Differential Assembly and Shifts of the Rhizosphere Bacterial Community by a Dual Transgenic Glyphosate-Tolerant Soybean Line with and without Glyphosate Application
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials, Field Design, and Sampling Methods
2.2. Basic Physicochemical Properties of Soils and Plants: Key Soil Enzyme Activities
2.3. Culturable Nitrogen-Fixing Bacteria in Rhizospheric Soil and Nodulation Effect of Soil Nitrogen-Fixing Bacteria
2.4. DNA Extraction from Soil Samples and PCR Amplification of 16S rDNA Amplicon Sequencing
2.5. OTU Analysis of 16S rDNA Amplicon Sequencing Data
2.6. Alpha Diversity, Beta Diversity, Taxonomy, Functional Analysis, and Statistical Analysis
3. Results
3.1. Basic Physicochemical Properties of Rhizospheric Soil and Plants
3.2. Key Enzyme Activities in Surrounding Soil and Rhizospheric Soil
3.3. Culturable Nitrogen-Fixing Bacteria and Nodulation Effect in Rhizospheric Soil
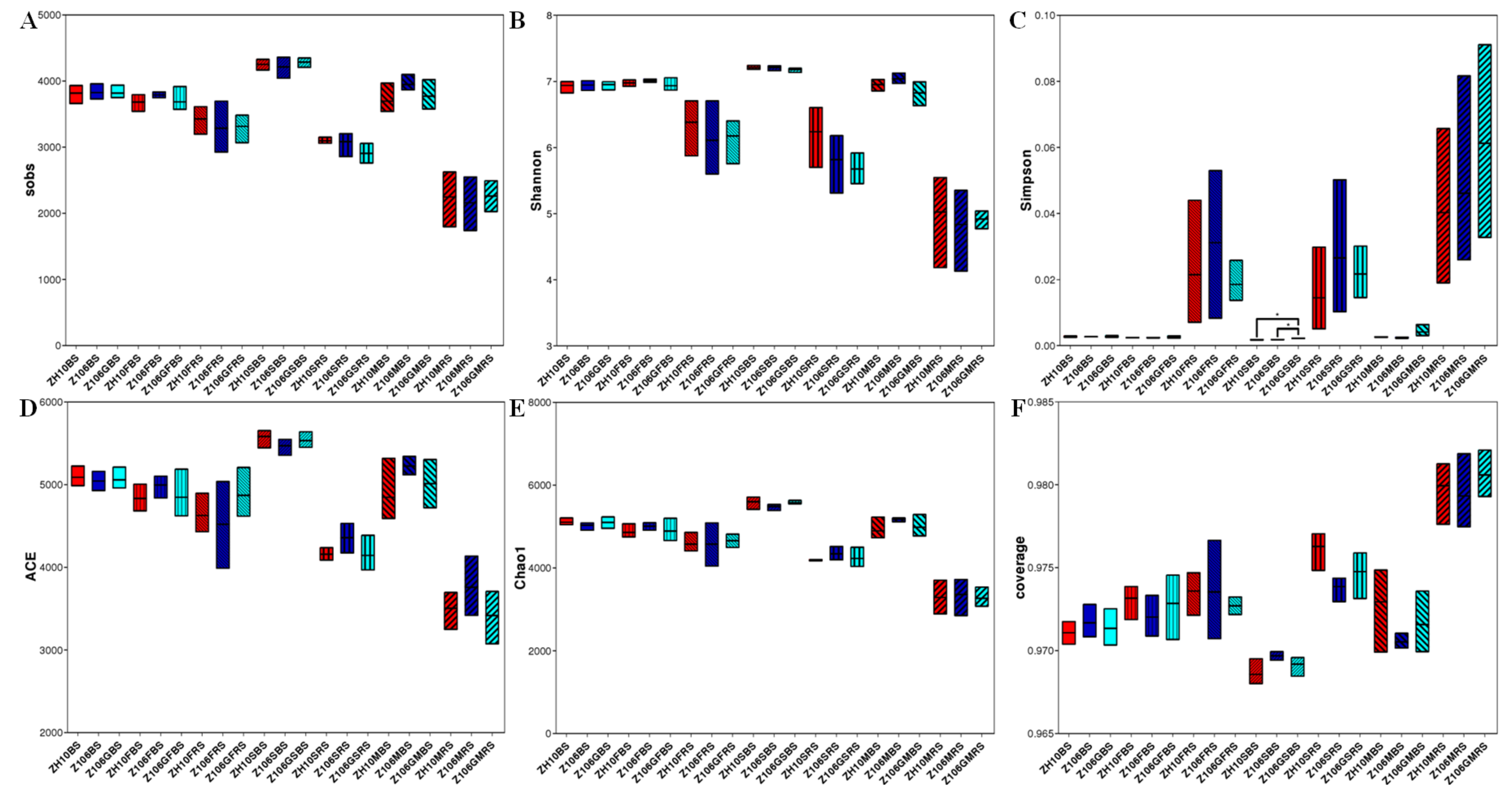
3.4. Alpha Diversity of Rhizosphere Bacterial Communities
3.5. Beta Diversity of Rhizosphere Bacterial Communities
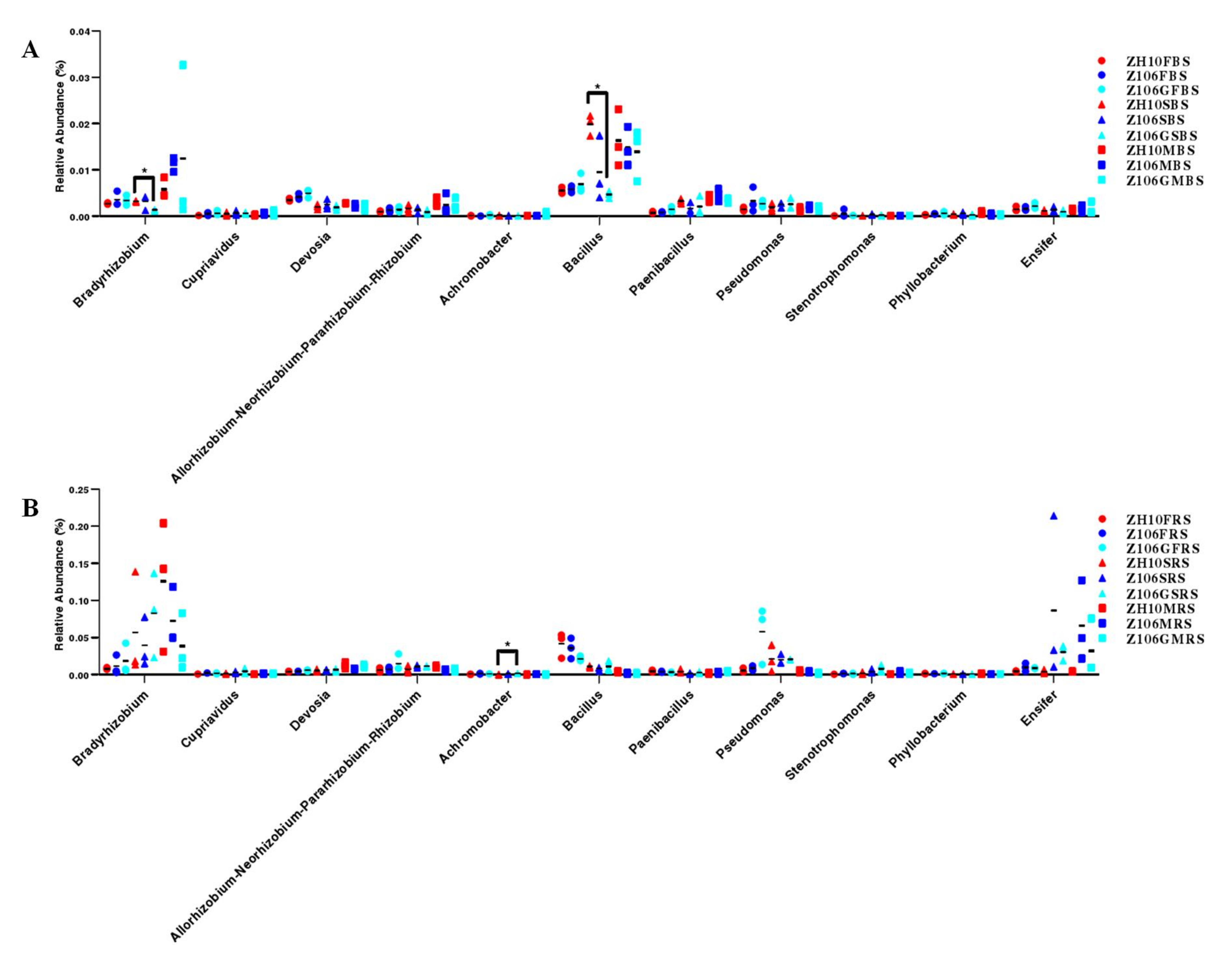
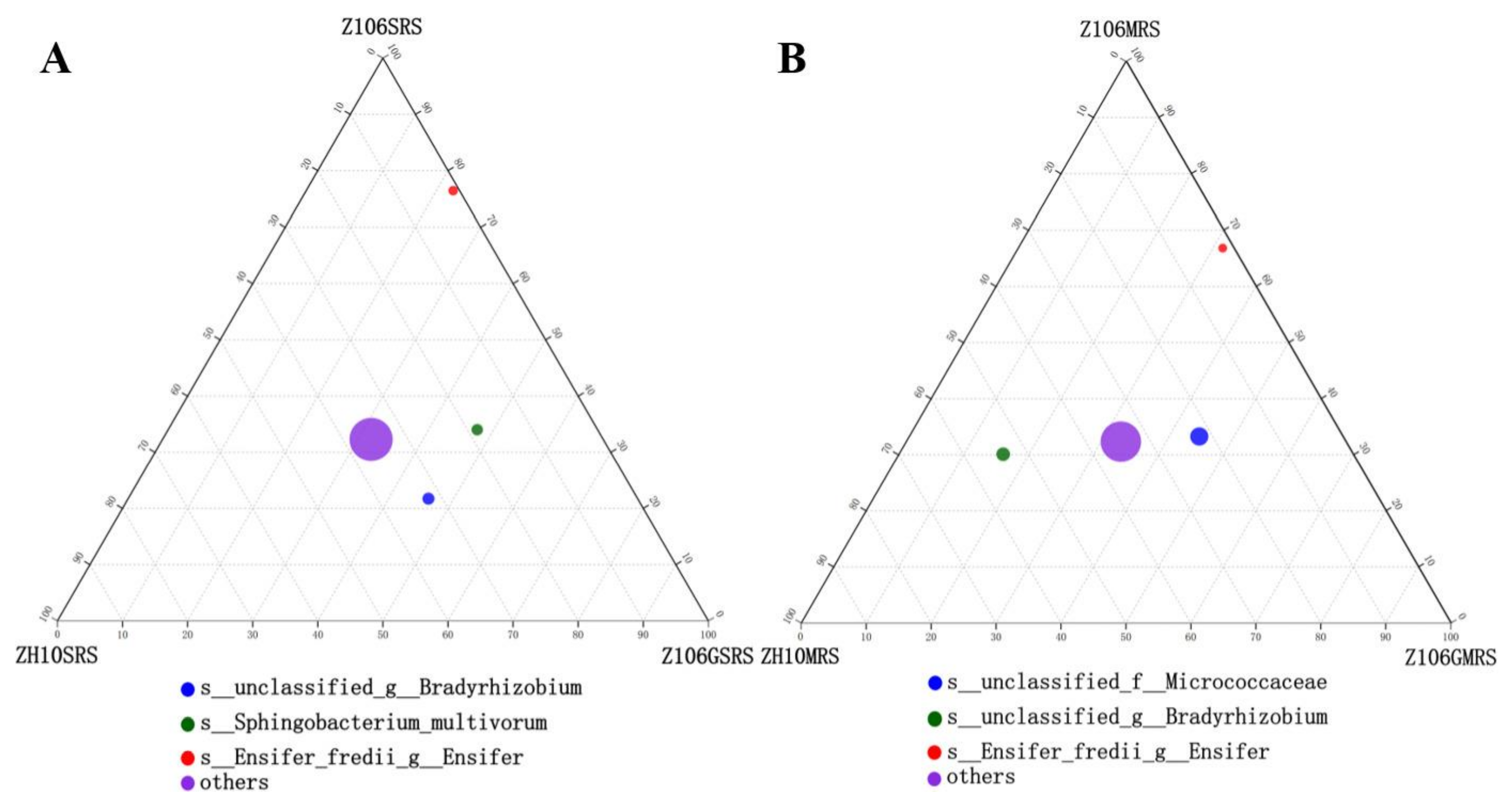
3.6. Taxonomic Analysis of Rhizosphere Bacterial Communities
3.7. Functional Composition of Microbial Community in Soil Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brookes, G.; Barfoot, P. GM crop technology use 1996–2018: Farm income and production impacts. GM Crop. Food 2020, 11, 242–261. [Google Scholar] [CrossRef] [PubMed]
- Brookes, G. Genetically modified (GM) crop use in Colombia: Farm level economic and environmental contributions. GM Crop. Food 2020, 11, 140–153. [Google Scholar] [CrossRef] [PubMed]
- ISAAA. Global Status of Commercialized Biotech/GM Crops in 2018: Biotech Crops Continue to Help Meet the Challenges of Increased Population and Climate Change. 2019. Available online: https://www.isaaa.org/resources/publications/briefs/54/default.asp (accessed on 30 March 2021).
