In Vitro Induction of Autotetraploids in the Subtropical Fruit Tree Cherimoya (Annona cherimola Mill.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Disinfection
2.2. Induction of Autopolyploid Shoots
2.3. Ploidy Analysis by Flow Cytometry
2.4. Multiplication and Rooting of Regenerated Shoots, and Acclimatization of Rooted Plantlets
2.5. Chromosome Observation
2.6. Characterization of the Leaves of Autotetraploid Cherimoyas
2.7. Statistical Analysis
3. Results
3.1. Induction of Autopolyploid Shoots
3.2. Ploidy Analysis by Flow Cytometry and Selection of Autotetraploid Shoots
3.3. Establishment of Autotetraploid Lines and Verification of Ploidy Level by Cytological Analysis
3.4. Characterization of the Leaves of Autotetraploid Cherimoyas
4. Discussion
4.1. Method of Polyploidy Induction
4.2. Rate of Autotetraploid Induction and Tetraploid Survival
4.3. Characterization of the Leaves of Autotetraploid Cherimoyas
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BAP | 6-Benzylaminopurine |
| GA3 | Gibberellic acid |
| IBA | Indole-3-butyric acid |
| MS | Murashige and Skoog medium |
References
- Encina, C.L.; Barceló-Muñoz, A.; Herrero-Castaño, A.; Pliego-Alfaro, F. In vitro morphogenesis of juvenile Annona cherimola Mill. bud explants. J. Hortic. Sci. 1994, 69, 1053–1059. [Google Scholar] [CrossRef]
- Padilla, I.M.G.; Encina, C.L. Micropropagation of adult cherimoya (Annona cherimola Mill.) cv. Fino de Jete. Vitr. Cell. Dev. Biol.-Plant 2004, 40, 210–214. [Google Scholar] [CrossRef]
- Jordan, M. Multiple shoot formation and rhizogenesis from cherimoya (Annona cherimola Mill.) hypocotyls and petiole explants. Gartenbauwissenschaft 1988, 53, 234–237. [Google Scholar]
- Lemos, E.E.P.; Blake, J. Micropropagation of juvenile and mature Annona muricata L. J. Hortic. Sci. 1996, 71, 395–403. [Google Scholar] [CrossRef]
- Rasai, S.; Kantharajah, A.; Dodd, W. The effect of growth regulators, source of explants and irradiance on in vitro regeneration of atemoya. Aust. J. Bot. 1994, 42, 333–340. [Google Scholar] [CrossRef]
- Lemos, E.E.P.; Blake, J. Micropropagation of juvenile and adult Annona squamosa. Plant Cell Tissue Organ Cult. 1996, 46, 77–79. [Google Scholar] [CrossRef]
- Padilla, I.M.G.; Encina, C.L. In vitro germination of cherimoya (Annona cherimola Mill.) seeds. Sci. Hortic. 2003, 97, 219–227. [Google Scholar] [CrossRef]
- Encina, C.L.; Carmona-Martín, E.; Arana-López, A.; Padilla, I.M.G. Biotechnology applied to Annona species: A review. Rev. Bras. Frutic. 2014, 36, 17–21. [Google Scholar] [CrossRef]
- Bowden, W.M. Chromosome numbers in the Annonaceae. Am. J. Bot. 1948, 35, 377–381. [Google Scholar] [CrossRef]
- Morawetz, W. Remarks on karyological differentiation patterns in tropical woody plants. Plant Syst. Evol. 1986, 152, 49–100. [Google Scholar] [CrossRef]
- Morawetz, W. Systematics and karyoevolution in Magnoliidae: Tetrameranthus as compared with other Annonaceae genera of the same chromosome number. Plant Syst. Evol. 1986, 154, 147–173. [Google Scholar] [CrossRef]
- Anuragi, H.; Dhaduk, H.L.; Kumar, S.; Dhruve, J.J.; Parekh, M.J.; Sakure, A.A. Molecular diversity of Annona species and proximate fruit composition of selected genotypes. 3 Biotech 2016, 6, 204. [Google Scholar] [CrossRef]
- Martin, C.; Viruel, M.A.; Lora, J.; Hormaza, J.I. Polyploidy in fruit tree crops of the genus Annona (Annonaceae). Front. Plant Sci. 2019, 10, 99. [Google Scholar] [CrossRef] [PubMed]
