Identification of Differentially Expressed Genes between “Honeycrisp” and “Golden Delicious” Apple Fruit Tissues Reveal Candidates for Crop Improvement
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Strategy
2.2. Differential Display
2.3. Cloning and Sequencing
2.4. Annotation of Differential Display Fragments
2.5. Quantitative Real Time PCR Validation of Differential Expression
2.6. Comparison of Expression with Previous Studies
3. Results and Discussion
3.1. Differential Display
3.2. Quantitative Reverse Transcription PCR Validation of Differential Expression
3.3. Comparison of Expression with Previous Studies
3.4. Differentially Expressed Homologs in Heterologous Systems
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Janssen, B.J.; Thodey, K.; Schaffer, R.J.; Alba, R.; Balakrishnan, L.; Bishop, R.; Bowen, J.H.; Crowhurst, R.N.; Gleave, A.P.; Ledger, S.; et al. Global gene expression analysis of apple fruit development from the floral bud to ripe fruit. BMC Plant Biol. 2008. [Google Scholar] [CrossRef] [PubMed]
- Kupferman, E. The Role of Ethylene in Determining Apple Harvest and Storage Life. Available online: http://postharvest.tfrec.wsu.edu/pages/N4I1C (accessed on 4 March 2016).
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acid Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Nosarzewski, M.; Archbold, D.D. Tissue-specific expression of SORBITOL DEHYDROGENASE in apple fruit during early development. J. Exp. Bot. 2007, 58, 1863–1872. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, W.; Xin, L.; Gao, Z.; Hou, Y.; Yu, X.; Zhang, Z.; Qu, S. Genomic variants of genes associated with three horticultural traits in apple revealed by genome re-sequencing. Hortic. Res. 2014, 1, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zsolt, A.; Deák, C.; Miskó, A.; Tóth, M.; Papp, I. Development of cDNA normalization system and preliminary transcription analysis of KCS genes in apple tissues. Acta Univ. Agric. Silvic. Mendel. Brun. 2014, 59, 9–12. [Google Scholar]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.B.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acid Res. 2009, 37, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acid Res. 2001, 29, 1–6. [Google Scholar] [CrossRef]
- Dixon, J.; Hewett, E.W. Factors affecting apple aroma/flavour volatile concentration: A Review. N. Z. J. Crop Hortic. Sci. 2000, 28, 155–173. [Google Scholar] [CrossRef]
- Goulao, L.F.; Santos, J.; de Sousa, I.; Oliveira, C.M. Patterns of enzymatic activity of cell wall-modifying enzymes during growth and ripening of apples. Postharvest Biol. Technol. 2007, 43, 307–318. [Google Scholar] [CrossRef]