- Feng, J.; Yang, F. The Regulation of Genetically Modified Food in China. Biotechnol. Law Rep. 2019, 38, 289–293. [Google Scholar] [CrossRef]
- Arpaia, S.; Christiaens, O.; Giddings, K.; Jones, H.; Mezzetti, B.; Moronta-Barrios, F.; Perry, J.N.; Sweet, J.B.; Taning, C.N.T.; Smagghe, G.; et al. Biosafety of GM Crop Plants Expressing dsRNA: Data Requirements and EU Regulatory Considerations. Front. Plant Sci. 2020, 11, 940. [Google Scholar] [CrossRef]
- Brookes, G.; Barfoot, P. Environmental impacts of genetically modified (GM) crop use 1996–2018: Impacts on pesticide use and carbon emissions. GM Crop. Food 2020, 11, 215–241. [Google Scholar] [CrossRef] [PubMed]
- Mertens, M.; Hoss, S.; Neumann, G.; Afzal, J.; Reichenbecher, W. Glyphosate, a chelating agent-relevant for ecological risk assessment? Environ. Sci. Pollut. Res. Int. 2018, 25, 5298–5317. [Google Scholar] [CrossRef] [Green Version]
- Landrigan, P.J.; Belpoggi, F. The need for independent research on the health effects of glyphosate-based herbicides. Environ. Health 2018, 17, 51. [Google Scholar] [CrossRef]
- Herman, R.A.; Ekmay, R.D.; Schafer, B.W.; Song, P.; Fast, B.J.; Papineni, S.; Shan, G.; Juberg, D.R. Food and feed safety of DAS-444O6-6 herbicide-tolerant soybean. Regul. Toxicol. Pharmacol. 2018, 94, 70–74. [Google Scholar] [CrossRef]
- Stenoien, C.; Nail, K.R.; Zalucki, J.M.; Parry, H.; Oberhauser, K.S.; Zalucki, M.P. Monarchs in decline: A collateral landscape-level effect of modern agriculture. Insect Sci. 2018, 25, 528–541. [Google Scholar] [CrossRef]
- Hoagland, R.E.; Duke, S.O. Biochemical Effects Of Glyphosate [N-(Phosphonomethyl)Glycine]. ACS Symp. Ser. 1982, 181, 175–205. [Google Scholar]
