Phylogenetics of Taxus Using the Internal Transcribed Spacers of Nuclear Ribosomal DNA and Plastid trnL-F Regions
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimens
2.2. DNA Extraction and Sequencing
2.3. Sequence Analysis and Phylogenetic Reconstruction
2.4. Leaf Impressions
3. Results
4. Discussion
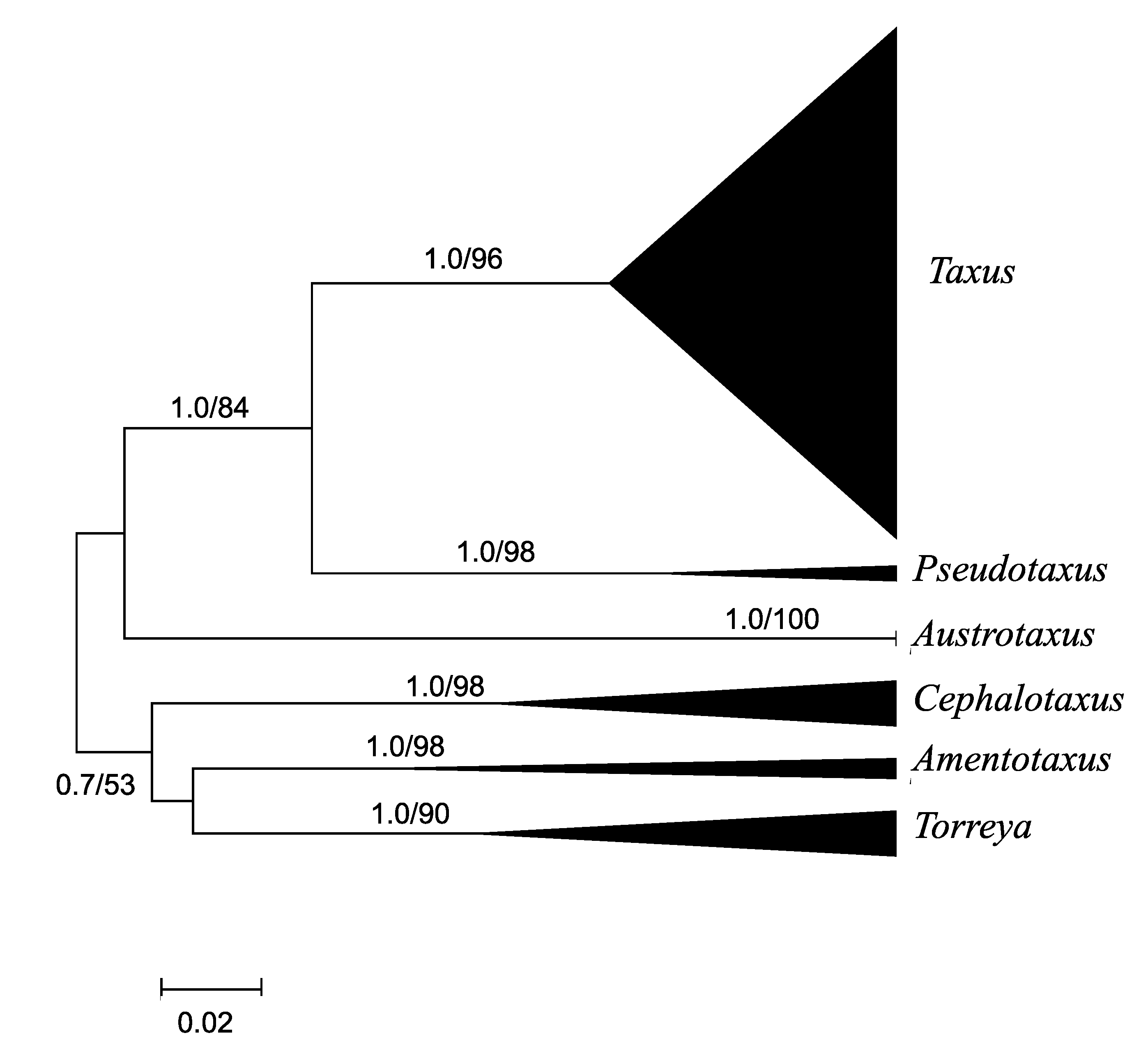
4.1. Taxaceae Phylogeny
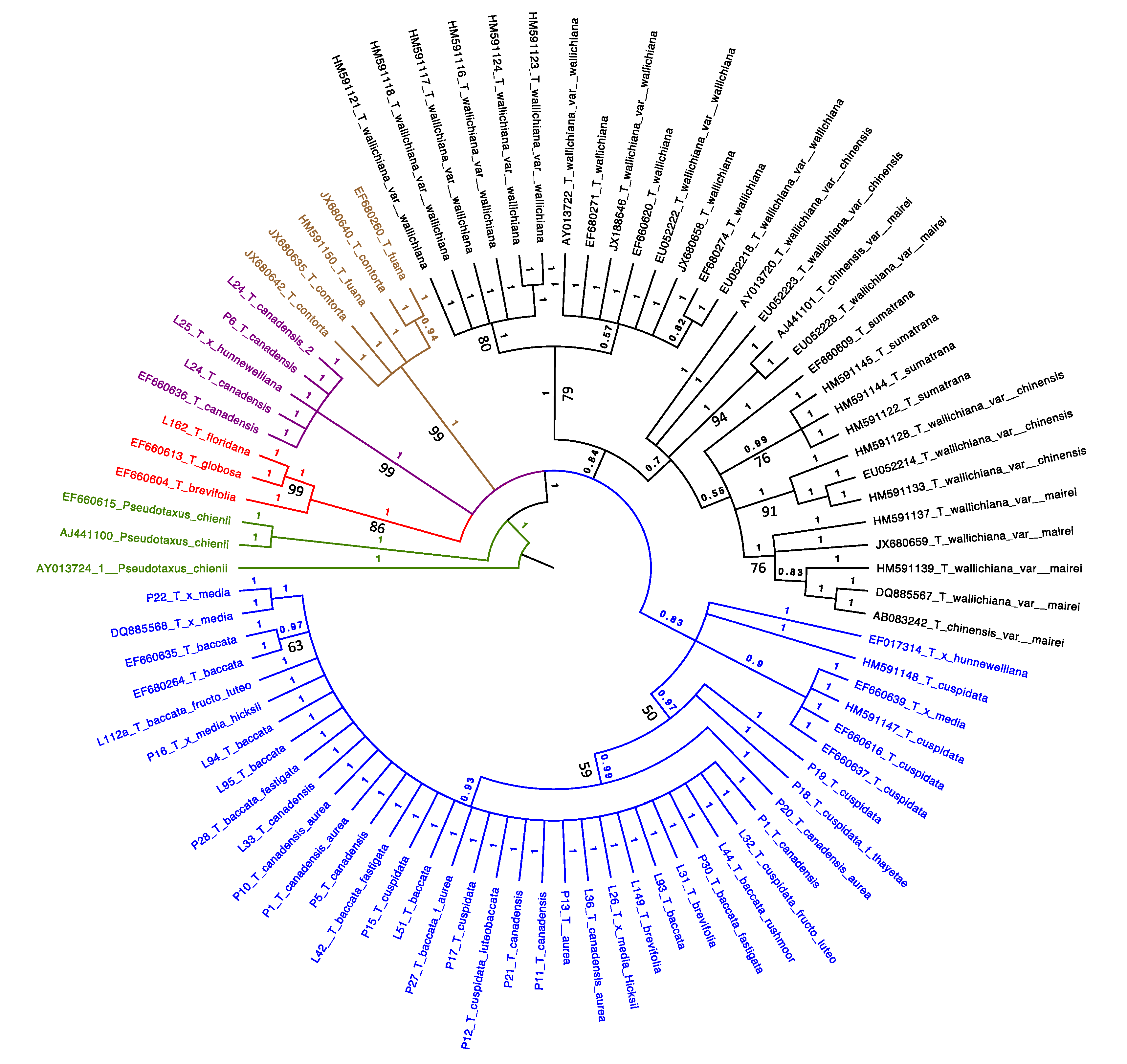
4.2. Taxus Phylogeny
4.3. Species Delimitation
4.4. Hybridization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Collins, D.; Mill, R.R.; Möller, M. Species separation of Taxus baccata, T. canadensis, and T. cuspidata (Taxaceae) and origins of their reputed hybrids inferred from RAPD and cpDNA data. Am. J. Bot. 2003, 90, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Pilger, R. Taxaceae; W. Engelmann: Leipzig, Germany, 1903. [Google Scholar]
- Elwes, H.J.; Henry, A. The Trees of Great Britain; SR Publishers Ltd.: Edinburgh, Scotland, 1906; Volume 1, pp. 98–126. [Google Scholar]
- Dempsey, D.; Hook, I. Yew (Taxus) species—Chemical and morphological variations. Pharm. Biol. 2000, 38, 274–280. [Google Scholar] [CrossRef]
- Dempsey, D. Factors Affecting Paclitaxel Content in Yew. Ph.D. Thesis, Trinity College Dublin, The University of Dublin, Dublin, Ireland, 2000. [Google Scholar]
