, isolation, and expression analysis of heat shock transcription factors in the diploid woodland strawberry Fragaria vesca. Front. Plant Sci. 2015, 6, 736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Gu, C.; Xuan, L.; Hua, J.; Shi, Q.; Fan, W.; Yin, Y.; Yu, F. Identification of suitable reference genes in Taxodium ‘Zhongshanshan’ under abiotic stresses. Trees 2017, 31, 1519–1530. [Google Scholar] [CrossRef]
- Shu, B.; Yu, H.; Dai, J.; Xie, Z.; Qian, W.; Lin, J. Stability evaluation of reference genes for real-time quantitative PCR normalization in Spodoptera frugiperda (Lepidoptera: Noctuidae). J. Integr. Agric. 2021, 20, 2471–2482. [Google Scholar] [CrossRef]
- Jia, Y.; Liu, S.-C.; Zhao, J.; Chen, X.-X.; Sun, L.-X.; Yu, X.-F.; Li, X. Reference gene selection and validation by qRT-PCR during flower development and in different organs of Primula forbesii. J. Hortic. Sci. Biotechnol. 2019, 95, 383–394. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, Z.; Feng, M.; Chen, J.; Qin, M.; Wang, W.; Bao, Y.; Xu, Q.; Ye, Y.; Ma, C. The circadian-controlled PIF8-BBX28 module regulates petal senescence in rose flowers by governing mitochondrial ROS homeostasis at night. Plant Cell. 2021, 33, 2716–2735. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Y.; Lu, Y.; Wang, G.; Liu, Y.; Zhang, Y.; Tang, H. Stable Reference Gene Selection for qRT-PCR Normalization in Strawberry (Fragaria × ananassa) Leaves under Different Stress and Light-Quality Conditions. Horticulturae 2021, 7, 452. https://doi.org/10.3390/horticulturae7110452
Ye Y, Lu Y, Wang G, Liu Y, Zhang Y, Tang H. Stable Reference Gene Selection for qRT-PCR Normalization in Strawberry (Fragaria × ananassa) Leaves under Different Stress and Light-Quality Conditions. Horticulturae. 2021; 7(11):452. https://doi.org/10.3390/horticulturae7110452
Chicago/Turabian StyleYe, Yuntian, Yang Lu, Guangyi Wang, Yongqiang Liu, Yunting Zhang, and Haoru Tang. 2021. "Stable Reference Gene Selection for qRT-PCR Normalization in Strawberry (Fragaria × ananassa) Leaves under Different Stress and Light-Quality Conditions" Horticulturae 7, no. 11: 452. https://doi.org/10.3390/horticulturae7110452
APA StyleYe, Y., Lu, Y., Wang, G., Liu, Y., Zhang, Y., & Tang, H. (2021). Stable Reference Gene Selection for qRT-PCR Normalization in Strawberry (Fragaria × ananassa) Leaves under Different Stress and Light-Quality Conditions. Horticulturae, 7(11), 452. https://doi.org/10.3390/horticulturae7110452





