Overexpression of IlMYB108 from Iris laevigata Confers Enhanced Drought and Salt Tolerance in Nicotiana tabacum
Abstract
1. Introduction
2. Materials and Methods
2.1. Plants Materials
2.2. Cloning of IlMYB108 and Phylogenetic Analysis
2.3. Subcellular Localization
2.4. Plant Transformation
2.5. Salt and Drought Stress Treatment
2.6. Physiological Performance of Tobacco Under Salt and Drought Stress
2.7. Physiological Analysis of Tobacco Under Salt and Drought Stress
2.8. Reverse Transcription PCR (RT-PCR) and RT-qPCR
2.9. Statistical Analyses
3. Results
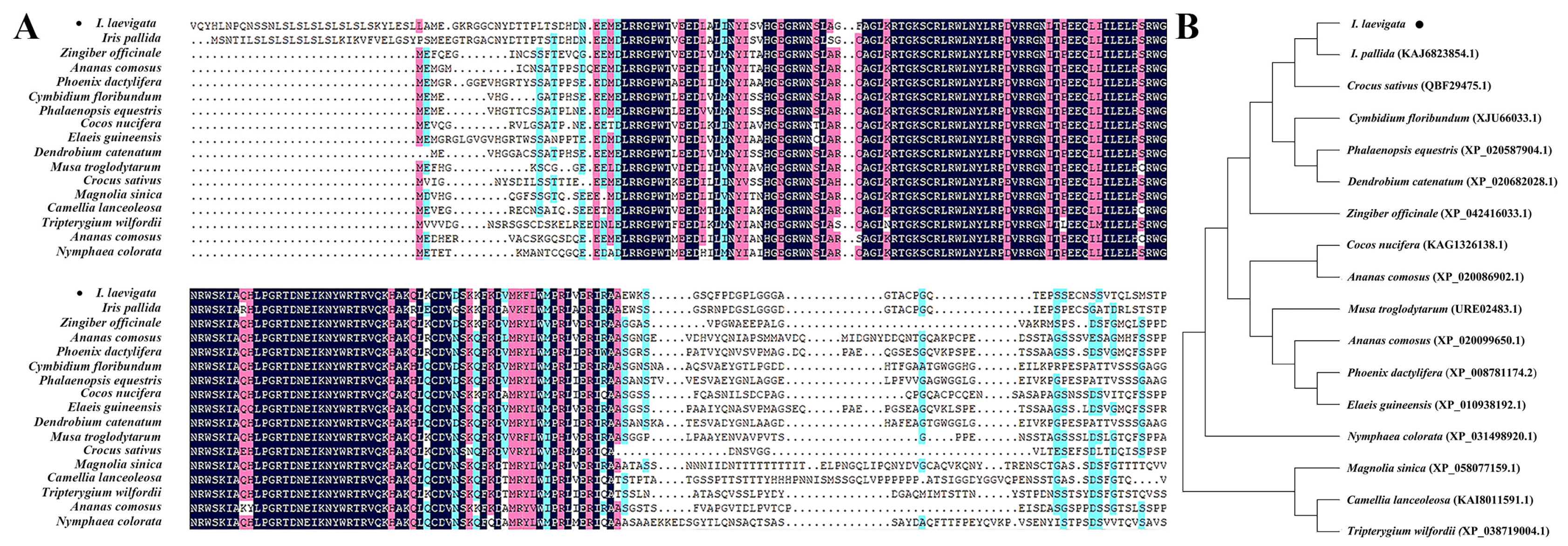
3.1. IlMYB108 Gene Cloning and Phylogenetic Tree Analysis
3.2. Expression of IlMYB108 in I. laevigata
3.3. Subcellular Localization of IlMYB108
3.4. Overexpressing IlMYB108 in Tobacco
3.5. Root Length Analysis of Overexpressed Tobacco Seeds Under Salt and Drought Stress
3.6. Overexpressing IlMYB108 in Tobacco Enhances Resistance to Salt and Drought Stress
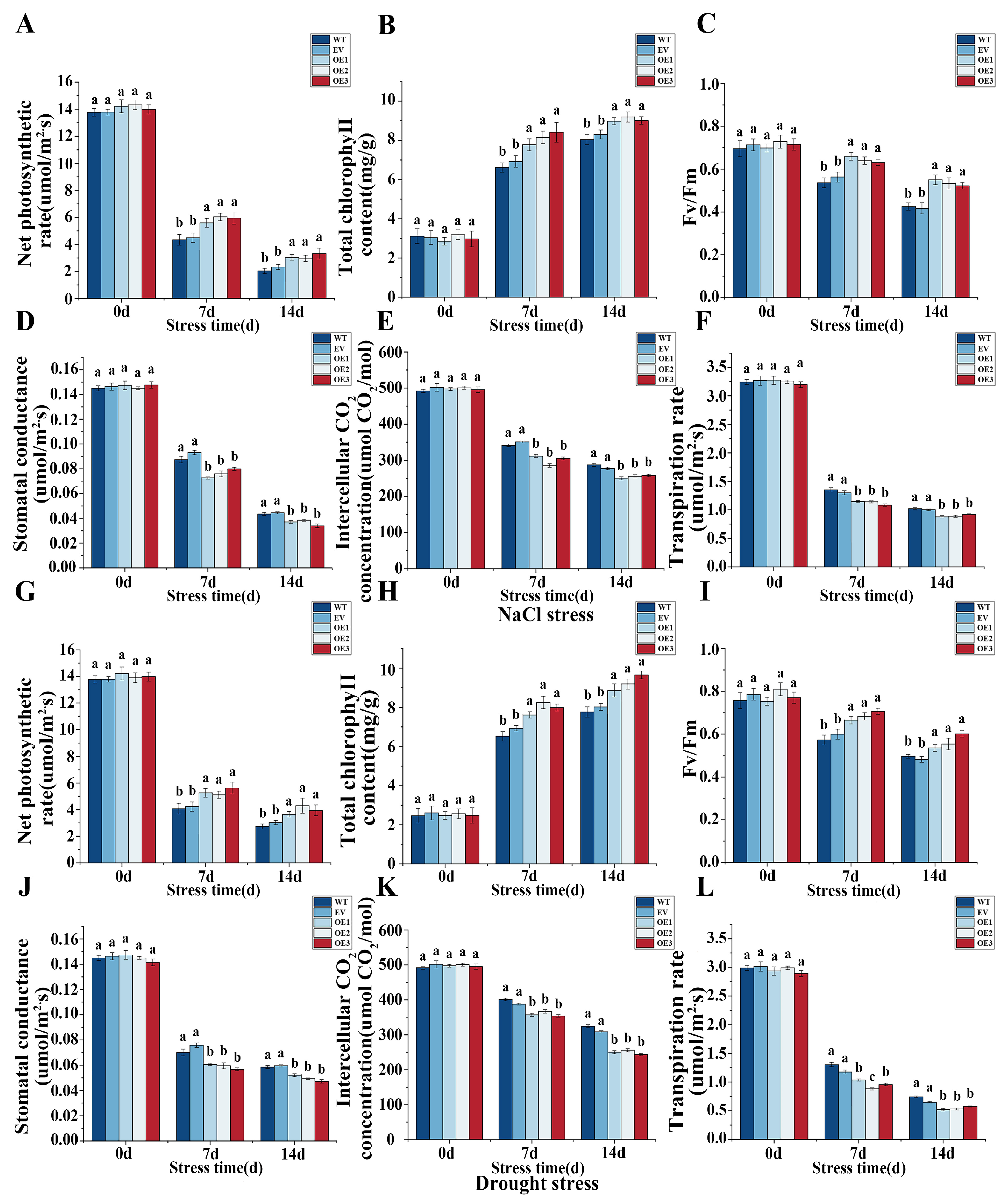
3.7. Impact of Overexpressing IlMYB108 on Photosynthesis Under Salt and Drought Stress
3.8. The Involvement of ROS Regulation in IlMYB108 Enhanced Salt and Drought Stress Resistance
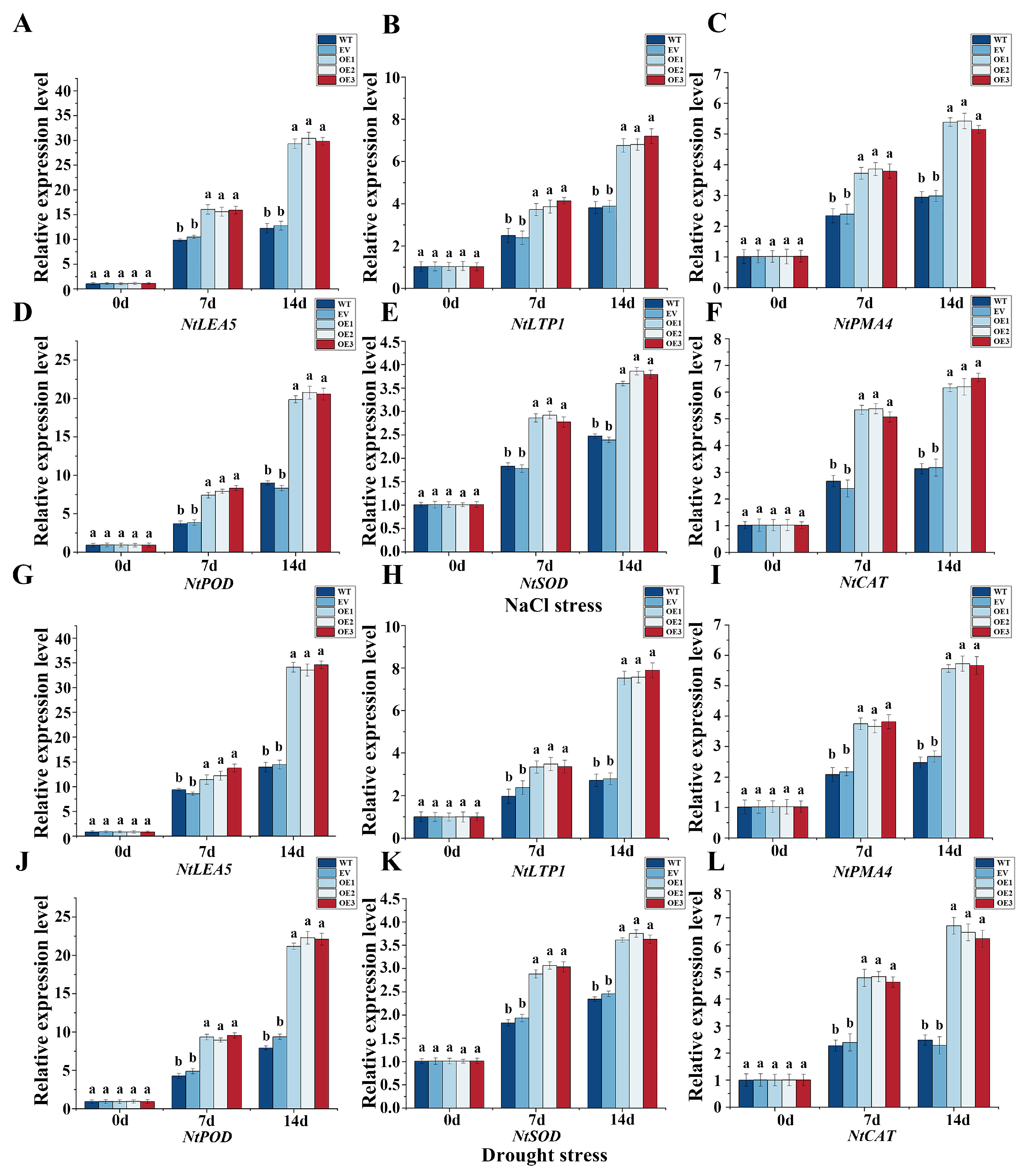
3.9. The IlMYB108 on Expression Levels of Stress-Related Genes NtLEA5, NtLTP1, NtPMA4, NtPOD, NtSOD and NtCAT Under Salt and Drought Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.; Wang, P. Tackling abiotic stress in plants: Recent insights and trends. Stress Biol. 2025, 5, 8. [Google Scholar] [CrossRef]
- Nawaz, M.; Sun, J.; Shabbir, S.; Khattak, W.A.; Ren, G.; Nie, X.; Bo, Y.; Javed, Q.; Du, D.; Sonne, C. A review of plants strategies to resist biotic and abiotic environmental stressors. Sci. Total Environ. 2023, 900, 165832. [Google Scholar] [CrossRef]
- Su, X.; Yao, L.; Wang, X.; Zhang, Y.; Zhang, G.; Li, X. Mechanisms for cell survival during abiotic stress: Focusing on plasma membrane. Stress Biol. 2025, 5, 1. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, J.; Li, R.; Ge, Y.; Li, Y.; Li, R. Plants’ response to abiotic stress: Mechanisms and strategies. Int. J. Mol. Sci. 2023, 24, 10915. [Google Scholar] [CrossRef]