- Cope, E.A. Taxaceae: The genera and cultivated species. Bot. Rev. 1998, 64, 291–322. [Google Scholar] [CrossRef]
- Farjon, A. World Checklist and Bibliography of Conifers, 2nd ed.; Royal Botanic Gardens, Kew: Richmond, UK, 1998. [Google Scholar]
- Spjut, R.W. A phytogeographical analysis of Taxus (Taxaceae) based on leaf anatomical characters. J. Bot. Res. Inst. Tex. 2007, 1, 291–332. [Google Scholar]
- The Plant List. Version 1.1. Published on the Internet. 2013. Available online: http://www.theplantlist.org/ (accessed on 3 September 2018).
- Li, J.; Davis, C.C.; Del Tredici, P.; Donoghue, M.J. Phylogeny and biogeography of Taxus (Taxaceae) inferred from sequences of the internal transcribed spacer region of nuclear ribosomal DNA. Harv. Pap. Bot. 2001, 6, 267–274. [Google Scholar]
- Appendino, G. The phytochemistry of the yew tree. Nat. Prod. Rep. 1995, 12, 349–360. [Google Scholar] [CrossRef]
- Lanker, U.; Malik, A.R.; Gupta, N.K.; Butola, J.S. Natural regeneration status of the endangered medicinal plant, Taxus baccata Hook. F. syn. T. wallichiana, in northwest Himalaya. Int. J. Biodivers. Sci. Ecosyst. Serv. Manag. 2010, 6, 20–27. [Google Scholar] [CrossRef]
