An Update on the Function, Biosynthesis and Regulation of Floral Volatile Terpenoids
Abstract
:1. Introduction
2. Biological Functions of FVTs
| Latin Name | Family | Main FVT compounds | Genes | Ref. |
|---|---|---|---|---|
| Actinidia deliciosa ‘Hayward’ | Actinidiaceae | (E,E)-α-farnesene, (E)-β-ocimene, (+)-germacrene D | [16] | |
| Albizia julibrissin | Leguminosae | α-farnesene, (Z, E)-β-farnesene | AjTPS2, AjTPS5, AjTPS7, AjTPS9, AjTPS10 | [17] |
| Camellia spp. | Theaceae | linalool and its oxides, geraniol, α-farnesene, hedycaryol | CbTPS1, ChTPS1, CbTPS18, CbTPS25, CbTPS28, CbTPS33, CbTPS35 CsTPS29, CbTPS47, CbTPS48, CbTPS51, CbTPS52 | [18,19,20] |
| Cananga odorata var. fruticosa | Annonaceae | linalool | CoTPS1, CoTPS2, CoTPS3, CoTPS4 | [21] |
| Chimonanthus praecox L. | Calycanthaceae | linalool, trans-β-ocimene, β-caryophyllene | CpTPS1, CpTPS9, CpTPS10, CpTPS14, CpTPS16, CpTPS4, CpTPS9, CpTPS42 | [22,23,24,25] |
| Datura wrightii | Solanaceae | linalool and its enantiomers | [26] | |
| Eurya japonica Thunb | Theaceae | α-pinene, linalool | [27] | |
| Gardenia jasminoides | Rubiaceae | farnesene, Z-3-hexenyl tiglate, indole | [28] | |
| Gelsemium sempervirens (L.) J. St.-Hil. | Gelsemiaceae | (Z)-α-ocimene, α-farnesene | [29] | |
| Gossypium hirsutum | Malvaceae | (3S)-linalool | GhTPS12 | [30,31] |
| Jasminum spp. | Oleaceae | 𝛼-farnesene, linalool, β-ocimene, germacrene-D | [32,33,34,35,36] | |
| Laurus nobilis | Lauraceae | sesquiterpenes, γ-cadinene, δ-cadinene | [37] | |
| Lonicera japonica | Caprifoliaceae | linalool | [38] | |
| Magnolia champaca | Magnoliaceae | (R)-linalool, linalool and its oxides | [39] | |
| Malus domestica | Rosaceae | (E)-linalool oxide | [40] | |
| Murraya paniculata | Rutaceae | E-β-ocimene, linalool, α-cubebene | [41,42] | |
| Myrtus communis L. | Myrtaceae | α-pinene, linalool, 1,8-cineole | [43] | |
| Osmanthus fragrans | Oleaceae | linalool and its derivatives, α-ionone, β-ionone | OfTPS1, OfTPS2, OfTPS3 | [44,45,46,47] |
| Paeonia spp. | Paeoniaceae | β-caryophyllene, linalool | [48,49] | |
| Psidium guajava | Myrtaceae | α-cadinol, β-caryophyllene, nerolidol | [50] | |
| Rosa spp. | Rosaceae | geraniol, linalool, nerolidol, myrcene, ocimene, citronellol | NEROLIDOL SYNTHASE (NES), RcLIN-NERS1, RcLIN-NERS2 | [51,52,53,54,55,56] |
| Styrax japonicas spp. | Styracaceae | linalool, α-pincnc, gcrmacrcnc D | [57] | |
| Syringa oblata Lindl. | Oleaceae | D-limonene | [58,59] | |
| Penstemon digitalis | Plantaginaceae | linalool and its enantiomers, cis-and trans-β-ocimene | [60,61,62] | |
| Alstroemeria spp. | Alstroemeriaceae | (E)-caryophyllene, α-caryophyllene | [63] | |
| Anthurium ‘Mystral’ | Araceae | eucalyptol, β/α-pinene, β-phellandrene, β-Myrcene | [64] | |
| Antirhinum majus | Plantaginaceae | nerolidol, linalool, (E)-β-ocimene, myrcene | [65,66] | |
| Arabidopsis thaliana | Brassicaceae | α-copaene, α-caryophyllene, β-elemene | AtTPS21, AtTPS11, and other 40 terpenoid synthase genes | [11,67,68,69,70] |
| Aristolochia gigantea | Aristolochiaceae | linalool, (Z,E)-α-farnesene, geraniol | [71] | |
| Caladenia plicata | Orchidaceae | β-citronellol | [72] | |
| Cannabis sativa | Cannabaceae | (+)-α-pinene, (−)-limonene, β-caryophyllene | [73] | |
| Chrysanthemum indicum | Asteraceae | 1,8-cineole, germacrene D, camphor | [74,75] | |
| Citrus L. | Rutaceae | linalool, β-myrcene, α-myrcene, limonene | [76] | |
| Clarkia breweri | Onagraceae | S-linalool, Linalool, linalool oxide | linalool synthase (LIS) gene | [77,78] |
| Clematis florida cv. ‘Kaiser’ | Ranunculaceae | linalool, linalool oxide, nerolidol | CfTPS1, CfTPS2, CfTPS3 | [79] |
| Cymbidium spp. | Orchidaceae | (E)-β-farrnesene, nerolidol, linalool | CgTPS7 | [80,81] |
| Dendrobium officinale | Orchidaceae | α-thujene, linalool, α-terpineol | DoTPS10 | [82,83,84] |
| Dianthus caryophyllus L. | Caryophyllaceae | caryophyllene, caryophyllene oxide, linalool | [85,86,87] | |
| Freesia hybrida. “Shiny Gold” | Iridaceae | linalool, β-ocimene, D-limonene | FhTPS1, FhTPS2, FhTPS3, FhTPS4, FhTPS5, FhTPS6, FhTPS7, FhTPS8 | [88,89,90] |
| Gymnadenia conopsea (L.) R. Br. | Orchidaceae | β-myrcene, α-terpineol, (+)-cyclosativene, α-santalene, trans-α-bergamotene, (Z,E)-α-farnesene, (E,E)-α-farnesene | [91] | |
| Hedychium coronarium | Zingiberaceae | β-ocimene, 1,8-cineole, linalool | HcTPS1, HcTPS3, HcTPS5, HcTPS6, HcTPS7, HcTPS8, HcTPS10, HcTPS11, HcTPS21 | [92,93,94,95,96] |
| Hippeastrum spp. | Amaryllidaceae | eucalyptol, (Z)-β-ocimene | [97] | |
| Lathyrus odoratus | Leguminosae | α-bergamotene, linalool, (−)-α-cubebene | [98] | |
| Lavandula spp. | Lamiaceae | linalool acetate, linalool, lavandulyl acetate, α/β-Pinene | LaLIMS, LaLINS | [98,99,100,101,102] |
| Lilium spp. | Liliaceae | linalool, myrcene, (E)-β-ocimene, α-pinene, limonene | LoTPS1, LoTPS2, LoTPS3, LoTPS4 | [103,104,105] |
| Maxillaria tenuifolia | Orchidaceae | β-caryophyllene, α-copaene, delta-decalacton | [106] | |
| Mentha citrata | Lamiaceae | linalool and its enantiomers | [107] | |
| Mimulus spp. | Phrymaceae | (E)-β-ocimene, d-limonene, β-myrcene | OCIMENE SYNTHASE (OS) gene | [108,109,110] |
| Narcissus spp. | Amaryllidaceae | myrcene, eucalyptol, linalool | [111,112] | |
| Nicotiana spp. | Solanaceae | (E)-α-bergamotene, (E)-β-ocimene, 1,8-cineole | NaTPS25, NaTPS38 | [113,114,115,116,117] |
| Nymphaea subg. Hydrocallis | Nymphaeaceae | linalool, farnesene, nerolidol | [118] | |
| Ocimum basilicum L. | Lamiaceae | linalool | [119] | |
| Petunia hybrida | Solanaceae | germacrene D, β-cadinene | PhTPS1, PhTPS2, PhTPS3, PhTPS4 | [120] |
| Passiflora edulis Sims | Passifloraceae | linalool | PeTPS2, PeTPS3, PeTPS4, PeTPS24 | [14] |
| Phalaenopsis spp. | Orchidaceae | α-pinene, trans-β-ocimene, linalool, geraniol and their derivatives | PbTPS5, PbTPS7, PbTPS9, PbTPS10, PbTPS3, PbTPS4 | [121,122,123] |
| Plectranthus amboinicus (Lour.) Spreng | Lamiaceae | linalool, nerolidol | [124] | |
| Polianthes tuberosa L. | Amaryllidaceae | germacrene D, 1, 8- cineole, α-terpineol | [125,126,127,128] | |
| Rheum nobile | Polygonaceae | α-pinene | [129] | |
| Salvia officinalis | Labiatae | myrcene, (+)-neomenthol, 1,8-cineole | [130] | |
| Tanacetum vulgare | Asteraceae | α-pinene, 3-hexen-1-ol-acetate | [131] |
2.1. Attraction of Pollinators
2.2. Enhancement of Plant Resistance
3. Complexity of FVT Biosynthesis and Emission
3.1. Spatio–Temporal Regulation
3.2. Luminous Intensity
3.3. Radiation
3.4. Composition of the Atmosphere
3.5. Ambient Temperature and Relative Humidity
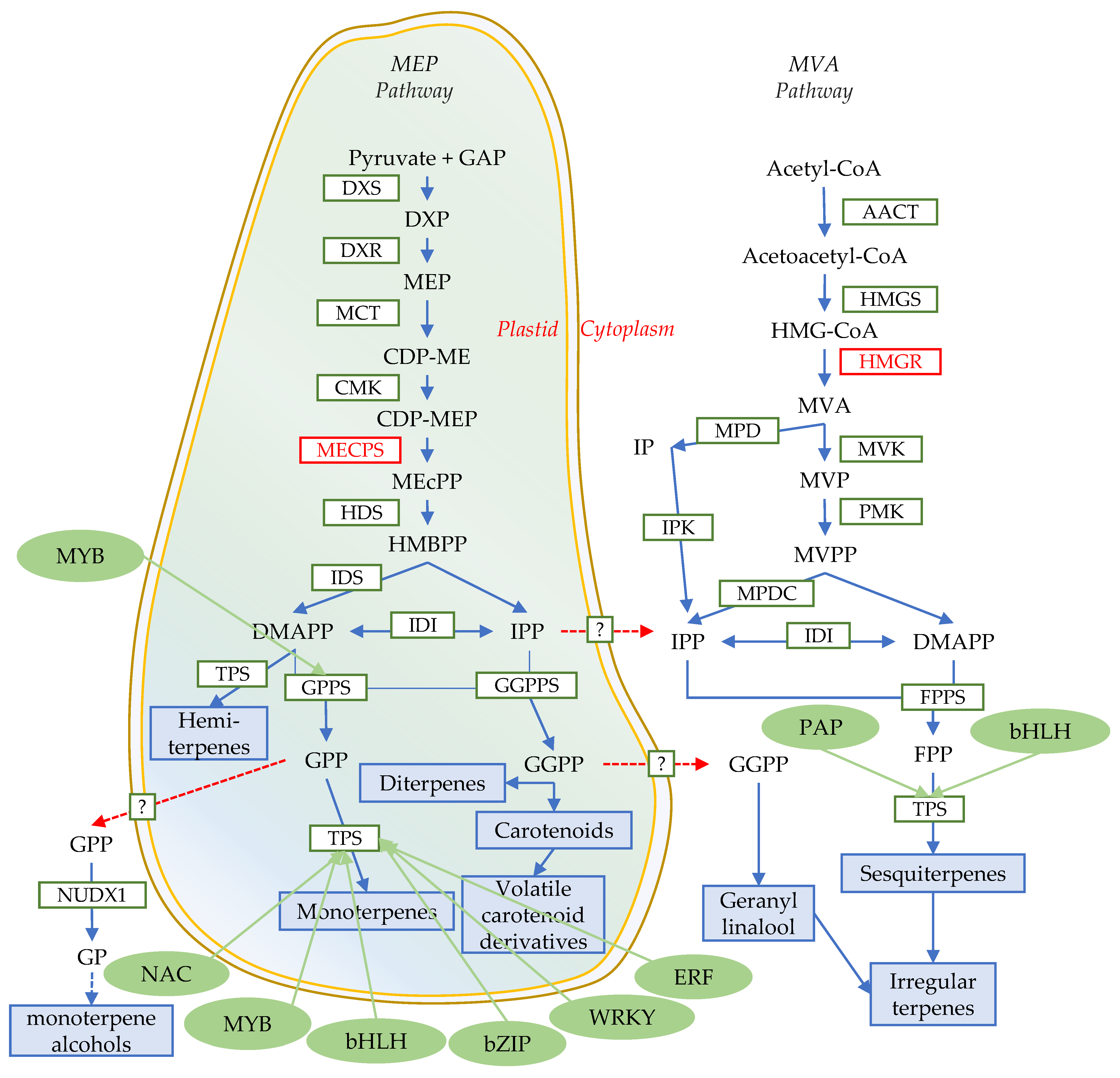
4. Biosynthesis of FVTs
5. Transcriptional Regulation and Modification of Terpene Skeletons
5.1. Transcriptional Regulation
5.1.1. MYB
5.1.2. bHLH
5.1.3. WRKY
5.1.4. Others
5.2. Modification of Terpene Skeletons
6. Intersection of Synthetic Pathways That Influence Flower Fragrance and Color
7. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Lu, X.; Tang, K.; Li, P. Plant metabolic engineering strategies for the production of pharmaceutical terpenoids. Front. Plant Sci. 2016, 7, 1647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabitov, A.; Gaweł-Bęben, K.; Sakipova, Z.; Strzępek-Gomółka, M.; Hoian, U.; Satbayeva, E.; Głowniak, K.; Ludwiczuk, A. Rosa platyacantha schrenk from Kazakhstan-Natural source of bioactive compounds with cosmetic significance. Molecules 2021, 26, 2578. [Google Scholar] [CrossRef]
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef]
- Farré-Armengol, G.; Fernández-Martínez, M.; Filella, I.; Junker, R.R.; Peñuelas, J. Deciphering the biotic and climatic factors that influence floral scents: A systematic review of floral volatile emissions. Front. Plant Sci. 2020, 11, 1154. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, J.T.; Eriksson, R.; Gershenzon, J.; Stahl, B. Diversity and distribution of floral scent. Bot. Rev. 2006, 72, 1–120. [Google Scholar] [CrossRef]