- Du, B.; Haensch, R.; Alfarraj, S.; Rennenberg, H. Strategies of plants to overcome abiotic and biotic stresses. Biol. Rev. 2024, 99, 1524–1536. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Jiao, C.; Sun, X.; Li, X. A MYB transcription factor atlas provides insights into the evolution of environmental adaptations in plants. Int. J. Mol. Sci. 2023, 24, 2566. [Google Scholar] [CrossRef]
- Wu, X.; Xia, M.; Su, P.; Zhang, Y.; Tu, L.; Zhao, H.; Gao, W.; Huang, L.; Hu, Y. MYB transcription factors in plants: A comprehensive review of their discovery, structure, classification, functional diversity and regulatory mechanism. Int. J. Biol. Macromol. 2024, 282, 136652. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhou, H.; Zhang, Y.; Zhao, Y.; Zhang, Y.; Feng, X.; Lin, H. Diverse roles of MYB transcription factors in plants. J. Integr. Plant Biol. 2025, 67, 539–562. [Google Scholar] [CrossRef]
- Wu, Y.; Wen, J.; Xia, Y.; Zhang, L.; Du, H. Evolution and functional diversification of R2R3-MYB transcription factors in plants. Hortic. Res. 2022, 9, uhac058. [Google Scholar] [CrossRef]
- Liang, Y.K.; Dubos, C.; Dodd, I.C.; Holroyd, G.H.; Hetherington, A.M.; Campbell, M.M. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr. Biol. 2005, 15, 1201–1206. [Google Scholar] [CrossRef]
- Cao, Z.H.; Zhang, S.Z.; Wang, R.K.; Zhang, R.F.; Hao, Y.J. Genome wide analysis of the apple myb transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS ONE 2013, 8, e69955. [Google Scholar] [CrossRef]
- Lee, S.B.; Kim, H.U.; Suh, M.C. MYB94 and MYB96 additively activate cuticular wax biosynthesis in Arabidopsis. Plant Cell Physiol. 2016, 57, 2300–2311. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, L.; Shi, Q.; Ren, Z. SlMYB102, an R2R3-type MYB gene, confers salt tolerance in transgenic tomato. Plant Sci. 2020, 291, 110356. [Google Scholar] [CrossRef] [PubMed]