- Gomes, M.P.; Smedbol, E.; Chalifour, A.; Henault-Ethier, L.; Labrecque, M.; Lepage, L.; Lucotte, M.; Juneau, P. Alteration of plant physiology by glyphosate and its by-product aminomethylphosphonic acid: An overview. J. Exp. Bot. 2014, 65, 4691–4703. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Wen, Z.; Fazal, A.; Hua, X.; Xu, X.; Yin, T.; Qi, J.; Yang, R.; Lu, G.; Hong, Z.; et al. Impact of a G2-EPSPS & GAT Dual Transgenic Glyphosate-Resistant Soybean Line on the Soil Microbial Community under Field Conditions Affected by Glyphosate Application. Microbes Environ. 2020, 35, ME20056. [Google Scholar] [CrossRef]
- Duke, S.O.; Powles, S.B. Glyphosate: A once-in-a-century herbicide. Pest Manag. Sci. 2008, 64, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Guo, Y.; Hong, H.; Jin, L.; Zhang, L.; Chang, R.Z.; Lu, W.; Lin, M.; Qiu, L.J. Co-expression of G2-EPSPS and glyphosate acetyltransferase GAT genes conferring high tolerance to glyphosate in soybean. Front. Plant Sci. 2015, 6, 847. [Google Scholar] [CrossRef] [Green Version]
- Castle, L.A.; Siehl, D.L.; Gorton, R.; Patten, P.A.; Chen, Y.H.; Bertain, S.; Cho, H.J.; Duck, N.; Wong, J.; Liu, D.; et al. Discovery and directed evolution of a glyphosate tolerance gene. Science 2004, 304, 1151–1154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmalenberger, A.; Schwieger, F.; Tebbe, C.C. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 2001, 67, 3557–3563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckers, B.; Op De Beeck, M.; Thijs, S.; Truyens, S.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Performance of 16s rDNA Primer Pairs in the Study of Rhizosphere and Endosphere Bacterial Microbiomes in Metabarcoding Studies. Front. Microbiol. 2016, 7, 650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, S.; Jaggi, V.; Gangola, S.; Singh, A.; Sah, V.K.; Sahgal, M. Contrasting rhizosphere bacterial communities of healthy and wilted Dalbergia sissoo Roxb. forests. Rhizosphere 2021, 17, 100295. [Google Scholar] [CrossRef]
- Guo, B.; Guo, Y.; Hong, H.; Qiu, L.J. Identification of Genomic Insertion and Flanking Sequence of G2-EPSPS and GAT Transgenes in Soybean Using Whole Genome Sequencing Method. Front Plant Sci. 2016, 7, 1009. [Google Scholar] [CrossRef] [Green Version]