- Solovchenko, A.E.; Merzlyak, M.N.; Pogosyan, S.I. Light-induced decrease of reflectance provides an insight in the photoprotective mechanisms of ripening apple fruit. Plant Sci. 2010, 178, 281–288. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Crifò, T.; Puglisi, I.; Petrone, G.; Recupero, G.R.; Piero, A.R.L. Expression analysis in response to low temperature stress in blood oranges: Implication of the flavonoid biosynthetic pathway. Gene 2011, 476, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.J.; Vogel, M.O.; Viehhauser, A. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 2010, 245, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.Y.; Vrebalov, J.; Alba, R.; Lee, J.; McQuinn, R.; Chung, J.D.; Klein, P.; Giovannoni, J. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 2010, 64, 936–947. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Xu, C.-J.; Li, X.; Ferguson, I.; Chen, K. Accumulation of lignin in relation to change in activities of lignification enzymes in loquat fruit flesh after harvest. Postharvest Biol. Technol. 2006, 40, 163–169. [Google Scholar] [CrossRef]
- Rose, J.K.C.; Bennett, A.B. Cooperative disassembly of the cellulose–xyloglucan network of plant cell walls: Parallels between cell expansion and fruit ripening. Trends Plant Sci. 1999, 4, 176–183. [Google Scholar] [CrossRef]
- Louveau, T.; Leitao, C.; Green, S.; Hamiaux, C.; van der Rest, B.; Dechy-Cabaret, O.; Atkinson, R.G.; Chervin, C. Predicting the substrate specificity of a glycosyltransferase implicated in the production of phenolic volatiles in tomato fruit. FEBS J. 2011, 278, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Puupponen-Pimiä, R.; Nohnek, L.; Meier, C.; Kähkönen, M.; Heinonen, M.; Hopia, A.; Oksman-Caldentey, K.M. Antimicrobial properties of phenolic compounds from berries. J. Appl. Microbiol. 2001, 90, 494–507. [Google Scholar] [CrossRef] [PubMed]
- Flachowsky, H.; Halbwirth, H.; Treutter, D.; Richter, K.; Hanke, M.V.; Szankowski, I.; Gosch, C.; Stich, K.; Fischer, T.C. Silencing of flavanone-3-hydroxylase in apple (Malus × domestica Borkh.) leads to accumulation of flavanones, but not to reduced fire blight susceptibility. Plant Physiol. Biochem. 2012, 51, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Çakir, B.; Agasse, A.; Gaillard, C.; Saumonneau, A.; Delrot, S.; Atanassova, R. A grape ASR protein involved in sugar and abscisic acid signaling. Plant Cell 2003, 15, 2165–2180. [Google Scholar] [CrossRef] [PubMed]
- Golan, I.; Guadalupe, P.; Dominguez, Z.K.; Shkolnik-Inbar, D.; Carrari, F.; Bar-Zvi, D. Tomato ABSCISIC ACID STRESS RIPENING (ASR) gene family revisited. PLoS ONE 2014, 9, e107117. [Google Scholar]