- Raguso, R.A. More lessons from linalool: Insights gained from a ubiquitous floral volatile. Curr. Opin. Plant Biol. 2016, 32, 31–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pichersky, E.; Raguso, R.A. Why do plants produce so many terpenoid compounds? New Phytol. 2018, 220, 692–702. [Google Scholar] [CrossRef]
- Brokl, M.; Fauconnier, M.L.; Benini, C.; Lognay, G.; Jardin, P.d.; Focant, J.F. Improvement of ylang-ylang essential oil characterization by GC × GC-TOFMS. Molecules 2013, 18, 1783–1797. [Google Scholar] [CrossRef] [Green Version]
- Raguso, R.A. Wake up and smell the roses: The ecology and evolution of floral scent. Annu. Rev. Ecol. Evol. Syst. 2008, 39, 549–569. [Google Scholar] [CrossRef]
- Unsicker, S.B.; Kunert, G.; Gershenzon, J. Protective perfumes: The role of vegetative volatiles in plant defense against herbivores. Curr. Opin. Plant Biol. 2009, 12, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Sanchez-Moreiras, A.M.; Abel, C.; Sohrabi, R.; Lee, S.; Gershenzon, J.; Tholl, D. The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-β-caryophyllene, is a defense against a bacterial pathogen. New Phytol. 2012, 193, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Ilc, T.; Halter, D.; Miesch, L.; Lauvoisard, F.; Kriegshauser, L.; Ilg, A.; Baltenweck, R.; Hugueney, P.; Werck-Reichhart, D.; Duchêne, E.; et al. A grapevine cytochrome P450 generates the precursor of wine lactone, a key odorant in wine. New Phytol. 2017, 213, 264–274. [Google Scholar] [CrossRef]
- Denby, C.M.; Li, R.A.; Vu, V.T.; Costello, Z.; Lin, W.; Chan, L.J.G.; Williams, J.; Donaldson, B.; Bamforth, C.W.; Petzold, C.J.; et al. Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer. Nat. Commun. 2018, 9, 965. [Google Scholar] [CrossRef]
- Xia, Z.; Huang, D.; Zhang, S.; Wang, W.; Ma, F.; Wu, B.; Xu, Y.; Xu, B.; Chen, D.; Zou, M.; et al. Chromosome-scale genome assembly provides insights into the evolution and flavor synthesis of passion fruit (Passiflora edulis Sims). Hortic. Res. 2021, 8, 14. [Google Scholar] [CrossRef]
- Wu, L.; Zhao, Y.; Zhang, Q.; Chen, Y.; Gao, M.; Wang, Y. Overexpression of the 3-hydroxy-3-methylglutaryl-CoA synthase gene LcHMGS effectively increases the yield of monoterpenes and sesquiterpenes. Tree Physiol. 2020, 40, 1095–1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nieuwenhuizen, N.J.; Wang, M.Y.; Matich, A.J.; Green, S.A.; Chen, X.; Yauk, Y.K.; Beuning, L.L.; Nagegowda, D.A.; Dudareva, N.; Atkinson, R.G. Two terpene synthases are responsible for the major sesquiterpenes emitted from the flowers of kiwifruit (Actinidia deliciosa). J. Exp. Bot. 2009, 60, 3203–3219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Yang, M.; Yang, X.; Ma, X.; Fu, J. Five TPSs are responsible for volatile terpenoid biosynthesis in Albizia julibrissin. J. Plant Physiol. 2021, 258–259, 153358. [Google Scholar] [CrossRef] [PubMed]
- Hattan, J.; Shindo, K.; Ito, T.; Shibuya, Y.; Watanabe, A.; Tagaki, C.; Ohno, F.; Sasaki, T.; Ishii, J.; Kondo, A.; et al. Identification of a novel hedycaryol synthase gene isolated from Camellia brevistyla flowers and floral scent of Camellia cultivars. Planta 2016, 243, 959–972. [Google Scholar] [CrossRef] [PubMed]
- Hattan, J.I.; Shindo, K.; Sasaki, T.; Ohno, F.; Tokuda, H.; Ishikawa, K.; Misawa, N. Identification of novel sesquiterpene synthase genes that mediate the biosynthesis of valerianol, which was an unknown ingredient of tea. Sci. Rep. 2018, 8, 12474. [Google Scholar] [CrossRef]
- Zhou, H.C.; Shamala, L.F.; Yi, X.K.; Yan, Z.; Wei, S. Analysis of terpene synthase family genes in Camellia sinensis with an emphasis on abiotic stress conditions. Sci. Rep. 2020, 10, 933. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Kim, M.J.; Dhandapani, S.; Tjhang, J.G.; Yin, J.L.; Wong, L.; Sarojam, R.; Chua, N.H.; Jang, I.C. The floral transcriptome of ylang ylang (Cananga odorata var fruticosa) uncovers biosynthetic pathways for volatile organic compounds and a multifunctional and novel sesquiterpene synthase. J. Exp. Bot. 2015, 66, 3959–3975. [Google Scholar] [CrossRef] [Green Version]
- Tian, J.P.; Ma, Z.Y.; Zhao, K.G.; Zhang, J.; Xiang, L.; Chen, L.Q. Transcriptomic and proteomic approaches to explore the differences in monoterpene and benzenoid biosynthesis between scented and unscented genotypes of wintersweet. Physiol. Plant 2019, 166, 478–493. [Google Scholar] [CrossRef]
- Shang, J.; Tian, J.; Cheng, H.; Yan, Q.; Li, L.; Jamal, A.; Xu, Z.; Xiang, L.; Saski, C.A.; Jin, S.; et al. The chromosome-level wintersweet (Chimonanthus praecox) genome provides insights into floral scent biosynthesis and flowering in winter. Genome. Biol. 2020, 21, 200. [Google Scholar] [CrossRef]
- Kamran, H.M.; Hussain, S.B.; Junzhong, S.; Xiang, L.; Chen, L.Q. Identification and molecular characterization of geranyl diphosphate synthase (GPPS) genes in wintersweet flower. Plants 2020, 9, 666. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.Z.; Lin, X.; Li, X.; Yang, N.; Chen, L. Molecular cloning and functional characterization of CpMYC2 and CpBHLH13 transcription factors from wintersweet (Chimonanthus praecox L.). Plants 2020, 9, 785. [Google Scholar] [CrossRef]
- Reisenman, C.E.; Riffell, J.A.; Bernays, E.A.; Hildebrand, J.G. Antagonistic effects of floral scent in an insect-plant interaction. Proc. Biol. Sci. 2010, 277, 2371–2379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Zheng, P.; Aoki, D.; Miyake, T.; Yagami, S.; Matsushita, Y.; Fukushima, K.; Nakagawa, M. Sexual and temporal variations in floral scent in the subdioecious shrub Eurya japonica Thunb. Ecol. Evol. 2018, 8, 8266–8272. [Google Scholar] [CrossRef]
- Kanlayavattanakul, M.; Lourith, N. Volatile profile and sensory property of Gardenia jasminoides aroma extracts. J. Cosmet. Sci. 2015, 66, 371–377. [Google Scholar] [PubMed]
- Obi, J.B.; Golonka, A.M.; Blackwell, A.; Vazquez, I.; Wolfram, N. Floral scent variation in the heterostylous species Gelsemium sempervirens. Molecules 2019, 24, 2818. [Google Scholar]
- Huang, X.Z.; Xiao, Y.T.; Köllner, T.G.; Jing, W.X.; Kou, J.F.; Chen, J.Y.; Liu, D.F.; Gu, S.H.; Wu, J.X.; Zhang, Y.J.; et al. The terpene synthase gene family in Gossypium hirsutum harbors a linalool synthase GhTPS12 implicated in direct defence responses against herbivores. Plant Cell Environ. 2018, 41, 261–274. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Fan, R.; Ye, X.; Lin, R.; Luo, Y.; Fang, N.; Zhong, H.; Chen, S. The transcriptome of flower development provides insight into floral scent formation in Freesia hybrida. Plant Growth Regul. 2018, 86, 93–104. [Google Scholar] [CrossRef]
- Barman, M.; Kotamreddy, J.N.R.; Agarwal, A.; Mitra, A. Enhanced emission of linalool from floral scent volatile bouquet in Jasminum auriculatum variants developed via gamma irradiation. Ind. Crops. Prod. 2020, 152, 112545. [Google Scholar] [CrossRef]
- Barman, M.; Mitra, A. Floral maturation and changing air temperatures influence scent volatiles biosynthesis and emission in Jasminum auriculatum Vahl. Environ. Exp. Bot. 2021, 181, 104296. [Google Scholar] [CrossRef]
- Bera, P.; Mukherjee, C.; Mitra, A. Enzymatic production and emission of floral scent volatiles in Jasminum sambac. Plant Sci. 2017, 256, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Pragadheesh, V.S.; Chanotiya, C.S.; Rastogi, S.; Shasany, A.K. Scent from Jasminum grandiflorum flowers: Investigation of the change in linalool enantiomers at various developmental stages using chemical and molecular methods. Phytochem. 2017, 140, 83–94. [Google Scholar] [CrossRef]