- Lu, G.H.; Hua, X.M.; Cheng, J.; Zhu, Y.L.; Wang, G.H.; Pang, Y.J.; Yang, R.W.; Zhang, L.; Shou, H.X.; Wang, X.M.; et al. Impact of Glyphosate on the Rhizosphere Microbial Communities of An EPSPS-Transgenic Soybean Line ZUTS31 by Metagenome Sequencing. Curr. Genom. 2018, 19, 36–49. [Google Scholar] [CrossRef]
- Lu, G.H.; Tang, C.Y.; Hua, X.M.; Cheng, J.; Wang, G.H.; Zhu, Y.L.; Zhang, L.Y.; Shou, H.X.; Qi, J.L.; Yang, Y.H. Effects of an EPSPS-transgenic soybean line ZUTS31 on root-associated bacterial communities during field growth. PLoS ONE 2018, 13, e0192008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, G.H.; Zhu, Y.L.; Kong, L.R.; Cheng, J.; Tang, C.Y.; Hua, X.M.; Meng, F.F.; Pang, Y.J.; Yang, R.W.; Qi, J.L.; et al. Impact of a Glyphosate-Tolerant Soybean Line on the Rhizobacteria, Revealed by Illumina MiSeq. J. Microbiol. Biotechnol. 2017, 27, 561–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inceoglu, O.; Salles, J.F.; van Overbeek, L.; van Elsas, J.D. Effects of plant genotype and growth stage on the betaproteobacterial communities associated with different potato cultivars in two fields. Appl. Environ. Microbiol. 2010, 76, 3675–3684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, Z.L.; Yang, M.K.; Du, M.H.; Zhong, Z.Z.; Lu, Y.T.; Wang, G.H.; Hua, X.M.; Fazal, A.; Mu, C.H.; Yan, S.F.; et al. Enrichments/Derichments of Root-Associated Bacteria Related to Plant Growth and Nutrition Caused by the Growth of an EPSPS-Transgenic Maize Line in the Field. Front. Microbiol. 2019, 10, 1335. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Faraway, J.J. Backscoring in Principal Coordinates Analysis. J. Comput. Graph. Stat. 2012, 21, 394–412. [Google Scholar] [CrossRef] [Green Version]