- Manna, M.; Thakur, T.; Chirom, O.; Mandlik, R.; Deshmukh, R.; Salvi, P. Transcription factors as key molecular target to strengthen the drought stress tolerance in plants. Physiol. Plant. 2021, 172, 847–868. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Ge, X.; Wang, Z.; Wang, Y.; Wang, P.; Shi, L.; Wang, B.; Zhang, A.; Li, F.; Wu, J. The cotton MYB33 gene is a hub gene regulating the trade-off between plant growth and defense in Verticillium dahliae infection. J. Adv. Res. 2024, 61, 1–17. [Google Scholar] [CrossRef]
- Wang, Z.; Peng, Z.; Khan, S.; Qayyum, A.; Rehman, A.; Du, X. Unveiling the power of MYB transcription factors: Master regulators of multi-stress responses and development in cotton. Int. J. Biol. Macromol. 2024, 276, 133885. [Google Scholar] [CrossRef]
- Li, S.; Liu, G.; Wu, Z.; Zhang, D.; Gan, C.; Li, B.; Liu, J.; Ni, Z.; Sun, Q.; Liang, R. The TaMYB44-TaMYB1 module regulates grain amylose biosynthesis and flour viscosity in wheat. Plant Commun. 2025, 6, 101461. [Google Scholar] [CrossRef]
- Chen, Y.; Hui, S.; Li, H.; Jiao, G.; Cao, R.; Zhou, L.; Wang, J.; Mawia, A.M.; Yang, L.; Wu, Y.; et al. A MYB61-SWB9-KOs module regulates grain chalkiness via gibberellin biosynthesis in rice endosperm. Plant Biotechnol. J. 2025, 23, 2793–2808. [Google Scholar] [CrossRef]
- Sun, W.; Ma, Z.; Chen, H.; Liu, M. MYB gene family in potato (Solanum tuberosum L.): Genome-wide identification of hormone-responsive reveals their potential functions in growth and development. Int. J. Mol. Sci. 2019, 20, 4847. [Google Scholar] [CrossRef]
- Shi, G.; Liu, G.; Liu, H.; Xu, N.; Yang, Q.; Song, Z.; Ye, W.; Wang, L. WRKY transcriptional factor IlWRKY70 from Iris laevigata enhances drought and salinity tolerances in Nicotiana tabacum. Int. J. Mol. Sci. 2023, 24, 16174. [Google Scholar] [CrossRef]
- Liu, G.; Li, F.; Shi, G.; Wang, L.; Wang, L.; Fan, L. Identification of mads-box transcription factors in Iris laevigata and functional assessment of IlSEP3 and IlSVP during flowering. Int. J. Mol. Sci. 2022, 23, 9950. [Google Scholar] [CrossRef]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple functions of MYB transcription factors in abiotic stress responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef]