- Joulain, D. Jasminum grandiflorum flowers-phytochemical complexity and its capture in extracts: A review. Flavour Fragr. J. 2021, 36, 526–553. [Google Scholar] [CrossRef]
- Yahyaa, M.; Matsuba, Y.; Brandt, W.; Doron-Faigenboim, A.; Bar, E.; McClain, A.; Davidovich-Rikanati, R.; Lewinsohn, E.; Pichersky, E.; Ibdah, M. Identification, functional characterization, and evolution of terpene synthases from a basal dicot. Plant Physiol. 2015, 169, 1683–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyake, T.; Yamaoka, R.; Yahara, T. Floral scents of hawkmoth-pollinated flowers in Japan. J. Plant Res. 1998, 111, 199–205. [Google Scholar] [CrossRef]
- Dhandapani, S.; Jin, J.; Sridhar, V.; Sarojam, R.; Chua, N.H.; Jang, I.C. Integrated metabolome and transcriptome analysis of Magnolia champaca identifies biosynthetic pathways for floral volatile organic compounds. BMC Genom. 2017, 18, 463. [Google Scholar] [CrossRef]
- Rachersberger, M.; Cordeiro, G.D.; Schäffler, I.; Dötterl, S. Honeybee pollinators use visual and floral scent cues to find apple (Malus domestica) flowers. J. Agric. Food Chem. 2019, 67, 13221–13227. [Google Scholar] [CrossRef]
- Paul, I.; Chatterjee, A.; Maiti, S.; Bhadoria, P.B.S.; Mitra, A. Dynamic trajectories of volatile and non-volatile specialised metabolites in ‘overnight’ fragrant flowers of Murraya paniculata. Plant Biol. 2019, 21, 899–910. [Google Scholar] [CrossRef] [PubMed]
- Paul, I.; Mitra, A.; Bhadoria, P. Seasonal and diel variations in scent composition of ephemeral Murraya paniculata (linn.) Jack flowers are contributed by separate volatile components. Biochem. Syst. Ecol. 2020, 89, 104004. [Google Scholar] [CrossRef]
- Bouzabata, A.; Cabral, C.; Gonçalves, M.J.; Cruz, M.T.; Bighelli, A.; Cavaleiro, C.; Casanova, J.; Tomi, F.; Salgueiro, L. Myrtus communis L. as source of a bioactive and safe essential oil. Food Chem. Toxicol. 2015, 75, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Liu, C.; Zheng, R.; Cai, X.; Luo, J.; Zou, J.; Wang, C. Emission and accumulation of monoterpene and the key terpene synthase (TPS) associated with monoterpene biosynthesis in Osmanthus fragrans lour. Front. Plant Sci. 2016, 6, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Hou, D.; Zhang, C.; Bao, Z.; Zhao, H.; Hu, S. The emission of the floral scent of four Osmanthus fragrans cultivars in response to different temperatures. Molecules 2017, 22, 430. [Google Scholar] [CrossRef] [Green Version]
- Ding, W.; Ouyang, Q.; Li, Y.; Shi, T.; Li, L.; Yang, X.; Ji, K.; Wang, L.; Yue, Y. Genome-wide investigation of WRKY transcription factors in sweet osmanthus and their potential regulation of aroma synthesis. Tree Physiol. 2020, 40, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Zhu, Z.; Wang, Y.; Hu, S.; Xi, W.; Xiao, W.; Qu, X.; Zhong, L.; Fu, Q.; Wang, C. UGT85A84 catalyzes the glycosylation of aromatic monoterpenes in Osmanthus fragrans Lour. flowers. Front. Plant Sci. 2019, 10, 1376. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Hu, Z.H.; Leng, P.S.; Cheng, F.Y. Developmental and diurnal change of fragrance emission from ‘High noon’ flowers (Paeonia×lemonei ‘High noon’). In Proceedings of the Third Conference on Horticulture Science and Technology (CHST) 2012, Thika, Kenya, 31 July–3 August 2012; pp. 54–61. [Google Scholar]
- Song, C.; Wang, Q.; da Silva, J.A.T.; Yu, X. Identification of floral fragrances and analysis of fragrance patterns in Herbaceous peony cultivars. J. Am. Soc. Hort. Sci. 2018, 143, 248–258. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, C.C.; Rezende, J.L.; Silva, E.A.J.; Silva, F.G.; Stenico., L.; Crotti, A.E.M.; Esperandim, V.R.; Santiago, M.B.; Martins, C.H.G.; Miranda, M.L.D. Chemical composition and biological activities of essential oil from flowers of Psidium guajava (Myrtaceae). Braz. J. Biol. 2021, 81, 728–736. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Chen, C.; Li, T.; Wang, M.; Tao, J.; Zhao, D.; Sheng, L. Flowery odor formation revealed by differential expression of monoterpene biosynthetic genes and monoterpene accumulation in rose (Rosa rugosa Thunb.). Plant Physiol. Biochem. 2014, 75, 80–88. [Google Scholar] [CrossRef]
- Magnard, J.L.; Roccia, A.; Caissard, J.C.; Vergne, P.; Sun, P.; Hecquet, R.; Dubois, A.; Hibrand-Saint Oyant, L.; Jullien, F.; Baudino, S.; et al. Plant volatiles. Biosynthesis of monoterpene scent compounds in roses. Science 2015, 349, 81–83. [Google Scholar] [CrossRef] [Green Version]
- Magnard, J.L.; Bony, A.R.; Bettini, F.; Campanaro, A.; Blerot, B.; Baudino, S.; Jullien, F. Linalool and linalool nerolidol synthases in roses, several genes for little scent. Plant Physiol. Biochem. 2018, 127, 74–87. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, Z.J. The genomic floral language of rose. Nat. Genet. 2018, 50, 770–771. [Google Scholar] [CrossRef] [PubMed]
- Raymond, O.; Gouzy, J.; Just, J.; Badouin, H.; Verdenaud, M.; Lemainque, A.; Vergne, P.; Moja, S.; Choisne, N.; Pont, C.; et al. The rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 2018, 50, 772–777. [Google Scholar] [CrossRef] [PubMed]
- Dani, K.G.S.; Fineschi, S.; Michelozzi, M.; Trivellini, A.; Pollastri, S.; Loreto, F. Diversification of petal monoterpene profiles during floral development and senescence in wild roses: Relationships among geraniol content, petal colour, and floral lifespan. Oecologia 2020, 1–3. [Google Scholar] [CrossRef]
- Chen, C.; Cao, Y.; Chen, H.; Ni, M.; Yu, F. Floral scent compounds and emission patterns of three styrax species. Dendrobiology 2021, 85, 30–38. [Google Scholar] [CrossRef]
- Zheng, J.; Hu, Z.; Guan, X.; Dou, D.; Bai, G.; Wang, Y.; Guo, Y.; Li, W.; Leng, P. Transcriptome analysis of Syringa oblata Lindl. inflorescence identifies genes associated with pigment biosynthesis and scent metabolism. PLoS ONE 2015, 10, e0142542. [Google Scholar]
- Yan, Z.; Ying, Q.; Zheng, J.; Leng, P.; Hu, Z. Gene cloning and expression analysis of limonene synthase in Syringa oblata and S. oblata var. alba. J. For. Res. 2019, 30, 1301–1309. [Google Scholar] [CrossRef]
- Parachnowitsch, A.L.; Raguso, R.A.; Kessler, A. Phenotypic selection to increase floral scent emission, but not flower size or color in bee-pollinated Penstemon digitalis. New Phytol. 2012, 195, 667–675. [Google Scholar] [CrossRef]
- Burdon, R.C.F.; Raguso, R.A.; Gegear, R.J.; Pierce, E.C.; Kessler, A.; Parachnowitsch, A.L. Scented nectar and the challenge of measuring honest signals in pollination. J. Ecol. 2020, 108, 2132–2144. [Google Scholar] [CrossRef]
- Burdon, R.C.F.; Junker, R.R.; Scofield, D.G.; Parachnowitsch, A.L. Bacteria colonising Penstemon digitalis show volatile and tissue-specific responses to a natural concentration range of the floral volatile linalool. Chemoecology 2018, 28, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aros, D.; Gonzalez, V.; Allemann, R.K.; Müller, C.T.; Rosati, C.; Rogers, H.J. Volatile emissions of scented Alstroemeria genotypes are dominated by terpenes, and a myrcene synthase gene is highly expressed in scented Alstroemeria flowers. J. Exp. Bot. 2012, 63, 2739–2752. [Google Scholar] [CrossRef] [Green Version]
- Wei, Q.; Xia, Q.; Wang, Y.; Chen, W.; Liu, C.; Zeng, R.; Xie, L.; Yi, M.; Guo, H. Profiling of volatile compounds and associated gene expression in two Anthurium cultivars and their F1 hybrid progenies. Molecules 2021, 26, 2902. [Google Scholar] [CrossRef]
- Dudareva, N.; Martin, D.; Kish, C.M.; Kolosova, N.; Gorenstein, N.; Fäldt, J.; Miller, B.; Bohlmann, J. (E)-beta-ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: Function and expression of three terpene synthase genes of a new terpene synthase subfamily. Plant Cell 2003, 15, 1227–1241. [Google Scholar] [CrossRef] [Green Version]