- Eubel, H.; Meyer, E.H.; Taylor, N.L.; Bussell, J.D.; O’Toole, N.O.; Heazlewood, J.L.; Castleden, I.; Small, I.D.; Smith, S.M.; Millar, A.H. Novel Proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol. 2008, 148, 1809–1829. [Google Scholar] [CrossRef] [PubMed]
- Kamada, T.; Nito, K.; Hayashi, H.; Mano, S.; Hayashi, M.; Nishimura, M. Functional differentiation of peroxisomes revealed by expression profiles of peroxisomal genes in Arabidopsis thaliana. Plant Cell Physiol. 2003, 44, 1275–1289. [Google Scholar] [CrossRef] [PubMed]
- Wiszniewski, A.A.G.; Zhou, W.; Smith, S.M.; Bussell, J.D. Identification of two Arabidopsis genes encoding a peroxisomal oxidoreductase-like protein and an acyl-CoA synthetase-like protein that are required for responses to pro-auxins. Plant Mol. Biol. 2009, 69, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Goda, H.; Sawa, S.; Asami, T.; Fujioka, S.; Shimada, Y.; Yoshida, S. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2004, 134, 1555–1573. [Google Scholar] [CrossRef] [PubMed]
- Schweighofer, A.; Hirt, H.; Meskiene, I. Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 2004, 9, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Joshi, D.; Yadav, P.K.; Gupta, A.K.; Bhatt, T.K. Role of ubiquitin-mediated degradation system in plant biology. Front. Plant Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Ascencio-Ibanez, J.T.; Sozzani, R.; Lee, T.J.; Chu, T.M.; Wolfinger, R.D.; Cella, R.; Hanley-Bowdoin, L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008, 148, 436–454. [Google Scholar] [CrossRef] [PubMed]
- Sierla, M.E.; Feys, B.J. Characterization of Arabidopsis thaliana orthologues of GAAP, a Golgi-localized anti-apoptotic protein. In Proceedings of the 20th International Conference on Arabidopsis Research, Edinburgh, UK, 30 June–4 July 2009.
- Cordle, A.R.; Irish, E.E.; Cheng, C.L. Gene expression associated with apogamy commitment in Ceratopteris richardii. Sex. Plant Reprod. 2012, 25, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Oxidant and antioxidant signalling in plants: A re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ. 2005, 28, 1056–1071. [Google Scholar] [CrossRef]
- Catoni, E.; Desimone, M.; Hilpert, M.; Wipf, D.; Kunze, R.; Schneider, A.; Flügge, U.I.; Schumacher, K.; Frommer, W.B. Expression pattern of a nuclear encoded mitochondrial arginine-ornithine translocator gene from Arabidopsis. BMC Plant Biol. 2003. [Google Scholar] [CrossRef]