- Rohart, F.; Gautier, B.; Singh, A.; Le Cao, K.A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, N.E.; Ferry, M. ggtern: Ternary Diagrams Using ggplot2. J. Stat. Softw. 2018, 87, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Sui, Q.; Liu, C.; Zhang, J.; Dong, H.; Zhu, Z.; Wang, Y. Response of nitrite accumulation and microbial community to free ammonia and dissolved oxygen treatment of high ammonium wastewater. Appl. Microbiol. Biotechnol. 2016, 100, 4177–4187. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- de Luca, M.J.; Hungria, M. Plant densities and modulation of symbiotic nitrogen fixation in soybean. Sci. Agric. 2014, 71, 181–187. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.H.; Chen, W.H.; Wang, E.T.; Chen, W.F.; Yan, J.; Han, X.Z.; Tian, C.F.; Sui, X.H.; Singh, R.P.; Jiang, G.M.; et al. Rhizobial biogeography and inoculation application to soybean in four regions across China. J. Appl. Microbiol. 2018, 125, 853–866. [Google Scholar] [CrossRef]
- Han, Q.; Ma, Q.; Chen, Y.; Tian, B.; Xu, L.; Bai, Y.; Chen, W.; Li, X. Variation in rhizosphere microbial communities and its association with the symbiotic efficiency of rhizobia in soybean. ISME J. 2020, 14, 1915–1928. [Google Scholar] [CrossRef] [Green Version]
- Lane, M.; Lorenz, N.; Saxena, J.; Ramsier, C.; Dick, R.P. Microbial activity, community structure and potassium dynamics in rhizosphere soil of soybean plants treated with glyphosate. Pedobiologia 2012, 55, 153–159. [Google Scholar] [CrossRef]
- Albareda, M.; Rodriguez-Navarro, D.N.; Temprano, F.J. Use of Sinorhizobium (Ensifer) fredii for soybean inoculants in South Spain. Eur. J. Agron. 2009, 30, 205–211. [Google Scholar] [CrossRef]
- Krishnan, H.B. NolX of Sinorhizobium fredii USDA257, a type III-secreted protein involved in host range determination, Iis localized in the infection threads of cowpea (Vigna unguiculata [L.] Walp) and soybean (Glycine max [L.] Merr.) nodules. J. Bacteriol. 2002, 184, 831–839. [Google Scholar] [CrossRef] [Green Version]