- Nagegowda, D.A.; Gutensohn, M.; Wilkerson, C.G.; Dudareva, N. Two nearly identical terpene synthases catalyze the formation of nerolidol and linalool in snapdragon flowers. Plant J. 2008, 55, 224–239. [Google Scholar] [CrossRef]
- Aubourg, S.; Lecharny, A.; Bohlmann, J. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genomics 2002, 267, 730–745. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Tholl, D.; D’Auria, J.C.; Farooq, A.; Pichersky, E.; Gershenzon, J. Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell. 2003, 15, 481–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tholl, D.; Chen, F.; Petri, J.; Gershenzon, J.; Pichersky, E. Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J. 2005, 42, 757–771. [Google Scholar] [CrossRef] [PubMed]
- Hong, G.J.; Xue, X.Y.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, K.R.; More, M.; Hipolito, J.; Charlemagne, S.; Schlumpberger, B.O.; Raguso, R.A. Spatial and temporal variation in volatile composition suggests olfactory division of labor within the trap flowers of Aristolochia gigantea. Flora 2016, 232, 153–168. [Google Scholar] [CrossRef] [Green Version]
- Wong, D.C.J.; Pichersky, E.; Peakall, R. The biosynthesis of unusual floral volatiles and blends involved in orchid pollination by deception: Current progress and future prospects. Front. Plant Sci. 2017, 8, 1955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Booth, J.K.; Yuen, M.M.S.; Jancsik, S.; Madilao, L.L.; Page, J.E.; Bohlmann, J. Terpene synthases and terpene variation in Cannabis sativa. Plant Physiol. 2020, 184, 130–147. [Google Scholar] [CrossRef] [PubMed]
- Hwang, E.S.; Kim, G.H. Safety evaluation of Chrysanthemum indicum L. flower oil by assessing acute oral toxicity, micronucleus abnormalities, and mutagenicity. Prev. Nutr. Food Sci. 2013, 18, 111–116. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Xian, J.; Wei, W.; Xu, C.; Yang, J.; Zhan, R.; Ma, D. Volatile metabolic profiling and functional characterization of four terpene synthases reveal terpenoid diversity in different tissues of Chrysanthemum indicum L. Phytochemistry 2021, 185, 112687. [Google Scholar] [CrossRef] [PubMed]
- Jabalpurwala, F.A.; Smoot, J.M.; Rouseff, R.L. A comparison of citrus blossom volatiles. Phytochemistry 2009, 70, 1428–1434. [Google Scholar] [CrossRef]
- Dudareva, N.; Cseke, L.; Blanc, V.M.; Pichersky, E. Evolution of floral scent in Clarkia: Novel patterns of S-linalool synthase gene expression in the Clarkia breweri flower. Plant Cell 1996, 8, 1137–1148. [Google Scholar]
- Lavy, M.; Zuker, A.; Lewinsohn, E.; Larkov, O.; Ravid, U.; Vainstein, A.; Weiss, D. Linalool and linalool oxide production in transgenic carnation flowers expressing the Clarkia breweri linalool synthase gene. Mol. Breed. 2002, 9, 103–111. [Google Scholar] [CrossRef]
- Jiang, Y.; Qian, R.; Zhang, W.; Wei, G.; Ma, X.; Zheng, J.; Köllner, T.G.; Chen, F. Composition and biosynthesis of scent compounds from sterile flowers of an ornamental plant Clematis florida cv. ‘Kaiser’. Molecules 2020, 25, 1711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramya, M.; Jang, S.; An, H.R.; Lee, S.Y.; Park, P.M.; Park, P.H. Volatile organic compounds from orchids: From synthesis and function to gene regulation. Int. J. Mol. Sci. 2020, 21, 1160. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.M.; Jang, E.J.; Hong, J.W.; Song, S.H.; Pak, C.H. A comparison of functional fragrant components of Cymbidium (Oriental Orchid) Species. Hortic. Sci. 2016, 34, 331–341. [Google Scholar]
- Li, N.; Dong, Y.; Lv, M.; Qian, L.; Sun, X.; Liu, L.; Cai, Y.; Fan, H. Combined analysis of volatile terpenoid metabolism and transcriptome reveals transcription factors related to terpene synthase in two cultivars of Dendrobium officinale flowers. Front. Genet. 2021, 12, 661296. [Google Scholar] [CrossRef]
- Zhao, C.; Yu, Z.; Silva, J.A.T.D.; He, C.; Wang, H.; Si, C.; Zhang, M.; Zeng, D.; Duan, J. Functional characterization of a Dendrobium officinale geraniol synthase DoGES1 involved in floral scent formation. Int. J. Mol. Sci. 2020, 21, 7005. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhao, C.; Zhang, G.; da Silva, J.A.T.; Duan, J. Genome-wide identification and expression profile of TPS gene family in Dendrobium officinale and the role of DoTPS10 in linalool biosynthesis. Int. J. Mol. Sci. 2020, 21, 5419. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, K.; Nakayama, M.; Yagi, M.; Onozaki, T.; Oyama-Okubo, N. Evaluation of wild Dianthus species as genetic resources for fragrant carnation breeding based on their floral scent composition. J. Jpn. Soc. Hortic. Sci. 2011, 80, 175–181. [Google Scholar] [CrossRef] [Green Version]
- Kishimoto, K.; Inamoto, K.; Ymaguchi, H. Component analysis and sensory evaluation of scent emitted from cut carnation flowers. Bull NARO Veg. & Flor. Sci. 2019, 3, 29–40. [Google Scholar]
- Kishimoto, K.; Shibuya, K. Scent emissions and expression of scent emission-related genes: A comparison between cut and intact carnation flowers. Sci. Hortic. 2021, 281, 109920. [Google Scholar] [CrossRef]
- Gao, F.; Liu, B.; Li, M.; Gao, X.; Fang, Q.; Liu, C.; Ding, H.; Wang, L.; Gao, X. Identification and characterization of terpene synthase genes accounting for volatile terpene emissions in flowers of Freesia × hybrida. J. Exp. Bot. 2018, 69, 4249–4265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivasan, A.; Ahn, M.S.; Jo, G.S.; Suh, J.N.; Seo, K.H.; Kim, W.H.; Kang, Y.I.; Lee, Y.R.; Choi, Y.J. Analysis of relative scent intensity, volatile compounds and gene expression in Freesia “Shiny Gold”. Plants 2020, 9, 1597. [Google Scholar] [CrossRef]
- Yang, Z.; Li, Y.; Gao, F.; Jin, W.; Li, S.; Kimani, S.; Yang, S.; Bao, T.; Gao, X.; Wang, L. MYB21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J. Exp. Bot. 2020, 71, 4140–4158. [Google Scholar] [CrossRef]
- Chapurlat, E.; Anderson, J.; Ågren, J.; Friberg, M.; Sletvold, N. Diel pattern of floral scent emission matches the relative importance of diurnal and nocturnal pollinators in populations of Gymnadenia conopsea. Ann. Bot. 2018, 121, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Yu, R.C.; Fan, Y.P. Characterization of two monoterpene synthases involved in floral scent formation in Hedychium coronarium. Planta 2014, 240, 745–762. [Google Scholar] [CrossRef]
- Li, X.Y.; Zheng, S.Y.; Yu, R.C.; Fan, Y.P. Promoters of HcTPS1 and HcTPS2 genes from Hedychium coronarium direct floral-specific, developmental-regulated and stress-inducible gene expression in transgenic tobacco. Plant Mol. Biol. Rep. 2014, 32, 864–880. [Google Scholar] [CrossRef]
- Yue, Y.; Yu, R.C.; Fan, Y.P. Transcriptome profiling provides new insights into the formation of floral scent in Hedychium coronarium. BMC Genom. 2015, 16, 470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbas, F.; Ke, Y.; Zhou, Y.; Yu, Y.; Waseem, M.; Ashraf, U.; Wang, C.; Wang, X.; Li, X.; Yue, Y.; et al. Genome-wide analysis reveals the potential role of MYB transcription factors in floral scent formation in Hedychium coronarium. Front. Plant Sci. 2021, 12, 623742. [Google Scholar] [CrossRef]
- Ke, Y.; Abbas, F.; Zhou, Y.; Yu, R.; Fan, Y. Auxin-responsive R2R3-MYB transcription factors HcMYB1 and HcMYB2 activate volatile biosynthesis in Hedychium coronarium flowers. Front. Plant Sci. 2021, 12, 710826. [Google Scholar] [CrossRef] [PubMed]
- Meerow, A.W.; Reed, S.T.; Dunn, C.; Schnell, E. Fragrance analysis of two scented Hippeastrum species. HortScience 2017, 52, 1853–1860. [Google Scholar] [CrossRef] [Green Version]
- Bao, T.; Shadrack, K.; Yang, S.; Xue, X.; Li, S.; Wang, N.; Wang, Q.; Wang, L.; Gao, X.; Cronk, Q. Functional characterization of terpene synthases accounting for the volatilized-terpene heterogeneity in Lathyrus odoratus cultivar flowers. Plant Cell Physiol. 2020, 61, 1733–1749. [Google Scholar] [PubMed]