- Picault, N. Plant mitochondria: From genome to function. In Plant Mitochondrial Carriers; Day, D.A., Millar, A.H., Whelan, J., Eds.; Kluwer Academic Publisher: Dordrecht, The Netherlands, 2004; Volume 17. [Google Scholar]
- Solomon, M.; Belenghi, B.; Delledonne, M.; Menachem, E.; Levine, A. The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 1999, 11, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Krüger, J.; Thomas, C.M.; Golstein, C.; Dixon, M.S.; Smoker, M.; Tang, S.; Mulder, L.; Jones, J.D.G. A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 2002, 296, 744–747. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.; Alba, R.; Schouten, H.; Soglio, V.; Gianfranceschi, L.; Serra, S.; Musacchi, S.; Sansavini, S.; Costa, G.; Fei, Z.; et al. Use of homologous and heterologous gene expression profiling tools to characterize transcription dynamics during apple fruit maturation and ripening. BMC Plant Biol. 2010, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Gosch, G.; Halbwirth, H.; Schneider, B.; Hölscher, D.; Stich, K. Cloning and heterologous expression of glycosyltransferases from Malus × domestica and Pyrus communis, which convert phloretin to phloretin 2′-O-glucoside (phloridzin). Plant Sci. 2010, 178, 299–306. [Google Scholar] [CrossRef]
- Blanco-Portales, R.; Medina-Escobar, N.; López-Ráez, J.A.; González-Reyes, J.A.; Villalba, J.M.; Moyano, E.; Caballero, J.L.; Muñoz-Blanco, J. Cloning, expression and immunolocalization pattern of a cinnamyl alcohol dehydrogenase gene from strawberry (Fragaria × ananassa cv. Chandler). J. Exp. Bot. 2002, 53, 1723–1734. [Google Scholar] [CrossRef] [PubMed]
- Salentijn, E.M.J.; Aharoni, A.; Schaart, J.G.; Boone, M.J.; Krens, F.A. Differential gene expression analysis of strawberry cultivars that differ in fruit firmness. Physiol. Plant. 2003, 118, 571–578. [Google Scholar] [CrossRef]
- Redgwell, R.J.; Fry, S.C. Xyloglucan endotransglycosylase activity increases during kiwifruit (Actinidia deliciosa) ripening (Implications for fruit softening). Plant Physiol. 1993, 103, 1399–1406. [Google Scholar] [PubMed]
- Cutillas-Iturralde, A.; Zarra, I.; Fry, S.C.; Lorences, E.P. Implication of persimmon fruit hemicellulose metabolism in the softening process. Importance of xyloglucan endotransglycosylase. Physiol. Plant. 2006, 91, 169–176. [Google Scholar] [CrossRef]
- Gutiérrez-Larraínzara, M.; Rúaa, J.; Carob, I.; de Castroa, C.; de Arriagaa, D.; García-Armestob, M.R.; del Valle, P. Evaluation of antimicrobial and antioxidant activities of natural phenolic compounds against foodborne pathogens and spoilage bacteria. Food Control 2012, 26, 555–563. [Google Scholar] [CrossRef]