- Feng, X.; Bai, S.; Zhou, L.; Song, Y.; Jia, S.; Guo, Q.; Zhang, C. Integrated analysis of transcriptome and metabolome provides insights into flavonoid biosynthesis of blueberry leaves in response to drought stress. Int. J. Mol. Sci. 2024, 25, 11135. [Google Scholar] [CrossRef]
- Song, P.; Yang, R.; Jiao, K.; Guo, B.; Zhang, L.; Li, Y.; Zhang, K.; Zhou, S.; Wu, X.; Li, X. FvMYB108, a MYB gene from Fragaria vesca, positively regulates cold and salt tolerance of Arabidopsis. Int. J. Mol. Sci. 2024, 25, 3405. [Google Scholar] [CrossRef]
- Du, B.; Liu, H.; Dong, K.; Wang, Y.; Zhang, Y. Over-expression of an R2R3 MYB gene, MdMYB108L, enhances tolerance to salt stress in transgenic plants. Int. J. Mol. Sci. 2022, 23, 9428. [Google Scholar] [CrossRef]
- Zhao, K.; Cheng, Z.; Guo, Q.; Yao, W.; Liu, H.; Zhou, B.; Jiang, T. Characterization of the poplar R2R3-MYB gene family and over-expression of PsnMYB108 confers salt tolerance in transgenic tobacco. Front. Plant Sci. 2020, 11, 571881. [Google Scholar] [CrossRef]
- Xu, L.; Xiang, G.; Sun, Q.; Ni, Y.; Jin, Z.; Gao, S.; Yao, Y. Melatonin enhances salt tolerance by promoting MYB108A-mediated ethylene biosynthesis in grapevines. Hortic. Res. 2019, 6, 114. [Google Scholar] [CrossRef]
- Wang, B.; Li, S.; Zou, L.; Guo, X.; Liang, J.; Liao, W.; Peng, M. Natural variation MeMYB108 associated with tolerance to stress-induced leaf abscission linked to enhanced protection against reactive oxygen species in cassava. Plant Cell Rep. 2022, 41, 1573–1587. [Google Scholar] [CrossRef]
- Fan, L.; Niu, Z.; Shi, G.; Song, Z.; Yang, Q.; Zhou, S.; Wang, L. WRKY22 transcription factor from Iris laevigata regulates flowering time and resistance to salt and drought. Plants 2024, 13, 1191. [Google Scholar] [CrossRef]
- Collings, D.A. Subcellular localization of transiently expressed fluorescent fusion proteins. Methods Mol Biol. 2013, 1069, 227–258. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, S.; Guan, C.; Kong, X.; Wang, Y.; Cui, Y.; Liu, B.; Zhou, Y.; Zhang, Y. Overexpressing the NAC transcription factor LpNAC13 from Lilium pumilum in tobacco negatively regulates the drought response and positively regulates the salt response. Plant Physiol. Biochem. 2020, 149, 96–110. [Google Scholar] [CrossRef]