- Thomas, P.; Farjon, A. Taxus wallichiana. The IUCN Red List of Threatened Species. 2011. Available online: http://dx.doi.org/10.2305/IUCN.UK.2011-2.RLTS.T46171879A9730085.en (accessed on 1 November 2018).
- Spector, T.; Thomas, P.; Determann, R. Taxus floridana. The IUCN Red List of Threatened Species. 2011. Available online: http://dx.doi.org/10.2305/IUCN.UK.2011-2.RLTS.T30965A9584928.en (accessed on 1 November 2018).
- Thomas, P.; Li, N.; Christian, T. Taxus chinensis. The IUCN Red List of Threatened Species. 2013. Available online: http://dx.doi.org/10.2305/IUCN.UK.2013-1.RLTS.T42548A2987120.en (accessed on 1 November 2018).
- Thomas, P. Taxus globosa. The IUCN Red List of Threatened Species. 2013. Available online: http://dx.doi.org/10.2305/IUCN.UK.2013-1.RLTS.T30724A2795235.en (accessed on 1 November 2018).
- Thomas, P. Taxus brevifolia. The IUCN Red List of Threatened Species. 2013. Available online: http://dx.doi.org/10.2305/IUCN.UK.2013-1.RLTS.T34041A2841142.en (accessed on 1 November 2018).
- Yang, L.; Christian, T.; Li, N. Taxus mairei. The IUCN Red List of Threatened Species. 2013. Available online: http://dx.doi.org/10.2305/IUCN.UK.2013-1.RLTS.T191659A1991533.en (accessed on 1 November 2018).