- Osorio, S.; Scossa, F.; Fernie, A.R. Molecular regulation of fruit ripening. Front. Plant Sci. 2013, 198, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pattison, R.J.; Csukasi, F.; Catalá, C. Mechanisms regulating auxin action during fruit development. Physiol. Plant. 2014, 151, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Watkins, C.B.; Nock, J.F.; Weis, S.A.; Jayanty, S.; Beaudry, R.M. Storage temperature, diphenylamine, and pre-storage delay effects on soft scald, soggy breakdown and bitter pit of “Honeycrisp” apples. Postharvest Biol. Technol. 2004, 32, 213–221. [Google Scholar] [CrossRef]
- Cherian, S.; Figueroa, C.R.; Nair, H. “Movers and Shakers” in the regulation of fruit ripening: A cross-dissection of climacteric versus non-climacteric fruit. J. Exp. Bot. 2014, 65, 4705–4722. [Google Scholar] [CrossRef] [PubMed]
- Ireland, H.S.; Yao, J.-L.; Tomes, S.; Sutherland, P.W.; Nieuwenhuizen, N.; Gunaseelan, K.; Winz, R.A.; David, K.M.; Schaffer, R.J. Apple SEPALLATA1/2-like genes control fruit flesh development and ripening. Plant J. 2013, 73, 1044–1056. [Google Scholar] [CrossRef] [PubMed]
- Soria-Guerra, R.E.; Rosales-Mendoza, S.; Gasic, K.; Wisniewski, M.E.; Band, M.; Korban, S.S. Gene expression is highly regulated in early developing fruit of apple. Plant Mol. Biol. Rep. 2011, 29, 885–897. [Google Scholar] [CrossRef]
- Zhu, Y.; Zheng, P.; Varanasi, V.; Shin, S.; Main, D.; Curry, E.; Mattheis, J.P. Multiple plant hormones and cell wall metabolism regulate apple fruit maturation patterns and texture attributes. Tree Genet. Genomes 2012, 8, 1389–1406. [Google Scholar] [CrossRef]
- Draghici, S.; Khatri, P.; Eklund, A.C.; Szallasi, Z. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 2006, 22, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Fung-Leung, W.-P.; Bittner, A.; Ngo, K.; Liu, X. Comparison of RNA-seq and microarray in transcriptome profiling of activated T cells. PLoS ONE 2014, 9, e78644. [Google Scholar] [CrossRef] [PubMed]
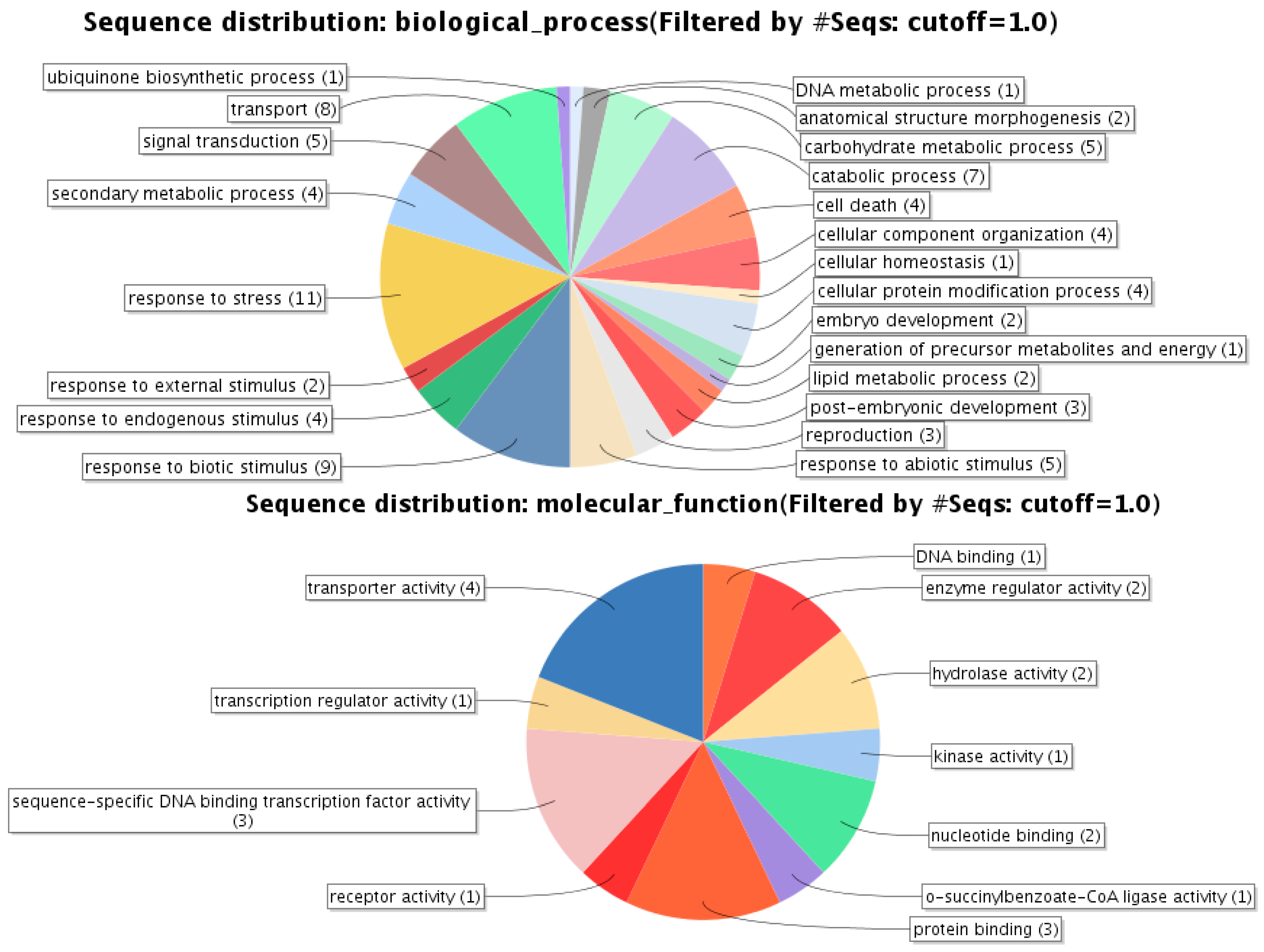
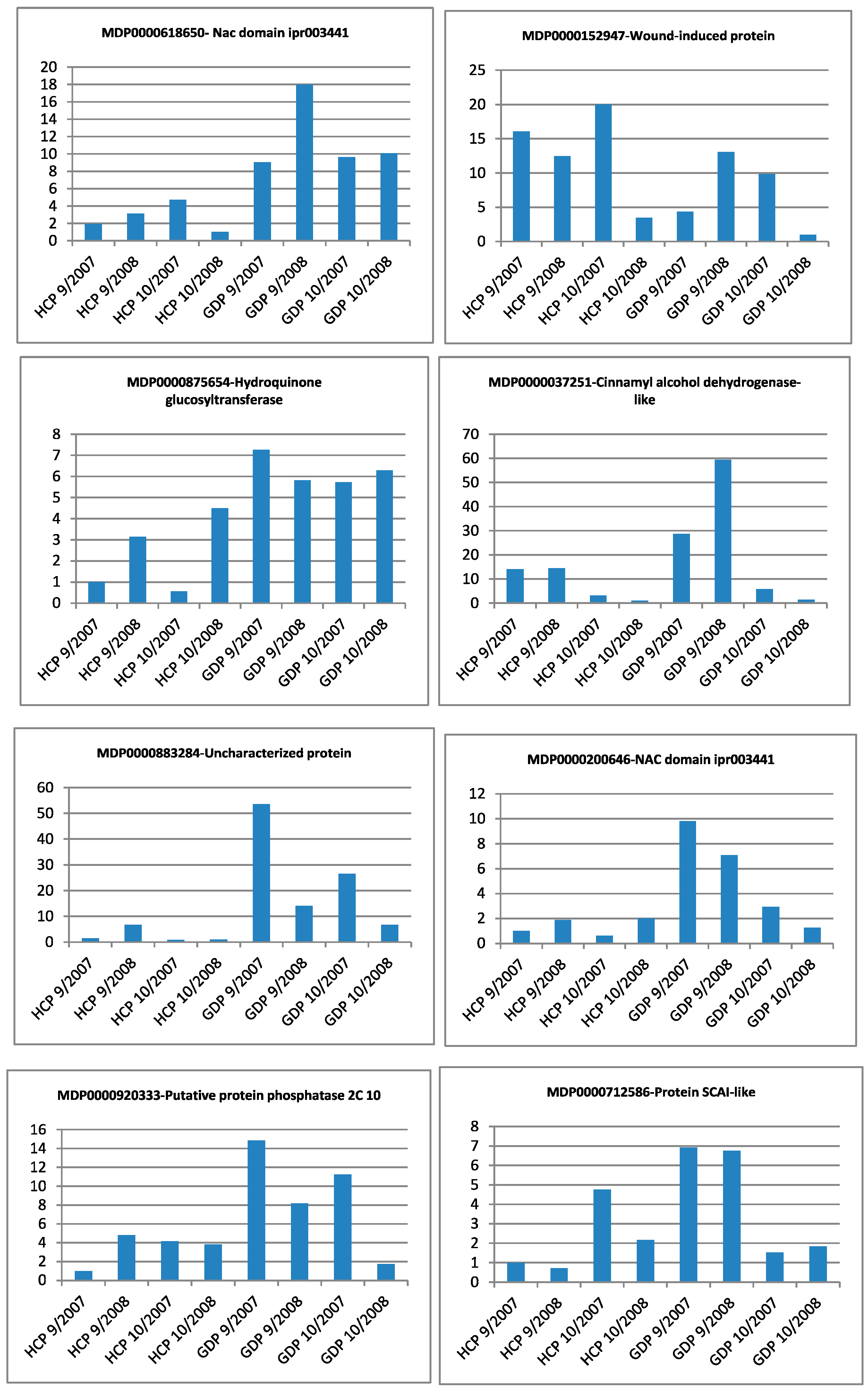
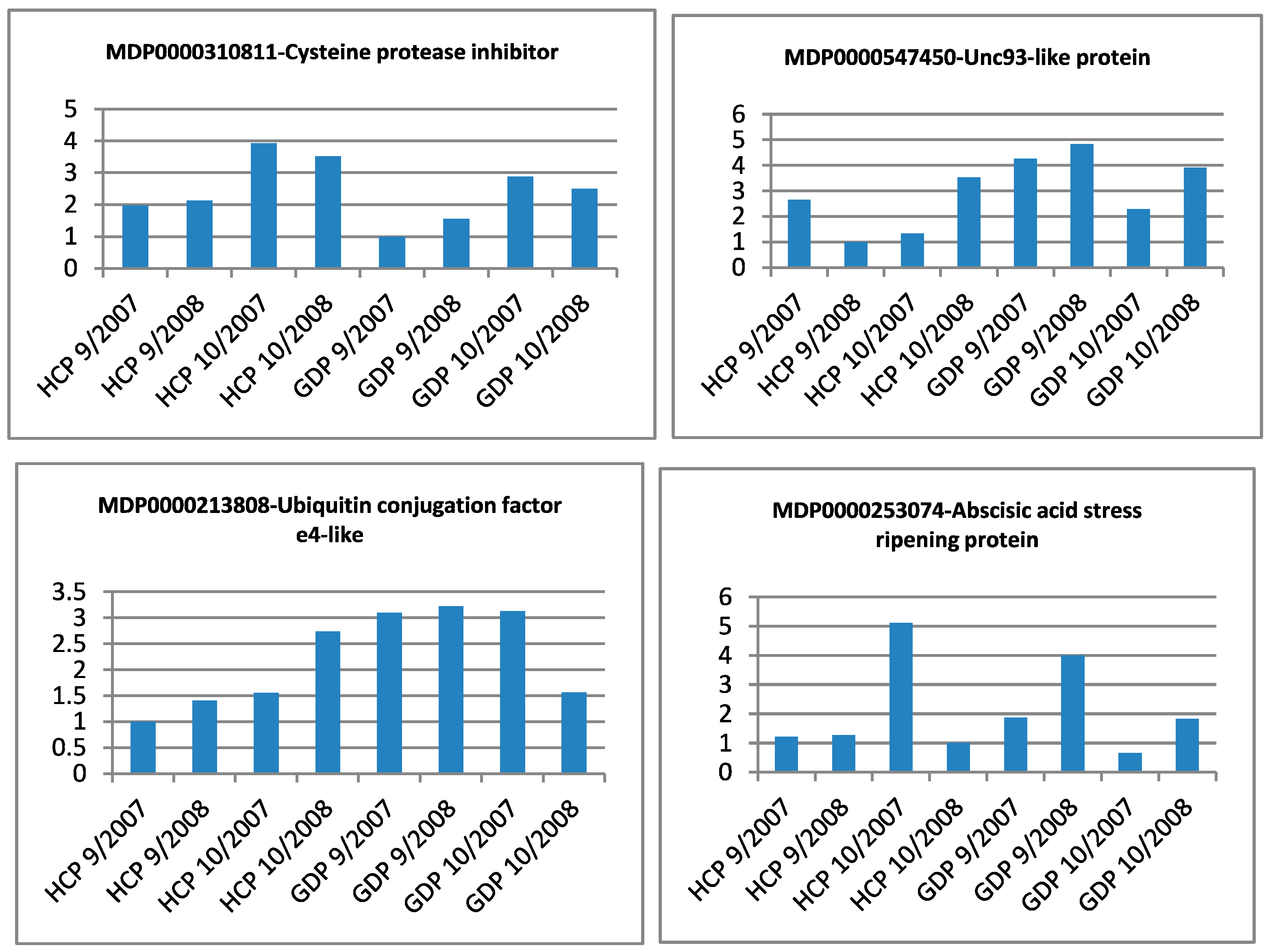
| Gene Identifier | Blast2GO Annotation | 129 DAA-2007 Season | 160 DAA-2007 Season | Cellular Localization | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GDE | GDP | HCE | HCP | GDE | GDP | HCE | HCP | |||
| MDP0000037251 | cinnamyl alcohol dehydrogenase-like protein | ++ | ++ | + | ++ | ++ | ++ | + | ++ | _ |
| MDP0000128468 | abscisic acid stress ripening protein homolog | ++ | ++ | ++ | + | ++ | ++ | ++ | + | M |
| MDP0000129664 | 3-ketoacyl-thiolase | - | - | + | + | - | - | + | + | _ |
| MDP0000138340 | NAC domain ipr003441 | ++ | ++ | + | + | ++ | ++ | + | + | _ |