- Wilson, T.M.; Poulson, A.; Packer, C.; Carlson, R.E.; Buch, R.M. Essential oil profile and yield of corolla, calyx, leaf, and whole flowering top of cultivated Lavandula angustifolia Mill. (Lamiaceae) from Utah. Molecules 2021, 26, 2343. [Google Scholar] [CrossRef]
- Aprotosoaie, A.C.; Gille, E.; Trifan, A.; Luca, V.S.; Miron, A. Essential oils of Lavandula genus: A systematic review of their chemistry. Phytochem. Rev. 2017, 16, 761–799. [Google Scholar] [CrossRef]
- Adal, A.M.; Sarker, L.S.; Malli, R.P.N.; Liang, P.; Mahmoud, S.S. RNA-Seq in the discovery of a sparsely expressed scent-determining monoterpene synthase in lavender (Lavandula). Planta 2019, 249, 271–290. [Google Scholar] [CrossRef]
- Guitton, Y.; Nicole, F.; Jullien, F.; Jean-Claude, C.; Saint-Marcoux, D.; Legendre, L.; Pasquier, B.; Moja, S. A comparative study of terpene composition in different clades of the genus Lavandula. Bot. Lett. 2018, 165, 494–505. [Google Scholar] [CrossRef]
- Du, F.; Wang, T.; Fan, J.M.; Liu, Z.Z.; Zong, J.X.; Fan, W.X.; Han, Y.H.; Grierson, D. Volatile composition and classification of Lilium flower aroma types and identification, polymorphisms, and alternative splicing of their monoterpene synthase genes. Hortic. Res. 2019, 6, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbas, F.; Ke, Y.; Zhou, Y.; Ashraf, U.; Li, X.; Yu, Y.; Yue, Y.; Ahmad, K.W.; Yu, R.; Fan, Y. Molecular cloning, characterization and expression analysis of LoTPS2 and LoTPS4 involved in floral scent formation in oriental hybrid Lilium variety ‘Siberia’. Phytochemistry 2020, 173, 112294. [Google Scholar] [CrossRef] [PubMed]
- Abbas, F.; Ke, Y.; Yu, R.; Fan, Y. Functional characterization and expression analysis of two terpene synthases involved in floral scent formation in Lilium ‘Siberia’. Planta 2019, 249, 71–93. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Ramya, M.; An, H.R.; Park, P.M.; Lee, S.Y.; Park, S.Y.; Park, P.H. Floral volatile compound accumulation and gene expression analysis of Maxillaria tenuifolia. Korean J. Hortic. Sci. Technol. 2019, 37, 756–766. [Google Scholar]
- Crowell, A.L.; Williams, D.C.; Davis, E.M.; Wildung, M.R.; Croteau, R. Molecular cloning and characterization of a new linalool synthase. Arch. Biochem. Biophys. 2002, 405, 112–121. [Google Scholar] [CrossRef]
- Peng, F.; Byers, K.J.R.P.; Bradshaw, H.D., Jr. Less is more: Independent loss-of-function OCIMENE SYNTHASE alleles parallel pollination syndrome diversification in monkeyflowers (Mimulus). Am. J. Bot. 2017, 104, 1055–1059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byers, K.J.; Vela, J.P.; Peng, F.; Riffell, J.A.; Bradshaw, H.D., Jr. Floral volatile alleles can contribute to pollinator-mediated reproductive isolation in monkeyflowers (Mimulus). Plant J. 2014, 80, 1031–1042. [Google Scholar] [CrossRef] [Green Version]
- Reddy, V.A.; Wang, Q.; Dhar, N.; Kumar, N.; Venkatesh, P.N.; Rajan, C.; Panicker, D.; Sridhar, V.; Mao, H.Z.; Sarojam, R. Spearmint R2R3-MYB transcription factor MsMYB negatively regulates monoterpene production and suppresses the expression of geranyl diphosphate synthase large subunit (MsGPPS.LSU). Plant Biotechnol. J. 2017, 15, 1105–1119. [Google Scholar] [CrossRef] [Green Version]
- Terry, M.I.; Ruiz-Hernández, V.; Águila, D.J.; Weiss, J.; Egea-Cortines, M. The effect of post-harvest conditions in Narcissus sp. cut flowers scent profile. Front. Plant Sci. 2021, 11, 540821. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ren, Y.; Zhang, D.; Chen, X.; Huang, J.; Xu, Y.; Aucapiña, C.B.; Zhang, Y.; Miao, Y. Transcriptome-based WGCNA analysis reveals regulated metabolite fluxes between floral color and scent in Narcissus tazetta flower. Int. J. Mol. Sci. 2021, 22, 8249. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Kügler, A.; McGale, E.; Haverkamp, A.; Knaden, M.; Guo, H.; Beran, F.; Yon, F.; Li, R.; Lackus, N.; et al. Tissue-specific emission of (E)-α-Bergamotene helps resolve the dilemma when pollinators are also herbivores. Curr. Biol. 2017, 27, 1336–1341. [Google Scholar] [CrossRef] [Green Version]
- Henry, L.K.; Thomas, S.T.; Widhalm, J.R.; Lynch, J.H.; Davis, T.C.; Kessler, S.A.; Bohlmann, J.; Noel, J.P.; Dudareva, N. Contribution of isopentenyl phosphate to plant terpenoid metabolism. Nat. Plants 2018, 4, 721–729. [Google Scholar] [CrossRef]
- Xu, S.; Kreitzer, C.; McGale, E.; Lackus, N.D.; Guo, H.; Köllner, T.G.; Schuman, M.C.; Baldwin, I.T.; Zhou, W. Allelic differences of clustered terpene synthases contribute to correlated intraspecific variation of floral and herbivory-induced volatiles in a wild tobacco. New Phytol. 2020, 228, 1083–1096. [Google Scholar] [CrossRef]
- Heiling, S.; Llorca, L.C.; Li, J.; Gase, K.; Schmidt, A.; Schäfer, M.; Schneider, B.; Halitschke, R.; Gaquerel, E.; Baldwin, I.T. Specific decorations of 17-hydroxygeranyllinalool diterpene glycosides solve the autotoxicity problem of chemical defense in Nicotiana attenuata. Plant Cell. 2021, 33, 1748–1770. [Google Scholar] [CrossRef]
- Roeder, S.; Hartmann, A.M.; Effmert, U.; Piechulla, B. Regulation of simultaneous synthesis of floral scent terpenoids by the 1, 8-cineole synthase of Nicotiana suaveolens. Plant Mol. Biol. 2007, 65, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Maia, A.C.D.; de Lima, C.T.; Navarro, D.M.D.A.F.; Chartier, M.; Giulietti, A.M.; Machado, I.C. The floral scents of Nymphaea subg. Hydrocallis (Nymphaeaceae), the New World night-blooming water lilies, and their relation with putative pollinators. Phytochemistry 2014, 103, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Filip, S.; Vidovi, S.; Vladi, J.; Pavli, B.; Zekovi, Z. Chemical composition and antioxidant properties of Ocimum basilicum L. extracts obtained by supercritical carbon dioxide extraction: Drug exhausting method. J. Supercrit. Fluids 2015, 109, 20–25. [Google Scholar] [CrossRef]
- Boachon, B.; Lynch, J.H.; Ray, S.; Yuan, J.; Caldo, K.M.P.; Junker, R.R.; Kessler, S.A.; Morgan, J.A.; Dudareva, N. Natural fumigation as a mechanism for volatile transport between flower organs. Nat. Chem. Biol. 2019, 15, 583–588. [Google Scholar] [CrossRef]
- Chuang, Y.C.; Hung, Y.C.; Tsai, W.C.; Chen, W.H.; Chen, H.H. Pbbhlh4 regulates floral monoterpene biosynthesis in Phalaenopsis orchids. J. Exp. Bot. 2018, 69, 4363–4377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Kuo, Y.W.; Chuang, Y.C.; Yang, Y.P.; Huang, L.M.; Jeng, M.F.; Chen, W.H.; Chen, H.H. Terpene synthase-b and terpene synthase-e/f genes produce monoterpenes for Phalaenopsis bellina floral scent. Front. Plant Sci. 2021, 12, 700958. [Google Scholar] [CrossRef] [PubMed]
- Chuang, Y.C.; Lee, M.C.; Chang, Y.L.; Chen, W.H.; Chen, H.H. Diurnal regulation of the floral scent emission by light and circadian rhythm in the Phalaenopsis orchids. Bot. Stud. 2017, 58, 50. [Google Scholar] [CrossRef] [PubMed]
- Ashaari, N.S.; Ab Rahim, M.H.; Sabri, S.; Lai, K.S.; Song, A.A.; Abdul Rahim, R.; Wan Abdullah, W.M.A.N.; Ong Abdullah, J. Functional characterization of a new terpene synthase from Plectranthus amboinicus. PLoS ONE 2020, 15, e0235416. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Chen, Y.; Ye, X.; Wu, J.; Lin, B.; Zhong, H. Transcriptome analysis of Polianthes tuberosa during floral scent formation. PLoS ONE 2018, 13, e0199261. [Google Scholar] [CrossRef]
- Kutty, N.N.; Mitra, A. Profiling of volatile and non-volatile metabolites in Polianthes tuberosa L. flowers reveals intraspecific variation among cultivars. Phytochemistry 2019, 162, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Kutty, N.N.; Ghissing, U.; Mitra, A. Revealing floral metabolite network in tuberose that underpins scent volatiles synthesis, storage and emission. Plant Mol. Biol. 2021, 106, 533–554. [Google Scholar] [CrossRef]