- Thomas, P. Taxus contorta. The IUCN Red List of Threatened Species. 2011. Available online: http://dx.doi.org/10.2305/IUCN.UK.2011-2.RLTS.T39147A10170545.en (accessed on 1 November 2018).
- Shah, A.; Li, D.-Z.; Gao, L.-M.; Li, H.-T.; Möller, M. Genetic diversity within and among populations of the endangered species Taxus fuana (Taxaceae) from Pakistan and implications for its conservation. Biochem. Syst. Ecol. 2008, 36, 183–193. [Google Scholar] [CrossRef]
- Chaw, S.M.; Long, H.; Wang, B.S.; Zharkikh, A.; Li, W.H. The phylogenetic position of Taxaceae based on 18S ribosomal—RNA sequences. J. Mol. Evol. 1993, 37, 624–630. [Google Scholar] [CrossRef]
- Cheng, Y.; Nicolson, R.G.; Tripp, K.; Chaw, S.-M. Phylogeny of Taxaceae and Cephalotaxaceae genera inferred from chloroplast matK gene and nuclear rDNA ITS region. Mol. Phylogenetics Evol. 2000, 14, 353–365. [Google Scholar] [CrossRef]
- Hao, D.C.; Huang, B.; Yang, L. Phylogenetic relationships of the genus Taxus inferred from chloroplast intergenic spacer and nuclear coding DNA. Biol. Pharm. Bull. 2008, 31, 260–265. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Appendino, G. Taxol (paclitaxel): Historical and ecological aspects. Fitoterapia 1993, 64, 2–25. [Google Scholar]
- Florin, R. Evolution in Cordaites and Conifers. Acta Horti Berginal 1951, 17, 259–388. [Google Scholar]
- Hartzell, H.J. The Yew Tree: A Thousand Whispers; Hulogosi: Eugene, OR, USA, 1991. [Google Scholar]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouve, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef]
- Sun, Y.; Skinner, D.Z.; Liang, G.H.; Hulbert, S.H. Phylogenetic analysis of sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theor. Appl. Genet. 1994, 89, 26–32. [Google Scholar] [CrossRef]
- Baldwin, B.G.; Sanderson, M.J.; Porter, J.M.; Wojciechowski, M.F.; Campbell, C.S.; Donoghue, M.J. The ITS region of nuclear ribosomal DNA—A valuable source of evidence on angiosperm phylogeny. Ann. Mo. Bot. Gard. 1995, 82, 247–277. [Google Scholar] [CrossRef]
- Wendel, J.F.; Schnabel, A.; Seelanan, T. An unusal ribosomal DNA-sequence from Gossypium gossypioides reveals ancient, cryptic, intergenomic introgression. Mol. Phylogenetics Evol. 1995, 4, 298–313. [Google Scholar] [CrossRef]
- Hodkinson, T.R.; Chase, M.W.; Takahashi, C.; Leitch, I.J.; Bennett, M.D.; Renvoize, S.A. The use of DNA sequencing (ITS and trnL-F), AFLP, and fluorescent in situ hybridization to study allopolyploid Miscanthus (Poaceae). Am. J. Bot. 2002, 89, 279–286. [Google Scholar] [CrossRef]
- Carolan, J.C.; Hook, I.L.; Chase, M.W.; Kadereit, J.W.; Hodkinson, T.R. Phylogenetics of Papaver and related genera based on DNA sequences from ITS nuclear ribosomal DNA and plastid trnL intron and trnL-F intergenic spacers. Ann. Bot. 2006, 98, 141–155. [Google Scholar] [CrossRef]
- Hao, D.C.; Xiao, P.G.; Huang, B.L.; Ge, G.B.; Yang, L. Interspecific relationships and origins of Taxaceae and Cephalotaxaceae revealed by partitioned Bayesian analyses of chloroplast and nuclear DNA sequences. Plant Syst. Evol. 2008, 276, 89–104. [Google Scholar] [CrossRef]
- Li, N.; Fu, L.K. Notes on gymnosperms 1. Taxonomic treatments of some Chinese conifers. Novon 1997, 7, 261–264. [Google Scholar]