| MDP0000152947 | wound-induced protein | + | ++ | + | + | + | + | + | + | _ |
| MDP0000161275 | mitochondrial substrate carrier family protein | + | + | + | ++ | + | + | + | ++ | _ |
| MDP0000166116 | acyl:CoA ligase acetate-coa synthetase-like protein | + | + | + | + | ++ | ++ | +++ | +++ | _ |
| MDP0000172863 | protein | - | - | - | - | - | + | - | - | S |
| MDP0000176723 | acyl:CoA ligase acetate-coa synthetase-like protein | ++ | ++ | + | + | ++ | ++ | + | + | _ |
| MDP0000180389 | disease resistance protein at3g14460-like | ++ | ++ | + | + | ++ | ++ | + | + | S |
| MDP0000200646 | NAC domain ipr003441 | + | + | + | + | + | + | ++ | ++ | _ |
| MDP0000213808 | probable ubiquitin conjugation factor e4-like | ++ | ++ | + | + | ++ | ++ | + | + | _ |
| MDP0000220601 | zinc finger CCCH domain-containing protein 53-like | + | + | + | + | + | + | + | ++ | _ |
| MDP0000232309 | transmembrane BAX inhibitor motif-containing protein 4 | + | + | + | + | ++ | ++ | + | + | _ |
| MDP0000233229 | Unknown protein | + | + | + | + | + | + | + | ++ | _ |
| MDP0000234325 | WWE protein-protein interaction domain family protein | - | - | + | - | - | - | + | - | _ |
| MDP0000237908 | metallothionein-like protein | ++ | ++ | + | + | ++ | ++ | + | + | _ |
| MDP0000253074 | abscisic acid stress ripening protein homolog | - | - | + | + | - | - | + | + | _ |
| MDP0000255887 | TIR-NBS-LRR resistance protein | - | + | - | - | - | + | - | - | _ |
| MDP0000273484 | SKP1-like protein | - | + | - | - | - | + | - | - | _ |
| MDP0000281279 | Unknown protein | ++ | + | + | + | ++ | + | + | + | _ |
| MDP0000286959 | dentin sialophosphoprotein | + | + | - | - | + | + | - | - | _ |
| MDP0000292888 | GYF domain-containing protein | - | - | - | + | - | - | - | + | C |
| MDP0000296716 | ethylene-responsive transcription factor RAP2-7-like | ++ | + | ++ | ++ | ++ | + | ++ | ++ | _ |
| MDP0000304285 | xanthine uracil permease family expressed | - | - | - | + | - | - | - | + | _ |
| MDP0000310811 | cysteine protease inhibitor | - | - | - | - | - | - | - | + | M |
| MDP0000316244 | probable ADP-ribosylation factor GTPase-activating protein AGD15-like | + | + | ++ | ++ | ++ | ++ | + | + | _ |
| MDP0000320533 | proteasome assembly chaperone | - | - | - | + | - | - | - | + | _ |
| MDP0000378585 | at4g03420 f9h3_4 | + | + | ++ | + | + | + | ++ | + | _ |
| MDP0000443265 | Unknown protein | - | - | - | - | - | - | + | - | C |
| MDP0000523205 | Unknown protein | - | - | - | - | - | + | - | - | _ |
| MDP0000547450 | UNC93-like protein | + | ++ | + | + | + | ++ | + | + | _ |