- Maiti, S.; Mitra, A. Elucidation of headspace volatilome in Polianthes tuberosa flower for identifying non-invasive biomarkers. Hortic. Environ. Biotech. 2019, 60, 269–280. [Google Scholar]
- Song., B.; Chen, G.; Stöcklin, J.; Peng, D.L.; Niu, Y.; Li, Z.M.; Sun, H. A new pollinating seed-consuming mutualism between Rheum nobile and a fly fungus gnat, Bradysia sp., involving pollinator attraction by a specific floral compound. New Phytol. 2014, 203, 1109–1118. [Google Scholar] [CrossRef]
- Ali, M.; Li, P.; She, G.; Chen, D.; Wan, X.; Zhao, J. Transcriptome and metabolite analyses reveal the complex metabolic genes involved in volatile terpenoid biosynthesis in garden sage (Salvia officinalis). Sci. Rep. 2017, 7, 16074. [Google Scholar] [CrossRef] [Green Version]
- Eilers, E.J.; Kleine, S.; Eckert, S.; Waldherr, S.; Müller, C. Flower production, headspace volatiles, pollen nutrients, and florivory in Tanacetum vulgare chemotypes. Front. Plant Sci. 2021, 11, 611877. [Google Scholar] [CrossRef] [PubMed]
- Krug, C.; Cordeiro, G.D.; Schäffler, I.; Silva, C.I.; Oliveira, R.; Schlindwein, C.; Dötterl, S.; Alves-Dos-Santos, I. Nocturnal bee pollinators are attracted to guarana flowers by their scents. Front. Plant Sci. 2018, 9, 1072. [Google Scholar] [CrossRef] [Green Version]
- Farré-Armengol, G.; Filella, I.; Llusià, J.; Peñuelas, J. β-Ocimene, a key floral and foliar volatile involved in multiple interactions between plants and other organisms. Molecules 2017, 22, 1148. [Google Scholar] [CrossRef] [Green Version]
- Abdalsamee, M.K.; Müller, C. Uncovering different parameters influencing florivory in a specialist herbivore. Ecol. Entomol. 2015, 40, 258–268. [Google Scholar] [CrossRef]
- Yu, X.D.; Pickett, J.; Ma, Y.Z.; Bruce, T.; Napier, J.; Jones, H.D.; Xia, L.Q. Metabolic engineering of plant-derived (E)-β-farnesene synthase genes for a novel type of aphid-resistant genetically modified crop plants. J. Integr. Plant Biol. 2012, 54, 282–299. [Google Scholar] [CrossRef]
- Li, J.; Hu, H.; Mao, J.; Yu, L.; Stoopen, G.; Wang, M.; Mumm, R.; De Ruijter, N.C.A.; Dicke, M.; Jongsma, M.A.; et al. Defense of pyrethrum flowers: Repelling herbivores and recruiting carnivores by producing aphid alarm pheromone. New Phytol. 2019, 223, 1607–1620. [Google Scholar] [CrossRef] [PubMed]
- Boachon, B.; Burdloff, Y.; Ruan, J.X.; Rojo, R.; Junker, R.R.; Vincent, B.; Nicolè, F.; Bringel, F.; Lesot, A.; Henry, L.; et al. A promiscuous CYP706A3 reduces terpene volatile emission from arabidopsis flowers, affecting florivores and the floral microbiome. Plant Cell. 2019, 31, 2947–2972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peñuelas, J.; Farré-Armengol, G.; Llusia, J.; Gargallo-Garriga, A.; Rico, L.; Sardans, J.; Terradas, J.; Filella, I. Removal of floral microbiota reduces floral terpene emissions. Sci. Rep. 2014, 4, 6727. [Google Scholar] [CrossRef] [Green Version]
- Saunier, A.; Mpamah, P.; Biasi, C.; Blande, J.D. Microorganisms in the phylloplane modulate the BVOC emissions of Brassica nigra leaves. Plant Signal Behav. 2020, 15, e1728468. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chen, J.; Wang, T.; Gao, C.; Li, Z.; Guo, L.; Xu, J.; Cheng, Y. Linking plant secondary metabolites and plant microbiomes: A review. Front. Plant Sci. 2021, 12, 621276. [Google Scholar] [CrossRef]
- Helletsgruber, C.; Dötterl, S.; Ruprecht, U.; Junker, R.R. Epiphytic bacteria alter floral scent emissions. J. Chem. Ecol. 2017, 43, 1073–1077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kessler, A. Introduction to a special feature issue-new insights into plant volatiles. New Phytol. 2018, 220, 655–658. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Stoopen, G.; Thoen, M.; Wiegers, G.; Jongsma, M.A. Chrysanthemum expressing a linalool synthase gene ‘smells good’, but ‘tastes bad’ to western flower thrips. Plant Biotechnol J. 2013, 11, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Dötterl, S.; Jahreiß, K.; Jhumur, U.S.; Jürgens, A. Temporal variation of flower scent in Silene otites (Caryophyllaceae): A species with a mixed pollination system. Bot. J. Linn. Soc. 2012, 169, 447–460. [Google Scholar] [CrossRef]
- Rodriguez-Saona, C.; Parra, L.; Quiroz, A.; Isaacs, R. Variation in highbush blueberry floral volatile profiles as a function of pollination status, cultivar, time of day and flower part: Implications for flower visitation by bees. Ann. Bot. 2011, 107, 1377–1390. [Google Scholar] [CrossRef] [PubMed]
- Muhlemann, J.K.; Waelti, M.O.; Widmer, A.; Schiestl, F.P. Postpollination changes in floral odor in Silene latifolia: Adaptive mechanisms for seed-predator avoidance? J. Chem. Ecol. 2006, 32, 1855–1860. [Google Scholar] [CrossRef]
- Dhandapani, S.; Jin, J.; Sridhar, V.; Chua, N.H.; Jang, I.C. CYP79D73 Participates in biosynthesis of floral scent compound 2-phenylethanol in Plumeria rubra. Plant Physiol. 2019, 180, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Al-Kateb, H.; Mottram, D.S. The relationship between growth stages and aroma composition of lemon basil Ocimum citriodorum Vis. Food Chem. 2014, 152, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Barman, M.; Mitra, A. Temporal relationship between emitted and endogenous floral scent volatiles in summer-and winter-blooming Jasminum species. Physiol. Plant. 2019, 166, 946–959. [Google Scholar] [CrossRef]
- Zhang, H.X.; Leng, P.S.; Hu, Z.H.; Zhao, J.; Wang, W.H.; Xu, F. The floral scent emitted from Lilium ‘Siberia’ at different flowering stages and diurnal variation. Acta. Hortic. Sin. 2013, 40, 693–702. [Google Scholar]
- Fu, X.; Chen, Y.; Mei, X.; Katsuno, T.; Kobayashi, E.; Dong, F.; Watanabe, N.; Yang, Z. Regulation of formation of volatile compounds of tea (Camellia sinensis) leaves by single light wavelength. Sci. Rep. 2015, 5, 16858. [Google Scholar] [CrossRef]
- Hu, Z.; Li, T.; Zheng, J.; Yang, K.; He, X.; Leng, P. Ca2+ signal contributing to the synthesis and emission of monoterpenes regulated by light intensity in Lilium ‘siberia’. Plant Physiol. Biochem. 2015, 91, 1–9. [Google Scholar] [CrossRef]
- Shamala, L.F.; Zhou, H.C.; Han, Z.X.; Wei, S. UV-B induces distinct transcriptional re-programing in UVR8-signal transduction, flavonoid, and terpenoids pathways in Camellia sinensis. Front. Plant Sci. 2020, 11, 234. [Google Scholar] [CrossRef] [Green Version]
- Blande, J.D.; Holopainen, J.K.; Niinemets, Ü. Plant volatiles in polluted atmospheres: Stress responses and signal degradation. Plant Cell Environ. 2014, 37, 1892–1904. [Google Scholar] [CrossRef] [Green Version]
- Farré-Armengol, G.; Peñuelas, J.; Li, T.; Yli-Pirilä, P.; Filella, I.; Llusia, J.; Blande, J.D. Ozone degrades floral scent and reduces pollinator attraction to flowers. New Phytol. 2016, 209, 152–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girling, R.D.; Lusebrink, I.; Farthing, E.; Newman, T.A.; Poppy, G.M. Diesel exhaust rapidly degrades floral odours used by honeybees. Sci. Rep. 2013, 3, 2779. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, M.A.; Burkle, L.A.; Manson, J.S.; Runyon, J.B.; Trowbridge, A.M.; Zientek, J. Global change effects on plant-insect interactions: The role of phytochemistry. Curr. Opin. Insect Sci. 2017, 23, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Burkle, L.A.; Runyon, J.B. Drought and leaf herbivory influence floral volatiles and pollinator attraction. Glob. Chang. Biol. 2016, 22, 1644–1654. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, M.K.; Campbell, D.R. Shifts in water availability mediate plant-pollinator interactions. New Phytol. 2017, 215, 792–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glenny, W.R.; Runyon, J.B.; Burkle, L.A. Drought and increased CO2 alter floral visual and olfactory traits with context-dependent effects on pollinator visitation. New Phytol. 2018, 220, 785–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sagae, M.; Oyama-Okubo, N.; Ando, T.; Marchesi, E.; Nakayama, M. Effect of temperature on the floral scent emission and endogenous volatile profile of Petunia axillaris. Biosci. Biotechnol. Biochem. 2008, 72, 110–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.; Zhang, H.; Leng, P.; Zhao, J.; Wang, W.; Wang, S. The emission of floral scent from Lilium “siberia” in response to light intensity and temperature. Acta Physiologiae Plantarum 2013, 35, 1691–1700. [Google Scholar] [CrossRef]