- Serbin, G.M.; Pinangé, D.S.B.; Machado, R.M.; Vasconcelos, S.; Amorim, B.S.; Clement, C.R. Relationship between fruit phenotypes and domestication in hexaploid populations of biribá (Annona mucosa) in Brazilian Amazonia. PeerJ 2023, 11, e14659. [Google Scholar] [CrossRef] [PubMed]
- Talavera, A.; Fernández-Pozo, N.; Matas, A.J.; Hormaza, J.I.; Bombarely, A. Genomics in neglected and underutilized fruit crops: A chromosome-scale genome sequence of cherimoya (Annona cherimola Mill., cv. “Fino de Jete”). Plants People Planet 2023, 5, 408–423. [Google Scholar] [CrossRef]
- Bretagnolle, F.; Thompson, J.D. Gametes with the somatic chromosome number: Mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytol. 1995, 129, 1–22. [Google Scholar] [CrossRef]
- Eng, W.-H.; Ho, W.S. Polyploidization using colchicine in horticultural plants: A review. Sci. Hortic. 2019, 246, 604–617. [Google Scholar] [CrossRef]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2015, 241, 281–296. [Google Scholar] [CrossRef] [PubMed]
- El-Nashar, Y.I.; Ammar, M.H. Mutagenic influences of colchicine on phenological and molecular diversity of Calendula officinalis L. Genet. Mol. Res. 2016, 15, 1–15. [Google Scholar] [CrossRef]
- Wu, J.H.; Ferguson, A.R.; Murray, B.G.; Jia, Y.; Datson, P.M.; Zhang, J. Induced polyploidy dramatically increases the size and alters the shape of fruit in Actinidia chinensis. Ann. Bot. 2012, 109, 169–179. [Google Scholar] [CrossRef]
- Park, S.M.; Wakana, A.; Kim, J.H.; Jeong, C.S. Male and female fertility in triploid grapes (Vitis complex) with special reference to the production of aneuploid plants. Vitis 2002, 41, 11–20. [Google Scholar]
- Aleza, P.; Juárez, J.; Ollitrault, P.; Navarro, L. Production of tetraploid plants of non-apomictic citrus genotypes. Plant Cell Rep. 2009, 28, 1837–1846. [Google Scholar] [CrossRef]
- Aleza, P.; Juárez, J.; Cuenca, J.; Ollitrault, P.; Navarro, L. Extensive citrus triploid hybrid production by 2x × 4x sexual hybridizations and parent-effect on the length of the juvenile phase. Plant Cell Rep. 2012, 31, 1723–1735. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, Z.; Zhi, S.; Xu, F. Breeding triploid plants: A review. Czech J. Genet. Plant Breed. 2016, 52, 41–54. [Google Scholar] [CrossRef]
- Li, M.; Guo, Y.; Liu, S.; Zhao, Y.; Pang, X.; Li, Y. Autotetraploidization in Ziziphus jujuba Mill. var. spinosa enhances salt tolerance conferred by active, diverse stress responses. Environ. Exp. Bot. 2019, 165, 92–107. [Google Scholar] [CrossRef]
- Sanford, J.C. Ploidy manipulations. In Methods in Fruit Breeding; Moore, J.N., Janick, J., Eds.; Purdue University Press: West Lafayette, IN, USA, 1983; pp. 100–123. [Google Scholar]
- Tossi, V.E.; Martínez Tosar, L.J.; Laino, L.E.; Iannicelli, J.; Regalado, J.J.; Escandón, A.S.; Baroli, I.; Causin, H.F.; Pitta-Álvarez, S.I. Impact of polyploidy on plant tolerance to abiotic and biotic stresses. Front. Plant Sci. 2022, 13, 869423. [Google Scholar] [CrossRef]
- Carmona-Martín, E.; Regalado, J.J.; Raghavan, L.; Encina, C.L. In vitro induction of autooctoploid asparagus genotypes. Plant Cell Tissue Organ Cult. 2015, 121, 249–254. [Google Scholar] [CrossRef]
- Regalado, J.J.; Carmona-Martín, E.; Castro, P.; Moreno, R.; Gil, J.; Encina, C.L. Study of the somaclonal variation produced by different methods of polyploidization in Asparagus officinalis L. Plant Cell Tissue Organ Cult. 2015, 122, 31–44. [Google Scholar] [CrossRef]
- Hansen, N.J.P.; Andersen, S.B. Efficient production of doubled haploid wheat plants by in vitro treatment of microspores with trifluralin or APM. Plant Breed. 1998, 117, 401–405. [Google Scholar] [CrossRef]