| MDP0000572242 | probable xyloglucan glycosyltransferase 12-like | + | ++ | + | + | + | ++ | + | + | _ |
| MDP0000580900 | porin voltage-dependent anion-selective channel protein | + | - | - | - | + | - | - | - | _ |
| MDP0000584042 | protein | + | + | - | - | + | + | - | - | S |
| MDP0000606453 | probable nitrite transporter at1g68570-like | - | - | - | - | - | - | + | - | _ |
| MDP0000618650 | NAC domain ipr003441 | - | - | - | + | - | - | - | + | _ |
| MDP0000689999 | protein disulfide isomerase | ++ | + | + | + | ++ | + | + | + | S |
| MDP0000697474 | reverse transcriptase | - | + | - | - | - | + | - | - | _ |
| MDP0000712586 | protein SCAI-like | + | + | ++ | ++ | ++ | ++ | + | + | S |
| MDP0000875654 | hydroquinone glucosyltransferase | ++ | ++ | + | + | + | + | + | + | _ |
| MDP0000876817 | Unknown protein | - | - | - | - | - | - | + | - | _ |
| MDP0000883284 | PREDICTED: uncharacterized protein LOC100248602 [Vitis vinifera] | + | + | - | - | + | + | - | - | S |
| MDP0000901731 | Unknown protein | - | - | - | + | - | - | - | + | S |
| MDP0000920333 | putative protein phosphatase 2C 10 | - | - | - | + | - | - | - | + | _ |
| Gene ID | Location | Gene | At Homolog | Sl Homolog | Potential Trait/Fruit Characteristic Influenced | Reference(s) |
|---|---|---|---|---|---|---|
| Transcriptional Regulation | ||||||
| MDP0000138340 | chr1:12,793,865..12,795,081 | NAC transcription factor-like 9 | AT4G35580 | XP_004239709.1 | Stress response | [15] |
| MDP0000200646 | chr1:12,796,087..12,797,315 | NAC transcription factor-like 9 | AT4G35580 | XP_004239709.1 | Stress response | [15] |
| MDP0000618650 | chr15:14,098,214..14,102,470 | NAC domain containing protein 75 | AT4G29230 | XP_004239596.1 | Stress response/general ripening regulation | [15] |
| MDP0000296716 | chr3:9,357,237..9,360,502 | AP2 transcription factor SlAP2d | AT2G28550 | NP_001234647.1 | Ethylene-regulated response | [16,17] |
| Secondary Biosynthetic Processes | ||||||
| MDP0000037251 | chr1:6,887,281..6,890,898 | Putative cinnamyl alcohol dehydrogenase 9 | AT4G39330 | XP_004250169.1 | Fruit firmness | [18] |
| MDP0000572242 | chr15:1,231,253..1,235,097 | Probable xyloglucan glycosyltransferase 12-like | AT4G07960 | XP_004238064.1 | Fruit firmness | [19] |
| MDP0000875654 | chr7:16,340,110..16,341,528 | Hydroquinone glucosyltransferase | AT4G01070 | XP_004231207.1 | Volatile/polyphenol glycosylation | [20,21] |
| MDP0000523205 | chr7:5,077,703..5,080,814 | Flavonoid biosynthesis oxidoreductase protein | AT4G10490 | NP_001233840.1 | Flavanoid synthesis | [22] |
| Signaling | ||||||