- Farré-Armengol, G.; Filella, I.; Llusià, J.; Niinemets, Ü.; Peñuelas, J. Changes in floral bouquets from compound-specific responses to increasing temperatures. Glob. Chang. Biol. 2014, 20, 3660–3669. [Google Scholar] [CrossRef] [Green Version]
- Campbell, D.R.; Sosenski, P.; Raguso, R.A. Phenotypic plasticity of floral volatiles in response to increasing drought stress. Ann. Bot. 2019, 123, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Rering, C.C.; Franco, J.G.; Yeater, K.M.; Mallinger, R.E. Drought stress alters floral volatiles and reduces floral rewards, pollinator activity, and seed set in a global plant. Ecosphere 2020, 11, e03254. [Google Scholar] [CrossRef]
- Kuppler, J.; Kotowska, M.M. A meta-analysis of responses in floral traits and flower-visitor interactions to water deficit. Glob. Chang. Biol. 2021, 27, 3095–3108. [Google Scholar] [CrossRef] [PubMed]
- Muhlemann, J.K.; Klempien, A.; Dudareva, N. Floral volatiles: From biosynthesis to function. Plant Cell Environ. 2014, 37, 1936–1949. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Chang, W.C.; Xiao, Y.; Liu, H.W.; Liu, P. Methylerythritol phosphate pathway of isoprenoid biosynthesis. Annu. Rev. Biochem. 2013, 82, 497–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudareva, N.; Andersson, S.; Orlova, I.; Gatto, N.; Reichelt, M.; Rhodes, D.; Boland, W.; Gershenzon, J. The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc. Natl. Acad. Sci. USA 2005, 102, 933–938. [Google Scholar] [CrossRef] [Green Version]
- Kopcsayova, D.; Vranova, E. Functional gene network of prenyltransferases in Arabidopsis thaliana. Molecules 2019, 24, 4556. [Google Scholar] [CrossRef] [Green Version]
- Tholl, D.; Kish, C.M.; Orlova, I.; Sherman, D.; Gershenzon, J.; Pichersky, E.; Dudareva, N. Formation of monoterpenes in Antirrhinum majus and Clarkia breweri flowers involves heterodimeric geranyl diphosphate synthases. Plant Cell. 2004, 16, 977–992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adal, A.M.; Mahmoud, S.S. Short-chain isoprenyl diphosphate synthases of lavender (Lavandula). Plant Mol. Biol. 2020, 102, 517–535. [Google Scholar] [CrossRef]
- Chen, Q.; Fan, D.; Wang, G. Heteromeric geranyl (geranyl) diphosphate synthase is involved in monoterpene biosynthesis in Arabidopsis flowers. Mol. Plant 2015, 8, 1434–1437. [Google Scholar] [CrossRef] [Green Version]
- Mendoza-Poudereux, I.; Kutzner, E.; Huber, C.; Segura, J.; Eisenreich, W.; Arrillaga, I. Metabolic cross-talk between pathways of terpenoid backbone biosynthesis in spike lavender. Plant Physiol. Biochem. 2015, 95, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Z.; Lei, Y.; Xiao, Y.; He, X.; Liang, J.; Jiang, J.; Dong, S.; Ke, H.; Leon, P.; Zerbe, P.; et al. Uncovering the functional residues of Arabidopsis isoprenoid biosynthesis enzyme HDS. Proc. Natl. Acad. Sci. USA 2020, 117, 355–361. [Google Scholar] [CrossRef]
- Pu, X.; Dong, X.; Li, Q.; Chen, Z.; Liu, L. An update on the function and regulation of methylerythritol phosphate and mevalonate pathways and their evolutionary dynamics. J. Integr. Plant Biol. 2021, 63, 1211–1226. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Concepcion, M.; Boronat, A. Breaking new ground in the regulation of the early steps of plant isoprenoid biosynthesis. Curr. Opin. Plant Biol. 2015, 25, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Kang, K.; Wang, P.; Li, M.; Huang, X. Transcriptome profiling of spike provides expression features of genes related to terpene biosynthesis in lavender. Sci. Rep. 2020, 10, 6933. [Google Scholar] [CrossRef]
- Vranová, E.; Coman, D.; Gruissem, W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef]
- Borghi, M.; Fernie, A.R.; Schiestl, F.P.; Bouwmeester, H.J. The sexual advantage of looking, smelling, and tasting good: The metabolic network that produces signals for pollinators. Trends Plant Sci. 2017, 22, 338–350. [Google Scholar] [CrossRef]
- Falara, V.; Akhtar, T.A.; Nguyen, T.T.; Spyropoulou, E.A.; Bleeker, P.M.; Schauvinhold, I.; Matsuba, Y.; Bonini, M.E.; Schilmiller, A.L.; Last, R.L.; et al. The tomato terpene synthase gene family. Plant Physiol. 2011, 157, 770–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.M.; Huang, H.; Chuang, Y.C.; Chen., W.H.; Wang, C.N.; Chen, H.H. Evolution of terpene synthases in Orchidaceae. Int. J. Mol. Sci. 2021, 22, 6947. [Google Scholar] [CrossRef] [PubMed]
- Karunanithi, P.S.; Zerbe, P. Terpene synthases as metabolic gatekeepers in the evolution of plant terpenoid chemical diversity. Front. Plant Sci. 2019, 10, 1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christianson, D.W. Structural and chemical biology of terpenoid cyclases. Chem. Rev. 2017, 117, 11570–11648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarker, L.S.; Adal, A.M.; Mahmoud, S.S. Diverse transcription factors control monoterpene synthase expression in lavender (Lavandula). Planta 2020, 251, 5. [Google Scholar] [CrossRef]
- Ramya, M.; Kwon, O.K.; An, H.R.; Park, P.M.; Baek, Y.S.; Park, P.H. Floral scent: Regulation and role of MYB transcription factors. Phytochem. Lett. 2017, 19, 114–120. [Google Scholar] [CrossRef]
- Van Moerkercke, A.; Steensma, P.; Schweizer, F.; Pollier, J.; Gariboldi, I.; Payne, R.; Vanden Bossche, R.; Miettinen, K.; Espoz, J.; Purnama, P.C.; et al. The bHLH transcription factor BIS 1 controls the iridoid branch of the monoterpenoid indole alkaloid pathway in Catharanthus roseus. Proc. Natl. Acad. Sci. USA 2015, 112, 8130–8135. [Google Scholar]
- Van Moerkercke, A.; Steensma, P.; Gariboldi, I.; Espoz, J.; Purnama, P.C.; Schweizer, F.; Miettinen, K.; Vanden, B.R.; De Clercq, R.; Memelink, J.; et al. The basic helix- loop- helix transcription factor BIS2 is essential for monoterpenoid indole alkaloid production in the medicinal plant Catharanthus roseus. Plant J. 2016, 88, 3–12. [Google Scholar]
- Alfieri, M.; Vaccaro, M.C.; Cappetta, E.; Ambrosone, A.; Tommasi, D.; Leone, A. Coactivation of MEP-biosynthetic genes and accumulation of abietane diterpenes in Salvia sclarea by heterologous expression of WRKY and MYC2 transcription factors. Sci. Rep. 2018, 8, 11009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, P.; Singh, S.K.; Patra, B.; Sui, X.; Pattanaik, S.; Yuan, L. A differentially regulated AP2/ERF transcription factor gene cluster acts downstream of a MAP kinase cascade to modulate terpenoid indole alkaloid biosynthesis in Catharanthus roseus. New Phytol. 2017, 213, 1107–1123. [Google Scholar] [CrossRef]
- Yang, Z.; Xie, C.; Huang, Y.; An, W.; Liu, S.; Huang, S.; Zheng, X. Metabolism and transcriptome profiling provides insight into the genes and transcription factors involved in monoterpene biosynthesis of borneol chemotype of Cinnamomum camphora induced by mechanical damage. PeerJ 2021, 9, e11465. [Google Scholar] [CrossRef]