- Eeckhaut, T.; Werbrouck, S.; Leus, L.W.H.; Van Bockstaele, E.; Debergh, P. Chemically induced polyploidization in Spathiphyllum wallisii Regel through somatic embryogenesis. Plant Cell Tissue Organ Cult. 2004, 78, 241–246. [Google Scholar] [CrossRef]
- Kadota, M.; Niimi, Y. In vitro induction of tetraploid plants from a diploid Japanese pear cultivar (Pyrus pyrifolia N. cv. Hosui). Plant Cell Rep. 2002, 21, 282–286. [Google Scholar] [CrossRef]
- Adaniya, S.; Shirai, D. In vitro induction of tetraploid ginger (Zingiber officinale Roscoe) and its pollen fertility and germinability. Sci. Hortic. 2001, 88, 277–287. [Google Scholar] [CrossRef]
- Shao, J.; Chen, C.; Deng, X. In vitro induction of tetraploid in pomegranate (Punica granatum). Plant Cell Tissue Organ Cult. 2003, 75, 241–246. [Google Scholar] [CrossRef]
- Gu, X.F.; Yang, A.F.; Meng, H.; Zhang, J.R. In vitro induction of tetraploid plants from diploid Zizyphus jujuba Mill. cv. Zhanhua. Plant Cell Rep. 2005, 24, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, B.; Qi, S.; Dong, M.; Wang, Z.; Li, Y.; Chen, S.; Li, B.; Zhang, J. Ploidy and hybridity effects on leaf size, cell size and related gene expression in triploids, diploids and their parents in Populus. Planta 2019, 249, 635–646. [Google Scholar] [CrossRef]
- Wu, J.; Sang, Y.R.; Zhou, Q.; Zhang, P.D. Colchicine in vitro tetraploid induction of Populus hopeiensis from leaf blades. Plant Cell Tissue Organ Cult. 2020, 141, 339–349. [Google Scholar] [CrossRef]
- Liu, G.F.; Li, Z.N.; Bao, M.Z. Colchicine-induced chromosome doubling in Platanus acerifolia and its effect on plant morphology. Euphytica 2007, 157, 145–154. [Google Scholar] [CrossRef]
- Ewald, D.; Ulrich, K.; Naujoks, G.; Schröder, M.B. Induction of tetraploid poplar and black locust plants using colchicine: Chloroplast number as an early marker for selecting polyploids in vitro. Plant Cell Tissue Organ Cult. 2009, 99, 353–357. [Google Scholar] [CrossRef]
- Silva, A.J.; Carvalho, C.R.; Clarindo, W.R. Chromosome set doubling and ploidy stability in synthetic auto- and allotetraploid of Eucalyptus: From in vitro condition to the field. Plant Cell Tissue Organ Cult. 2019, 138, 387–394. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, J.; Qiu, B.; Ma, Z.; Lu, T.; Kang, X.; Yang, J. Induction and characterization of tetraploid through zygotic chromosome doubling in Eucalyptus urophylla. Front. Plant Sci. 2022, 13, 870698. [Google Scholar] [CrossRef]
- Denaeghel, H.E.; Van Laere, K.; Leus, L.; Lootens, P.; Van Huylenbroeck, J.; Van Labeke, M.C. The variable effect of polyploidization on the phenotype in Escallonia. Front. Plant Sci. 2018, 9, 354. [Google Scholar] [CrossRef] [PubMed]
- Podwyszyńska, M.; Gabryszewska, E.; Dyki, B.; Stępowska, A.A.; Kowalski, A.; Jasiński, A. Phenotypic and genome size variation in synthetic tetraploids of daylily (Hemerocallis) in relation to their diploid counterparts. Euphytica 2015, 203, 1–16. [Google Scholar] [CrossRef]
- Regalado, J.J.; Carmona-Martín, E.; Querol, V.; Veléz, C.G.; Encina, C.L.; Pitta-Álvarez, S.I. Production of compact petunias through polyploidization. Plant Cell Tissue Organ Cult. 2017, 129, 61–71. [Google Scholar] [CrossRef]
- Rafiee, M.; Naderi, R.; Miri, S.M. In vitro polyploidy induction in primrose (Primula sp.) using colchicine. In Proceedings of the 4th International Conference on Applied Research in Chemistry, Science and Biology, Tehran, Iran, 2017; Available online: https://www.researchgate.net/publication/320616103 (accessed on 20 February 2021).