- Xiang, X.Y.; Chen, J.; Xu, W.X.; Qiu, J.R.; Song, L.; Wang, J.T.; Tang, R.; Chen, D.; Jiang, C.Z.; Huang, Z. Dehydration-induced WRKY transcriptional factor MfWRKY70 of Myrothamnus flabellifolia enhanced drought and salinity tolerance in Arabidopsis. Biomolecules 2021, 11, 327. [Google Scholar] [CrossRef]
- Wang, Y.; Cui, Y.; Liu, B.; Wang, Y.; Sun, S.; Wang, J.; Tan, M.; Yan, H.; Zhang, Y. Lilium pumilum stress-responsive NAC transcription factor LpNAC17 enhances salt stress tolerance in tobacco. Front. Plant Sci. 2022, 13, 993841. [Google Scholar] [CrossRef]
- Hu, Z.; Li, Y.; Yang, J.; Song, S.; Li, X.; Xiong, C.; Yi, P.; Liu, C.; Hu, R.; Huang, X. The positive impact of the NtTAS14-like1 gene on osmotic stress response in Nicotiana tabacum. Plant Cell Rep. 2023, 43, 25. [Google Scholar] [CrossRef]
- Zhang, Z.; Tao, S.; Zhou, B.; Zhang, X.; Zhao, Z. Plant stomatal conductance determined transpiration and photosynthesis both contribute to the enhanced negative air ion (NAI). Ecol. Indic. 2021, 130, 108114. [Google Scholar] [CrossRef]
- Jian, Z.; Tang, X.; Wang, H.; Xu, G. Evaluate the photosynthesis and chlorophyll fluorescence of Epimedium brevicornu maxim. Sci. Rep. 2022, 12, 19470. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]
- Anwar, T.; Shehzadi, A.; Qureshi, H.; Shah, M.N.; Danish, S.; Salmen, S.H.; Ansari, M.J. Alleviation of cadmium and drought stress in wheat by improving growth and chlorophyll contents amended with GA3 enriched deashed biochar. Sci. Rep. 2023, 13, 18503. [Google Scholar] [CrossRef]
- Hanson, A.D.; Zhao, Y. Plant physiology synthetic biology initiative. Plant Physiol. 2022, 190, 180–181. [Google Scholar] [CrossRef]
- Zhu, X.; Zhang, Y.; Du, Z.; Chen, X.; Zhou, X.; Kong, X.; Sun, W.; Chen, Z.; Chen, C.; Chen, M. Tender leaf and fully-expanded leaf exhibited distinct cuticle structure and wax lipid composition in Camellia sinensis cv Fuyun 6. Sci. Rep. 2018, 8, 14944. [Google Scholar] [CrossRef]
- Daudi, A.; O’Brien, J.A. Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio-Protocol 2012, 2, e263. [Google Scholar] [CrossRef]