- Temprano-Vera, F.; Rodriguez-Navarro, D.N.; Acosta-Jurado, S.; Perret, X.; Fossou, R.K.; Navarro-Gomez, P.; Zhen, T.; Yu, D.; An, Q.; Buendia-Claveria, A.M.; et al. Sinorhizobium fredii Strains HH103 and NGR234 Form Nitrogen Fixing Nodules With Diverse Wild Soybeans (Glycine soja) From Central China but Are Ineffective on Northern China Accessions. Front. Microbiol. 2018, 9, 2843. [Google Scholar] [CrossRef] [PubMed]
- Contador, C.A.; Lo, S.K.; Chan, S.H.J.; Lam, H.M. Metabolic Analyses of Nitrogen Fixation in the Soybean Microsymbiont Sinorhizobium fredii Using Constraint-Based Modeling. mSystems 2020, 5, e00516-19. [Google Scholar] [CrossRef] [Green Version]
- Canarini, A.; Kaiser, C.; Merchant, A.; Richter, A.; Wanek, W. Root Exudation of Primary Metabolites: Mechanisms and Their Roles in Plant Responses to Environmental Stimuli. Front. Plant Sci. 2019, 10, 157. [Google Scholar] [CrossRef] [Green Version]
- Meena, R.S.; Kumar, S.; Datta, R.; Lal, R.; Vijayakumar, V.; Brtnicky, M.; Sharma, M.P.; Yadav, G.S.; Jhariya, M.K.; Jangir, C.K.; et al. Impact of Agrochemicals on Soil Microbiota and Management: A Review. Land 2020, 9, 34. [Google Scholar] [CrossRef] [Green Version]
- Carini, P.; Marsden, P.J.; Leff, J.; Morgan, E.E.; Strickland, M.S.; Fierer, N. Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2017, 2, 16242. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Kuramae, E.E.; de Hollander, M.; Klinkhamer, P.G.; van Veen, J.A. Functional traits dominate the diversity-related selection of bacterial communities in the rhizosphere. ISME J. 2017, 11, 56–66. [Google Scholar] [CrossRef] [Green Version]

| Analysis | Trait | Flowering Stage (Mean ± SD) | Seed Filling Stage (Mean ± SD) | Maturing Stage (Mean ± SD) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ZH10 | Z106 | Z106G | ZH10 | Z106 | Z106G | ZH10 | Z106 | Z106G | ||
| Soil analysis | pH value | 7.4 ± 0.01 a | 7.29 ± 0.04 b | 7.42 ± 0.13 ab | 7.23 ± 0.23 | 7.3 ± 0.15 | 7.54 ± 0.09 | 7.34 ± 0.04 | 7.35 ± 0.06 | 7.36 ± 0.06 |