- Farjon, A. Handbook of the World’s Conifers; EJ Brill: Leiden, The Netherlands, 2010; Volume 1, pp. 1–1111. [Google Scholar]
- Doyle, J.; Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Gawel, N.J.; Jarret, R.L. A modifies CTAB DNA extraction procedure for Musa and Ipomoea. Plant Mol. Biol. Report. 1991, 9, 262–266. [Google Scholar] [CrossRef]
- Hodkinson, T.R.; Waldren, S.; Parnell, J.A.N.; Kelleher, C.T.; Salamin, K.; Salamin, N. DNA banking for plant breeding, biotechnology and biodiversity evaluation. J. Plant Res. 2007, 120, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Liston, A.; Robinson, W.A.; Oliphant, J.M.; Alvarez-Buylla, E.R. Length variation in the nuclear ribosomal DNA internal transcribed spacer region of non-flowering seed plants. Syst. Bot. 1996, 21, 109–120. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA 7: Molecular Evolutionary Genetics Analysis, version 7 for bigger data sets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenback, J.P. MrBayes 3: Bayesian phylogenetic interence under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Salamin, N.; Hodkinson, T.R.; Savolainen, V. Towards building the tree of life: A simulation study for all angiosperm genera. Syst. Biol. 2005, 54, 183–196. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. Popart: Full-feature software for haplotype nextwork construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Clement, M.J.; Snell, Q.; Walker, P.; Posada, D.; Crandall, K.A. TCS: Estimating gene genealogies. In Proceedings of the 16th International Parallel and Distributed Processing Symposium, Fort Lauderdale, FL, USA, 1 April 2002; pp. 1–7. [Google Scholar]
- Sarvella, P.; Meyer, J.R.; Owings, A. A scotch tape method for counting or measuring stomata. Crop Sci. 1961, 1, 81–82. [Google Scholar] [CrossRef]
- Bandelt, H.-J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef] [PubMed]
- Christenhusz, M.J.M.; Reveal, J.L.; Farjon, A.; Gardner, M.F.; Mill, R.R.; Chase, M.W. A new classification and linear sequence of extant gymnosperms. Phytotaxa 2011, 19, 55–70. [Google Scholar] [CrossRef]
- Price, R.A. Generic and familial relationships of the Taxaceae from rbcL and matK sequence comparisons. In Proceedings of the Fourth International Conifer Conference: Conifers for the Future? Wye College, UK, 30 September 2003; pp. 235–237. [Google Scholar]
- Fu, L.; Li, N.; Mill, R.R. Taxaceae. Flora China 1999, 4, 89–96. [Google Scholar]
- Auders, A.G.; Spicer, D.P. Royal Horticultural Society Encyclopedia of Conifers: A Comprehensive Guide to Cultivars and Species; Royal Horticultural Society: London, UK, 2012. [Google Scholar]
- Farjon, A. Handbook of the World’s Conifers; EJ Brill: Leiden, The Netherlands, 2010; Volume 2. [Google Scholar]
- Eckenwalder, J.E. Conifers of the World: The Complete Reference; Timber Press: Portland, OR, USA, 2009. [Google Scholar]
- Elpe, C.; Knopf, P.; Stutzel, T.; Schulz, C. Diversity and evolution of leaf anatomical characters in Taxaceae s.l.-fluorescence microscopy reveals new delimitating characters. J. Plant Res. 2018, 131, 125–141. [Google Scholar] [CrossRef]