- Roughani, A.; Miri, S.M.; Kashi, A.K.; Naserian Khiabani, B. Increasing the ploidy level in spinach (Spinacia oleracea L.) using mitotic inhibitors. Plant Cell Biotechnol. Mol. Biol. 2017, 18, 124–130. [Google Scholar]
- Sanglard, N.A.; Amaral-Silva, P.M.; Sattler, M.C.; de Oliveira, S.C.; Cesário, L.M.; Ferreira, A.; Paiva, L.V.; Davide, L.C.; Paiva, R.M.A.; Carvalho, C.H.S. Indirect somatic embryogenesis in Coffea with different ploidy levels: A revisiting and updating study. Plant Cell Tissue Organ Cult. 2019, 136, 255–267. [Google Scholar] [CrossRef]
- Švécarová, M.; Navrátilová, B.; Hašler, P.; Ondřej, V. Artificial induction of tetraploidy in Humulus lupulus L. using oryzalin. Acta Agrobot. 2019, 72, 1764. [Google Scholar] [CrossRef]
- Divya, U.K.; Kumari, S.S. Development of in vitro tetraploid plants of Hevea brasiliensis. Int. J. Plant Soil Sci. 2019, 28, 1–12. [Google Scholar] [CrossRef]
- Chakraborti, S.P.; Vijayan, K.; Roy, B.N.; Qadri, S.M.H. In vitro induction of tetraploidy in mulberry (Morus alba L.). Plant Cell Rep. 1998, 17, 799–803. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Z.; Ren, Y.; Feng, Y.; Guo, Q.; Dong, L.; Sun, Y.; Li, Y. Induction and early identification of tetraploid black locust by hypocotyl in vitro. Vitr. Cell. Dev. Biol.-Plant 2021, 57, 372–379. [Google Scholar] [CrossRef]
- Wen, Y.; Liu, H.; Meng, H.; Qiao, L.; Zhang, G.; Cheng, Z. In vitro induction and phenotypic variations of autotetraploid garlic (Allium sativum L.) with dwarfism. Front. Plant Sci. 2022, 13, 917910. [Google Scholar] [CrossRef]
- Thao, N.T.P.; Ureshino, K.; Miyajima, I.; Ozaki, Y.; Okubo, H. Induction of tetraploids in ornamental Alocasia through colchicine and oryzalin treatments. Plant Cell Tissue Organ Cult. 2003, 72, 19–25. [Google Scholar] [CrossRef]
- Abu-Qaoud, H.; Skirvin, R.M.; Chevreau, E. In vitro separation of chimeral pears into their component genotypes. Euphytica 1990, 48, 189–196. [Google Scholar] [CrossRef]
- Sinski, I.; Dal Bosco, D.; Pierozzi, N.I.; Garcia Maia, J.D.; Silva Ritschel, P.; Quecini, V. Improving in vitro induction of autopolyploidy in grapevine seedless cultivars. Euphytica 2014, 196, 299–311. [Google Scholar] [CrossRef]
- Liu, J.; Yang, D.; Li, X.; Jin, Z.; Li, J. In vitro inducted tetraploid Elsholtzia splendens Nakai ex F. Maek. alters polyphenol species and synthesis. Plants 2024, 13, 3374. [Google Scholar] [CrossRef] [PubMed]
- Grosser, J.W.; Kainth, D.; Dutt, M. Production of colchicine-induced autotetraploids in pummelo (Citrus grandis Osbeck) through indirect organogenesis. HortScience 2014, 49, 944–948. [Google Scholar] [CrossRef]
- Blasco, M.; Badenes, M.L.; Naval, M.M. Colchicine-induced polyploidy in loquat (Eriobotrya japonica (Thunb.) Lindl.). Plant Cell Tissue Organ Cult. 2015, 120, 453–461. [Google Scholar] [CrossRef]
- Stanys, V.; Weckman, A.; Staniene, G.; Duchovskis, P. In vitro induction of polyploidy in Japanese quince (Chaenomeles japonica). Plant Cell Tissue Organ Cult. 2006, 84, 263–268. [Google Scholar] [CrossRef]