| Water content (%) | 17.46 ± 0.55 | 18.32 ± 1.16 | 17.1 ± 0.19 | 14.96 ± 0.93 | 14.05 ± 1.04 | 15.28 ± 1.8 | 16.79 ± 0.29 | 16.5 ± 1.11 | 16.98 ± 0.64 | |
| C content (%) | 0.95 ± 0.05 b | 1.07 ± 0.1 ab | 1.27 ± 0.1 a | 0.96 ± 0.07 | 1.06 ± 0.06 | 1.07 ± 0.12 | 1.19 ± 0.09 | 1.19 ± 0.04 | 1.19 ± 0.1 | |
| N content (%) | 0.12 b | 0.14 ± 0.01 a | 0.15 ± 0.02 a | 0.13 ± 0.01 | 0.1 ± 0.06 | 0.12 ± 0.02 | 0.16 ± 0.01 a | 0.14 b | 0.13 ± 0.01 b | |
| Plant analysis | C content (%) | 40.8 ± 0.06 | 40.83 ± 0.45 | 39.98 ± 0.86 | 41.81 ± 0.81 | 41.49 ± 0.9 | 41.73 ± 0.24 | 45.37 ± 0.42 | 43.45 ± 1.86 | 44.83 ± 1.31 |
| N content (%) | 4.26 ± 0.43 | 4.08 ± 0.42 | 4.07 ± 0.18 | 3.94 ± 0.09 | 4.0 ± 0.43 | 3.96 ± 0.05 | 3.86 ± 0.16 | 3.27 ± 1.59 | 4.11 ± 0.37 | |
| Sampling Compartment | Trait | Flowering Stage (Mean ± SD) | Seed Filling Stage (Mean ± SD) | Maturing Stage (Mean ± SD) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ZH10 | Z106 | Z106G | ZH10 | Z106 | Z106G | ZH10 | Z106 | Z106G | ||
| Surrounding Soil | S-UE (U/g) | 300.82 ± 97.93 | 372.35 ± 27.1 | 380.89 ± 65.81 | 363.27 ± 78.73 | 320.04 ± 86.32 | 417.71 ± 134.68 | 194.61 ± 72.24 | 166.85 ± 58.46 | 180.19 ± 69.84 |
| S-NR (U/g) | 112.48 ± 6.86 | 105.81 ± 4.41 | 109.95 ± 1.77 | 74.33 ± 10.98 | 96.76 ± 12.37 | 89.67 ± 2.33 | 118.43 ± 4.03 | 119.43 ± 3.44 | 124.29 ± 7.95 | |
| S-NiR (U/g) | 24.54 ± 3.01 | 22.7 ± 4.26 | 20.65 ± 4.39 | 39.25 ± 4.36 | 40.62 ± 3.56 | 40.27 ± 3.69 | 37.52 ± 5.12 | 42.15 ± 5.24 | 42.68 ± 4.84 | |
| S-SC (U/g) | 26.38 ± 2.38 | 25.35 ± 2.87 | 25.27 ± 3.45 | 27.16 ± 10.42 | 25.07 ± 2.71 | 25.74 ± 0.89 | 27.83 ± 2.29 | 27.23 ± 1.89 | 26.23 ± 3.01 | |
| Rhizospheric Soil | S-UE (U/g) | 317.9 ± 94.48 | 411.31 ± 77.3 | 370.74 ± 28.84 | 501.51 ± 52.7 a | 274.67 ± 72.19 b | 468.42 ± 138.5 ab | 267.73 ± 30.93 | 302.96 ± 85.23 | 326.44 ± 108.87 |