- Ghimire, B.; Heo, K. Cladistic analysis of Taxaceae sl. Plant Syst. Evol. 2014, 300, 217–223. [Google Scholar] [CrossRef]
- Shah, A.; Li, D.-Z.; Möller, M.; Gao, L.-M.; Hollingsworth, M.L.; Gibby, M. Delimitation of Taxus fuana Nan Li & RR Mill (Taxaceae) based on morphological and molecular data. Taxon 2008, 57, 211–222. [Google Scholar]
- Kelleher, C.T.; Hodkinson, T.R.; Kelly, D.L.; Douglas, G.C. Characterisation of chloroplast DNA haplotypes to reveal the provenance and genetic structure of oaks in Ireland. For. Ecol. Manag. 2004, 189, 123–131. [Google Scholar] [CrossRef]
- McGrath, S.; Hodkinson, T.R.; Barth, S. Extremely high cytoplasmic diversity in natural and breeding populations of Lolium (Poaceae). Heredity 2007, 99, 531–544. [Google Scholar] [CrossRef][Green Version]
- Feng, Y.; Oh, S.-H.; Manos, P.S. Phylogeny and historical biogeography of the genus Platanus as inferred from nuclear and chloroplast DNA. Syst. Bot. 2005, 30, 786–799. [Google Scholar] [CrossRef]
- Snow, B.; Snow, D. Birds Berries; A.&C Black: London, UK, 2010. [Google Scholar]
- Williamson, R. The Great Yew Forest: The Natural History of Kingley Vale; MacMillan: London, UK, 1978. [Google Scholar]
- Lavabre, J.E.; García, D. Geographic consistency in the seed dispersal patterns of Taxus baccata L. in the Iberian Peninsula. For. Syst. 2015, 24, e040. [Google Scholar] [CrossRef]
- Hultén, E.; Fries, M. Atlas of North European Vascular Plants North of the Tropic of Cancer; Koeltz Scientific: Parsberg, Germany, 1986. [Google Scholar]
- Liu, J.; Möller, M.; Gao, L.-M.; Zhang, D.-Q.; Li, D.-Z. DNA barcoding for the discrimination of Eurasian yews (Taxus L., Taxaceae) and the discovery of cryptic species. Mol. Ecol. Resour. 2011, 11, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Spjut, R.W. Taxonomy and nomenclature of Taxus (Taxaceae). J. Bot. Res. Inst. Tex. 2007, 1, 203–289. [Google Scholar]
- Elder, J.F., Jr.; Turner, B.J. Concerted evolution of repetitive DNA sequences in eukaryotes. Q. Rev. Biol. 1995, 70, 297–320. [Google Scholar] [CrossRef] [PubMed]
- Teerawatananon, A.; Jacobs, S.W.; Hodkinson, T.R. Phylogenetics of Panicoideae (Poaceae) based on chloroplast and nuclear DNA sequences. Telopea 2011, 13, 115–142. [Google Scholar] [CrossRef]
- Wendel, J.F.; Doyle, J.J. Phylogenetic Incongruence: Window into Genome History and Molecular Evolution, in Molecular Systematics of Plants II; Springer: Boston, MA, USA, 1998; pp. 265–296. [Google Scholar]
- McGuire, W.P.; Rowinsky, E.K.; Rosenshein, N.B.; Grumbine, F.C.; Ettinger, D.S.; Armstrong, D.K.; Donehower, R.C. Taxol: A unique antineoplastic agent with significant activity in advanced ovarian epithelial neoplasms. Ann. Intern. Med. 1989, 111, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Holmes, F.A.; Walters, R.S.; Theriault, R.L.; Forman, A.D.; Newton, L.K.; Raber, M.N.; Buzdar, A.U.; Frye, D.K.; Hortobagyi, G.N. Phase II Trial of Taxol, an active drug in the treatment of metastatic breast cancer. J. Natl. Cancer Inst. 1991, 83, 1797–1805. [Google Scholar] [CrossRef] [PubMed]
- Bristol Myers Squibb. Package Insert for TAXOL® (Paclitaxel) Injection; Bristol Myers Squibb: New York, NY, USA, 2011. [Google Scholar]