- Shi, Q.H.; Liu, P.; Liu, M.J.; Wang, J.R.; Xu, J. A novel method for rapid in vivo induction of homogeneous polyploids via calluses in a woody fruit tree (Ziziphus jujuba Mill.). Plant Cell Tissue Organ Cult. 2015, 121, 423–433. [Google Scholar] [CrossRef]
- Xie, X.; Agüero, C.B.; Wang, Y.; Walker, M.A. In vitro induction of tetraploids in Vitis × Muscadinia hybrids. Plant Cell Tissue Organ Cult. 2015, 122, 675–683. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Galbraith, D.W.; Harkins, K.R.; Maddox, J.M.; Ayres, N.M.; Sharma, D.P.; Firoozabady, E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 1983, 220, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, T.W. Chemistry and Biochemistry of Plant Pigments, 2nd ed.; Academic Press: London, UK, 1976; 870p. [Google Scholar]
- Zhou, Y.G.; Yao, N.; Feng, H.; Bai, J.P.; Zhang, T.B.; He, J.Q. Variations of leaf shape coefficients of winter wheat. Agric. Res. Arid Areas 2017, 35, 1–7. [Google Scholar] [CrossRef]
- Rêgo, M.M.; Rêgo, E.R.; Bruckner, C.H.; Finger, F.L.; Otoni, W.C. In vitro induction of autotetraploids from diploid yellow passion fruit (Passiflora edulis Sims.) mediated by colchicine and oryzalin. Plant Cell Tissue Organ Cult. 2011, 107, 451–459. [Google Scholar] [CrossRef]
- Cui, Y.; Hou, L.; Li, X.; Huang, F.; Pang, X.; Li, Y. In vitro induction of tetraploid Ziziphus jujuba Mill. var. spinosa plants from leaf explants. Plant Cell Tissue Organ Cult. 2017, 131, 175–182. [Google Scholar] [CrossRef]
- Wu, J.-H.; Ferguson, A.R.; Murray, B.G. Manipulation of ploidy for kiwifruit breeding: In vitro chromosome doubling in diploid Actinidia chinensis Planch. Plant Cell Tissue Organ Cult. 2011, 106, 503–511. [Google Scholar] [CrossRef]
- Li, S.; Liu, X.; Liu, H.; Zhang, X.; Ye, Q.; Zhang, H. Induction, identification and genetics analysis of tetraploid Actinidia chinensis. R. Soc. Open Sci. 2019, 6, 191052. [Google Scholar] [CrossRef]
- Sun, Q.R.; Sun, H.Y.; Li, L.G.; Bell, R.L. In vitro colchicine-induced polyploid plantlet production and regeneration from leaf explants of the diploid pear (Pyrus communis L.) cultivar ‘Fertility’. J. Hortic. Sci. Biotechnol. 2009, 84, 548–552. [Google Scholar] [CrossRef]
- Liu, R.; Gao, C.; Jin, J.; Wang, Y.; Jia, X.; Ma, H.; Zhang, Y.; Zhang, H.; Qi, B.; Xu, J. Induction and identification of tetraploids of pear plants (Pyrus bretschneideri and Pyrus betulaefolia). Sci. Hortic. 2022, 304, 111322. [Google Scholar] [CrossRef]
- Kainth, D.; Grosser, J.W. Induction of autotetraploids in pummelo (Citrus grandis L. Osbeck) through colchicine treatment of meristematically active seeds in vitro. Proc. Fla. State Hortic. Soc. 2010, 123, 44–48. [Google Scholar]
- Wulandari, D.R.; Ermayanti, T.M.; Purwito, A.; Susanto, S.; Husni, A. In vitro induction of tetraploid pummelo ‘Nambangan’ (Citrus maxima (Burm.) Merr.) by colchicine treatment using germinated seed, shoot tip and cotyledonary node as explants. Ann. Bogor. 2015, 19, 29–36. [Google Scholar]
- Wu, H.W.; Mooney, P. Autotetraploid tangor plant regeneration from in vitro citrus somatic embryogenic callus treated with colchicine. Plant Cell Tissue Organ Cult. 2002, 70, 99–104. [Google Scholar] [CrossRef]