| S-NR (U/g) | 112.67 ± 10.24 | 129.71 ± 2.87 | 139.0 ± 14.29 | 123.24 ± 13.89 b | 155.33 ± 12.37 a | 120.29 ± 10.49 b | 157.33 ± 16.07 | 167.14 ± 15.95 | 184.76 ± 19.37 | |
| S-NiR (U/g) | 27.52 ± 11.17 | 23.56 ± 15.44 | 17.37 ± 13.94 | 38.21 ± 4.57 | 36.5 ± 2.41 | 32.28 ± 14.31 | 30.77 ± 2.66 | 40.98 ± 6.78 | 27.31 ± 11.14 | |
| S-SC (U/g) | 45.53 ± 4.1 | 44.71 ± 3.82 | 47.32 ± 3.58 | 42.57 ± 2.52 | 47.59 ± 8.22 | 55.49 ± 17.8 | 73.82 ± 3.4 c | 109.73 ± 10.1 a | 89.47 ± 1.98 b | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Wen, Z.; Hao, C.; Fazal, A.; Liao, Y.; Luo, F.; Yao, W.; Yin, T.; Yang, R.; Qi, J.; et al. Differential Assembly and Shifts of the Rhizosphere Bacterial Community by a Dual Transgenic Glyphosate-Tolerant Soybean Line with and without Glyphosate Application. Horticulturae 2021, 7, 374. https://doi.org/10.3390/horticulturae7100374
Yang M, Wen Z, Hao C, Fazal A, Liao Y, Luo F, Yao W, Yin T, Yang R, Qi J, et al. Differential Assembly and Shifts of the Rhizosphere Bacterial Community by a Dual Transgenic Glyphosate-Tolerant Soybean Line with and without Glyphosate Application. Horticulturae. 2021; 7(10):374. https://doi.org/10.3390/horticulturae7100374
Chicago/Turabian StyleYang, Minkai, Zhongling Wen, Chenyu Hao, Aliya Fazal, Yonghui Liao, Fuhe Luo, Weixuan Yao, Tongming Yin, Rongwu Yang, Jinliang Qi, and et al. 2021. "Differential Assembly and Shifts of the Rhizosphere Bacterial Community by a Dual Transgenic Glyphosate-Tolerant Soybean Line with and without Glyphosate Application" Horticulturae 7, no. 10: 374. https://doi.org/10.3390/horticulturae7100374
APA StyleYang, M., Wen, Z., Hao, C., Fazal, A., Liao, Y., Luo, F., Yao, W., Yin, T., Yang, R., Qi, J., Hong, Z., Lu, G., & Yang, Y. (2021). Differential Assembly and Shifts of the Rhizosphere Bacterial Community by a Dual Transgenic Glyphosate-Tolerant Soybean Line with and without Glyphosate Application. Horticulturae, 7(10), 374. https://doi.org/10.3390/horticulturae7100374