- Zhang, F.Y.; Xiang, L.E.; Yu, Q.; Zhang, H.X.; Zhang, T.X.; Zeng, J.L.; Geng, C.; Li, L.; Fu, X.Q.; Shen, Q.; et al. Artemisinin Biosynthesis Promoting Kinase 1 positively regulates artemisinin biosynthesis through phosphorylating Aab-ZIP1. J. Exp. Bot. 2018, 69, 1109–1123. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Yue, Y.Z.; Ding, W.J.; Chen, G.W.; Li, L.; Li, Y.L.; Shi, T.T.; Yang, X.L.; Wang, L.G. Genome-wide identification, classification, and expression profiling reveals R2R3-MYB transcription factors related to monoterpenoid biosynthesis in Osmanthus fragrans. Genes 2020, 11, 353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben Zvi, M.M.; Negre-Zakharov, F.; Masci, T.; Ovadis, M.; Shklarman, E.; Ben-Meir, H.; Tzfira, T.; Dudareva, N.; Vainstein, A. Interlinking showy traits: Co-engineering of scent and colour biosynthesis in flowers. Plant Biotechnol. J. 2008, 6, 403–415. [Google Scholar]
- Zvi, M.M.B.; Shklarman, E.; Masci, T.; Kalev, H.; Debener, T.; Shafir, S.; Ovadis, M.; Vainstein, A. PAP1 transcription factor enhances production of phenylpropanoid and terpenoid scent compounds in rose flowers. New Phytol. 2012, 195, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Duan, G.; Li, D.; Wu, J.; Liu, X.; Hong, B.; Yi, M.; Zhang, Z. Two-dimensional analysis provides molecular insight into flower scent of Lilium ‘Siberia’. Sci. Rep. 2018, 8, 5352. [Google Scholar] [CrossRef] [Green Version]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L. The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 2005, 5, 3. [Google Scholar] [CrossRef] [Green Version]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Hurst, H.C. Transcription factors 1: bZIP proteins. Protein Profile 1994, 1, 123–168. [Google Scholar] [PubMed]
- Hamberger, B.; Bak, S. Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2013, 368, 20120426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reed, J.; Osbourn, A. Engineering terpenoid production through transient expression in Nicotiana benthamiana. Plant Cell Rep. 2018, 37, 1431–1441. [Google Scholar] [CrossRef]
- Reed, J.; Stephenson, M.J.; Miettinen, K.; Brouwer, B.; Leveau, A.; Brett, P.; Goss, R.J.M.; Goossens, A.; O’Connell, M.A.; Osbourn, A. A translational synthetic biology platform for rapid access to gram-scale quantities of novel drug-like molecules. Metab. Eng. 2017, 42, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Yazaki, K.; Arimura, G.I.; Ohnishi, T. ‘Hidden’ terpenoids in plants: Their biosynthesis, localization and ecological roles. Plant Cell Physiol. 2017, 58, 1615–1621. [Google Scholar] [CrossRef] [Green Version]
- Abdollahi, F.; Alebrahim, M.T.; Ngov, C.; Lallemand, E.; Zheng, Y.; Villette, C.; Zumsteg, J.; André, F.; Navrot, N.; Werck-Reichhart, D.; et al. Innate promiscuity of the CYP706 family of P450 enzymes provides a suitable context for the evolution of dinitroaniline resistance in weed. New Phytol. 2021, 229, 3253–3268. [Google Scholar] [CrossRef]
- Tholl, D.; Lee, S. Terpene specialized metabolism in Arabidopsis thaliana. Arab. Book 2011, 9, e0143. [Google Scholar] [CrossRef] [Green Version]
- Sohrabi, R.; Huh, J.H.; Badieyan, S.; Rakotondraibe, L.H.; Kliebenstein, D.J.; Sobrado, P.; Tholl, D. In planta variation of volatile biosynthesis: An alternative biosynthetic route to the formation of the pathogen-induced volatile homoterpene DMNT via triterpene degradation in Arabidopsis roots. Plant Cell. 2015, 27, 874–890. [Google Scholar] [CrossRef] [PubMed]
- Donath, J.; Boland, W. Biosynthesis of acyclic homoterpenes: Enzyme selectivity and absolute configuration of the nerolidol precursor. Phytochemistry 1995, 39, 785–790. [Google Scholar] [CrossRef]
- Ginglinger, J.F.; Boachon, B.; Höfer, R.; Paetz, C.; Köllner, T.G.; Miesch, L.; Lugan, R.; Baltenweck, R.; Mutterer, J.; Ullmann, P.; et al. Gene coexpression analysis reveals complex metabolism of the monoterpene alcohol linalool in Arabidopsis flowers. Plant Cell 2013, 25, 4640–4657. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Li, J.; Dong, Y.; Hao, H.; Ling, Z.; Bai, H.; Wang, H.; Cui, H.; Shi, L. Time-series transcriptome provides insights into the gene regulation network involved in the volatile terpenoid metabolism during the flower development of lavender. BMC Plant Biol. 2019, 19, 313. [Google Scholar] [CrossRef] [Green Version]
- Boachon, B.; Junker, R.R.; Miesch, L.; Bassard, J.E.; Höfer, R.; Caillieaudeaux, R.; Seidel, D.E.; Lesot, A.; Heinrich, C.; Ginglinger, J.F.; et al. CYP76C1 (Cytochrome P450)-mediated linalool metabolism and the formation of volatile and soluble linalool oxides in Arabidopsis flowers: A strategy for defense against floral antagonists. Plant Cell 2015, 27, 2972–2990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seifert, A.; Antonovici, M.; Hauer, B.; Pleiss, J. An efficient route to selective bio-oxidation catalysts: An iterative approach comprising modeling, diversification, and screening, based on CYP102A1. Chembiochem. 2011, 12, 1346–1351. [Google Scholar] [CrossRef]
- Miettinen, K.; Dong, L.; Navrot, N.; Schneider, T.; Burlat, V.; Pollier, J.; Woittiez, L.; van der Krol, S.; Lugan, R.; Ilc, T.; et al. The seco-iridoid pathway from Catharanthus roseus. Nat. Commun. 2014, 5, 3606. [Google Scholar] [CrossRef] [Green Version]
- Kantsa, A.; Raguso, R.A.; Dyer, A.G.; Sgardelis, S.P.; Olesen, J.M.; Petanidou, T. Community-wide integration of floral colour and scent in a Mediterranean scrubland. Nat. Ecol. Evol. 2017, 1, 1502–1510. [Google Scholar] [CrossRef] [PubMed]
- Kantsa, A.; Raguso, R.A.; Dyer, A.G.; Olesen, J.M.; Tscheulin, T.; Petanidou, T. Disentangling the role of floral sensory stimuli in pollination networks. Nat. Commun. 2018, 9, 1041. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, J.M.R.B.V.; Ferreira, G.S.; Sanches, P.A.; Bento, J.M.S.; Sazima, M. What pollinators see does not match what they smell: Absence of color-fragrance association in the deceptive orchid Ionopsis utricularioides. Phytochemistry 2021, 182, 112591. [Google Scholar] [CrossRef]
- Rasouli, O.; Ahmadi, N.; Monfared, S.R.; Sefidkon, F. Physiological, phytochemicals and molecular analysis of color and scent of different landraces of Rosa damascena during flower development stages. Sci. Hortic. 2018, 231, 144–150. [Google Scholar] [CrossRef]
- Park, P.H.; Ramya, M.; An, H.R.; Park, P.M.; Lee, S.Y. Breeding of Cymbidium ‘Sale Bit’ with bright yellow flowers and floral scent. Korean J. Breed. Sci. 2019, 51, 258–262. [Google Scholar] [CrossRef] [Green Version]
- Adebesin, F.; Widhalm, J.R.; Boachon, B.; Lefèvre, F.; Pierman, B.; Lynch, J.H.; Alam, I.; Junqueira, B.; Benke, R.; Ray, S.; et al. Emission of volatile organic compounds from petunia flowers is facilitated by an ABC transporter. Science 2017, 356, 1386–1388. [Google Scholar] [CrossRef] [Green Version]
- Liao., P.; Ray, S.; Boachon, B.; Lynch, J.H.; Deshpande, A.; McAdam, S.; Morgan, J.A.; Dudareva, N. Cuticle thickness affects dynamics of volatile emission from petunia flowers. Nat. Chem. Biol. 2021, 17, 138–145. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, Z.; Hu, H.; Shi, S.; Yuan, X.; Yan, B.; Chen, L. An Update on the Function, Biosynthesis and Regulation of Floral Volatile Terpenoids. Horticulturae 2021, 7, 451. https://doi.org/10.3390/horticulturae7110451
Qiao Z, Hu H, Shi S, Yuan X, Yan B, Chen L. An Update on the Function, Biosynthesis and Regulation of Floral Volatile Terpenoids. Horticulturae. 2021; 7(11):451. https://doi.org/10.3390/horticulturae7110451
Chicago/Turabian StyleQiao, Zhenglin, Huizhen Hu, Senbao Shi, Xuemei Yuan, Bo Yan, and Longqing Chen. 2021. "An Update on the Function, Biosynthesis and Regulation of Floral Volatile Terpenoids" Horticulturae 7, no. 11: 451. https://doi.org/10.3390/horticulturae7110451
APA StyleQiao, Z., Hu, H., Shi, S., Yuan, X., Yan, B., & Chen, L. (2021). An Update on the Function, Biosynthesis and Regulation of Floral Volatile Terpenoids. Horticulturae, 7(11), 451. https://doi.org/10.3390/horticulturae7110451