- Yadav, S.; Gill, S.S.; Passricha, N.; Gill, R.; Badhwar, P.; Anjum, N.A.; Francisco, J.J.; Tuteja, N. Genome-wide analysis and transcriptional expression pattern-assessment of superoxide dismutase (SOD) in rice and Arabidopsis under abiotic stresses. Plant Gene 2019, 17, 100165. [Google Scholar] [CrossRef]
- Zhong, M.; Song, R.; Wang, Y.; Shu, S.; Sun, J.; Guo, S. TGase regulates salt stress tolerance through enhancing bound polyamines-mediated antioxidant enzymes activity in tomato. Environ. Exp. Bot. 2020, 179, 104191. [Google Scholar] [CrossRef]
- Pereira-Gomez, M.; Fajardo, A.; Echeverria, N.; Lopez-Tort, F.; Perbolianachis, P.; Costabile, A.; Aldunate, F.; Moreno, P.; Moratorio, G. Evaluation of SYBR Green real time PCR for detecting SARS-CoV-2 from clinical samples. J. Virol. Methods 2021, 289, 114035. [Google Scholar] [CrossRef]
- Flexas, J.; Medrano, H. Drought-inhibition of photosynthesis in C3 plants: Stomatal and non-stomatal limitations revisited. Ann. Bot. 2002, 89, 183–189. [Google Scholar] [CrossRef]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef]
- Lawlor, D.W.; Cornic, G. Photosynthetic carbon assimilation and associated metabolism in relation to water deficits in higher plants. Plant Cell Environ. 2002, 25, 275–294. [Google Scholar] [CrossRef]
- Yoo, J.H.; Park, C.Y.; Kim, J.C.; Heo, W.D.; Cheong, M.S.; Park, H.C.; Kim, M.C.; Moon, B.C.; Choi, M.S.; Kang, Y.H.; et al. Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. J. Biol. Chem. 2005, 280, 3697–3706. [Google Scholar] [CrossRef]
- Liu, J.; Osbourn, A.; Ma, P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef]
- Jiang, C.K.; Rao, G.Y. Insights into the diversification and evolution of R2R3-MYB transcription factors in plants. Plant Physiol. 2020, 183, 637–655. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Cheng, X.; Liu, X.; Wu, H.; Bi, H.; Xu, H. The wheat MYB transcription factor TaMYB31 is involved in drought stress responses in Arabidopsis. Front. Plant Sci. 2018, 9, 1426. [Google Scholar] [CrossRef]
- Chen, T.; Li, W.; Hu, X.; Guo, J.; Liu, A.; Zhang, B. A cotton MYB transcription factor, GBMYB5, is positively involved in plant adaptive response to drought stress. Plant Cell Physiol. 2015, 56, 917–929. [Google Scholar] [CrossRef]