- Elkassaby, Y.A.; Yanchuk, A.D. Genetic diversity, differentiation, and genetic diversity, differentiation, and inbreeding in Pacific yew from British Columbia. J. Hered. 1994, 85, 112–117. [Google Scholar] [CrossRef]
- Kwit, C.; Horvitz, C.C.; Platt, W.J. Conserving slow-growing, long-lived tree species: Input from the demography of a rare understory conifer, Taxus floridana. Conserv. Biol. 2004, 18, 432–443. [Google Scholar] [CrossRef]
- Mohapatra, K.P.; Sehgal, R.N.; Sharma, R.K.; Mohapatra, T. Genetic analysis and conservation of endangered medicinal tree species Taxus wallichiana in the Himalayan region. New For. 2009, 37, 109–121. [Google Scholar] [CrossRef]
- Zhang, J.-T.; Ru, W. Population characteristics of endangered species Taxus chinensis var. mairei and its conservation strategy in Shanxi, China. Popul. Ecol. 2010, 52, 407–416. [Google Scholar] [CrossRef]



| Name | Sample No. | Voucher or Living Specimen | GenBank Code ITS; trnL + trnL-F+ |
|---|---|---|---|
| Amentotaxus formosana | L66 | Edinburgh 19763745 | MK748450 |
| Austrotaxus spicata | L164 | Edinburgh CAGNC*69 | MK748455 |
| Cephalotaxus harringtonia ‘Fastigiata’ | P8 | XX.006542 Glasnevin Coughlan P8 | MK116531; MK731931 |
| Cephalotaxus oliveri | L75 | Edinburgh 951689 | _ |
| Cephalotaxus sinensis | L68 | Edinburgh 19081025 | _ |
| Podocarpus macrophyllus | P26 | 2005.0135 Glasnevin Coughlan P26 | MK731953 |
| Podocarpus salignus | P23 | XX.007572 Glasnevin Coughlan P23 | MK731946 |
| Taxus ‘Aurea’ | P13 | XX.006570 Glasnevin Coughlan P13 | MK731936 |
| T. baccata | L10 | Ranelagh Park, Dublin | MK783697 |
| T. baccata | L51 | Bedgebury 13/025 | MK783706 |
| T. baccata | L93 | Cornalack Lough Derg s.n. | MK748451 |
| T. baccata | L95 | Clorhane s.n. | MK748453 |
| T. baccata | T1 | Physic garden, TCD, s.n. | _ |
| T. baccata | L94 | Cornalack lough Derg s.n. | MK748452 |
| T. baccata ‘Fastigiata’ | P28 | 13638 Glasnevin Coughlan P28 | MK211157; MK731950 |
| T. baccata ‘Fructo Luteo’ | L112 | Mount Usher s.n. | _ |
| T. baccata ‘Amersfort’ | L108 | Mount Usher s.n. | _ |
| T. baccata ‘Fastigiata’ | P30 | Glasnevin Cemetery Coughlan P30 | MK731952 |
| T. baccata ‘Fastigiata’ | L42 | Bedgebury 13/108 | MK783704 |
| T. baccata ‘Fastigiata’ | L106 | Florence Court s.n. | _ |
| T. baccata ‘Grandis’ | P29 | Glasnevin Coughlan P29 | MK731951 |
| T. baccata ‘Rushmoor’ | L44 | Bedgebury 15/0287 | MK783705 |
| T. baccata f. aurea | P27 | XX.006574 Glasnevin Coughlan P27 | MK211154, MK211155, MK211156; MK731949 |
| T. brevifolia | P14 | 1885.006579 Glasnevin Coughlan P14 | MK123470; MK731937 |
| T. brevifolia | L31 | Glasnevin 1885.006579 | MK783701 |
| T. brevifolia | L149 | JFK Arboretum, New Ross 33 | MK748454 |
| T. canadensis | P5 | JFK Hook s.n. | MK116529; MK731929 |
| T. canadensis | P6 | Canada Hook s.n. | MK116530; MK731930 |
| T. canadensis | P11 | XX.006556 Glasnevin Coughlan P11 | MK168613, MK168614, MK168615; MK731934 |
| T. canadensis | P21 | XX.005502 Glasnevin Coughlan P21 | Mk21148, MK211149, MK211150; MK731944 |
| T. canadensis | L24 | Bedgebury 14/94 | MK272737; MK748448 |
| T. canadensis | L33 | Glasnevin XX.006556 | MK272738; MK748449 |
| T. canadensis | L145 | JFK Arboretum, New Ross 8.M.5 | _ |