| MDP0000128468 | chr16:4,935,225..4,935,891 | Abscisic acid stress ripening (ASR1) protein | No hit | NM_001247208.2 | Abiotic stress response, developmental response to sugar levels | [23,24] |
| MDP0000176723 | chr17:23,896,643..23,897,062 | Acyl:CoA ligase acetate-CoA synthase-like protein | AT5G16370 | XP_004231630.1 | Abiotic stress response/ripening | [25,26,27] |
| MDP0000920333 | chr10:29,789,438..29,789,994 | Putative protein phosphatase 2C-10 | AT1G34750 | XP_004237914.1 | Ripening/abiotic stress response | [28,29] |
| MDP0000213808 | chr17:4,103,927..4,109,528 | putative ubiquitin conjugation factor E4 | AT5G15400 | XP_004232186.1 | Ethylene production, development, ripening | [30] |
| Cell/Organ Development | ||||||
| MDP0000232309 | chr16:13,766,655..13,768,223 | BAX inhibitor-1 like protein | No hit | No hit | Cell elongation/fruit size | [31,32] |
| MDP0000547450 | chr7:26,032,235..26,033,783 | UNC93-like protein 1-like | AT1G18000 | XP_004235041.1 | Apogamy | [33] |
| Stress Response | ||||||
| MDP0000712586 | chr14:27,117,925..27,120,988 | SCAI-like protein | No Hit | No Hit | Redox homeostasis/respiration | [34] |
| MDP0000161275 | chr12:24,524,346..24,526,137 | Mitochondrial succinate-fumarate transporter 1-like | AT5G01340 | XP_004249636.1 | Hypoxia-induced fermentation/respiration | [35,36] |
| MDP0000310811 | chr7:3,004,269..3,005,959 | Cysteine proteinase inhibitor 6 | AT3G12490 | XP_004228480.1 | Abiotic stress tolerance | [37,38] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schaeffer, S.; Hendrickson, C.; Fox, R.; Dhingra, A. Identification of Differentially Expressed Genes between “Honeycrisp” and “Golden Delicious” Apple Fruit Tissues Reveal Candidates for Crop Improvement. Horticulturae 2016, 2, 11. https://doi.org/10.3390/horticulturae2030011
Schaeffer S, Hendrickson C, Fox R, Dhingra A. Identification of Differentially Expressed Genes between “Honeycrisp” and “Golden Delicious” Apple Fruit Tissues Reveal Candidates for Crop Improvement. Horticulturae. 2016; 2(3):11. https://doi.org/10.3390/horticulturae2030011
Chicago/Turabian StyleSchaeffer, Scott, Christopher Hendrickson, Rachel Fox, and Amit Dhingra. 2016. "Identification of Differentially Expressed Genes between “Honeycrisp” and “Golden Delicious” Apple Fruit Tissues Reveal Candidates for Crop Improvement" Horticulturae 2, no. 3: 11. https://doi.org/10.3390/horticulturae2030011
APA StyleSchaeffer, S., Hendrickson, C., Fox, R., & Dhingra, A. (2016). Identification of Differentially Expressed Genes between “Honeycrisp” and “Golden Delicious” Apple Fruit Tissues Reveal Candidates for Crop Improvement. Horticulturae, 2(3), 11. https://doi.org/10.3390/horticulturae2030011