- Lee, H.G.; Seo, P.J. MYB96 recruits the HDA15 protein to suppress negative regulators of ABA signaling in Arabidopsis. Nat. Commun. 2019, 10, 1713. [Google Scholar] [CrossRef]
- Simeoni, F.; Skirycz, A.; Simoni, L.; Castorina, G.; de Souza, L.P.; Fernie, A.R.; Alseekh, S.; Giavalisco, P.; Conti, L.; Tonelli, C.; et al. The AtMYB60 transcription factor regulates stomatal opening by modulating oxylipin synthesis in guard cells. Sci. Rep. 2022, 12, 533. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Yarra, R.; Yang, Y.; Liu, Y.; Yang, M.; Cao, H. The oil palm R2R3-MYB subfamily genes EgMYB111 and EgMYB157 improve multiple abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Rep. 2022, 41, 377–393. [Google Scholar] [CrossRef]
- Wang, Z.; Li, X.; Gao, X.R.; Dai, Z.R.; Peng, K.; Jia, L.C.; Wu, Y.K.; Liu, Q.C.; Zhai, H.; Gao, S.P.; et al. IbMYB73 targets abscisic acid-responsive IbGER5 to regulate root growth and stress tolerance in sweet potato. Plant Physiol. 2024, 194, 787–804. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Niu, Z.; Shi, G.; Wang, Z.; Fan, L.; Wang, L. Overexpression of IlMYB108 from Iris laevigata Confers Enhanced Drought and Salt Tolerance in Nicotiana tabacum. Horticulturae 2026, 12, 3. https://doi.org/10.3390/horticulturae12010003
Niu Z, Shi G, Wang Z, Fan L, Wang L. Overexpression of IlMYB108 from Iris laevigata Confers Enhanced Drought and Salt Tolerance in Nicotiana tabacum. Horticulturae. 2026; 12(1):3. https://doi.org/10.3390/horticulturae12010003
Chicago/Turabian StyleNiu, Zhaoqian, Gongfa Shi, Zhengyide Wang, Lijuan Fan, and Ling Wang. 2026. "Overexpression of IlMYB108 from Iris laevigata Confers Enhanced Drought and Salt Tolerance in Nicotiana tabacum" Horticulturae 12, no. 1: 3. https://doi.org/10.3390/horticulturae12010003
APA StyleNiu, Z., Shi, G., Wang, Z., Fan, L., & Wang, L. (2026). Overexpression of IlMYB108 from Iris laevigata Confers Enhanced Drought and Salt Tolerance in Nicotiana tabacum. Horticulturae, 12(1), 3. https://doi.org/10.3390/horticulturae12010003