| T. canadensis | L165 | Wendall Massachusetts s.n. | _ |
| T. canadensis | TH1 | Quebec, Canada | MK685277 |
| T. canadensis | TH2 | Quebec, Canada | MK685278 |
| T. canadensis ‘Aurea’ | P2 | Glasnevin Hook s.n. | MK123469; MK731928 |
| T. canadensis ‘Aurea’ | P10 | XX.006549 Glasnevin Coughlan P10 | MK168610, MK168611, MK168612; MK731933 |
| T. canadensis ‘Aurea’ | P20 | XX.006609 Glasnevin Coughlan P20 | MK731943 |
| T. canadensis ‘Aurea’ | L36 | Glasnevin XX.006549 | MK272740; MK783703 |
| T. cuspidata | L27 | Bedgebury 13/136 | MK783700 |
| T. cuspidata | P3 | JFK Hook s.n. | Mk116528; |
| T. cuspidata | P15 | XX.006597 Glasnevin Coughlan P15 | MK123471; MK731938 |
| T. cuspidata | P17 | 1911.006591 Glasnevin Coughlan P17 | MK123473; MK731940 |
| T. cuspidata | P19 | XX.006601 Glasnevin Coughlan P19 | MK168784, MK168785, MK168786; MK731942 |
| T. cuspidata ‘Fructo luteo’ | L32 | Glasnevin 12/30 Kew | MK783702 |
| T. cuspidata f. thayerae | P9 | 1952.006540 Glasnevin Coughlan P9 | MK168608, MK168609; MK731932 |
| T. cuspidata f. thayerae | P18 | 1952.006593 Glasnevin Coughlan P18 | MK168781, MK168782, MK168783; MK731941 |
| T. cuspidata var. luteobaccata | P12 | 1930.006571 Glasnevin Coughlan P12 | MK168616, MK168617, MK168618; MK731935 |
| T. floridana | L162 | Smith College Arboretum PULOG 7 | MK748456 |
| T. ×media ‘Hatfieldii’ | L40 | Bedgebury 13/280 | _ |
| Taxus canadensis | P1 | Glasnevin Hook s.n. | MK116527; MK731927 |
| Taxus ×hunnewelliana | L25 | Bedgebury 13/125 | MK783698 |
| Taxus ×media ‘Cuftoni’ | P22 | XX.007572 Glasnevin Coughlan P22 | MK211151; MK731945 |
| Taxus ×media ‘Hicksii’ | P16 | XX.006589 Glasnevin Coughlan P16 | MK123472; MK731939 |
| Taxus ×media ‘Hicksii’ | L26 | Bedgebury 13/006 | MK783699 |
| Torreya californica | P24 | 7746 Glasnevin Coughlan P24 | MK211152; MK731947 |
| Torreya jackii | L73 | Edinburgh 19970112 | _ |
| Torreya nucifera | L34 | Glasnevin 7734 | MK272739 |
| Unlabelled Torreya sp. | P25 | XX.007599 Glasnevin Coughlan P25 | MK211153; MK731948 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coughlan, P.; Carolan, J.C.; Hook, I.L.I.; Kilmartin, L.; Hodkinson, T.R. Phylogenetics of Taxus Using the Internal Transcribed Spacers of Nuclear Ribosomal DNA and Plastid trnL-F Regions. Horticulturae 2020, 6, 19. https://doi.org/10.3390/horticulturae6010019
Coughlan P, Carolan JC, Hook ILI, Kilmartin L, Hodkinson TR. Phylogenetics of Taxus Using the Internal Transcribed Spacers of Nuclear Ribosomal DNA and Plastid trnL-F Regions. Horticulturae. 2020; 6(1):19. https://doi.org/10.3390/horticulturae6010019
Chicago/Turabian StyleCoughlan, Patricia, James C. Carolan, Ingrid L. I. Hook, Lisa Kilmartin, and Trevor R. Hodkinson. 2020. "Phylogenetics of Taxus Using the Internal Transcribed Spacers of Nuclear Ribosomal DNA and Plastid trnL-F Regions" Horticulturae 6, no. 1: 19. https://doi.org/10.3390/horticulturae6010019
APA StyleCoughlan, P., Carolan, J. C., Hook, I. L. I., Kilmartin, L., & Hodkinson, T. R. (2020). Phylogenetics of Taxus Using the Internal Transcribed Spacers of Nuclear Ribosomal DNA and Plastid trnL-F Regions. Horticulturae, 6(1), 19. https://doi.org/10.3390/horticulturae6010019