- Elyazid, D.M.A.; El-Shereif, A.R. In vitro induction of polyploidy in Citrus reticulata Blanco. Am. J. Plant Sci. 2014, 5, 1679–1685. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, M.; Deng, X. Obtaining autotetraploids in vitro at a high frequency in Citrus sinensis. Plant Cell Tissue Organ Cult. 2007, 89, 211–216. [Google Scholar] [CrossRef]
- Jokari, S.; Shekafandeh, A.; Jowkar, A. In vitro tetraploidy induction in Mexican lime and sour orange and evaluation of their morphological and physiological characteristics. Plant Cell Tissue Organ Cult. 2022, 150, 651–668. [Google Scholar] [CrossRef]
- Bora, L.; Vijayakumar, R.M.; Ganga, M.; Ganesan, N.M.; Sarkar, M.; Kundu, M. Induction of polyploids in acid lime (Citrus aurantifolia) through colchicine treatment of in vitro derived shoot tip explants. Plant Breed. 2023, 142, 118–127. [Google Scholar] [CrossRef]
- Zeng, S.; Chen, C.; Hong, L.; Liu, J.; Deng, X. In vitro induction, regeneration and analysis of autotetraploids derived from protoplasts and callus treated with colchicine in Citrus. Plant Cell Tissue Organ Cult. 2006, 87, 85–93. [Google Scholar] [CrossRef]
- Cimen, B. Induction of polyploidy in C35 citrange through in vitro colchicine treatments of seed-derived explants. Int. J. Fruit Sci. 2020, 20, S1929–S1941. [Google Scholar] [CrossRef]
- Narukulla, V.; Lahane, Y.; Fiske, K.; Pandey, S.; Ziogas, V. Induction of polyploidy in Citrus rootstocks through in vitro colchicine treatment of seed-derived explants. Agronomy 2023, 13, 1442. [Google Scholar] [CrossRef]
- Notsuka, K.; Tsuru, T.; Shiraishi, M. Induced polyploid grapes via in vitro chromosome doubling. J. Jpn. Soc. Hortic. Sci. 2000, 69, 543–551. [Google Scholar] [CrossRef]
- Yang, X.M.; Cao, Z.Y.; An, L.Z.; Wang, Y.M.; Fang, X.W. In vitro tetraploid induction via colchicine treatment from diploid somatic embryos in grapevine (Vitis vinifera L.). Euphytica 2006, 152, 217–224. [Google Scholar] [CrossRef]
- Iannicelli, J.; Guariniello, J.; Tossi, V.E.; Regalado, J.J.; Di Ciaccio, L.; van Baren, C.M.; Pitta-Álvarez, S.I.; Escandón, A.S. The “polyploid effect” in the breeding of aromatic and medicinal species. Sci. Hortic. 2020, 260, 108854. [Google Scholar] [CrossRef]
- Lavania, U.C. Polyploidy, body size, and opportunities for genetic enhancement and fixation of heterozygosity in plants. Nucleus 2013, 56, 1–6. [Google Scholar] [CrossRef]
- Trojak-Goluch, A.; Kawka-Lipińska, M.; Wielgusz, K.; Praczyk, M. Polyploidy in industrial crops: Applications and perspectives in plant breeding. Agronomy 2021, 11, 2574. [Google Scholar] [CrossRef]
- Robinson, D.O.; Coate, J.E.; Singh, A.; Hong, L.; Bush, M.; Doyle, J.J.; Roeder, A.H.K. Ploidy and size at multiple scales in the Arabidopsis sepal. Plant Cell 2018, 30, 2308–2329. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, L.; Ye, T.; Zhao, J.; Wang, L.; Wei, H.; Liu, P.; Liu, M. Autotetraploidization alters morphology, photosynthesis, cytological characteristics and fruit quality in sour jujube (Ziziphus acidojujuba Cheng et Liu). Plants 2023, 12, 1106. [Google Scholar] [CrossRef]
- Bhuvaneswari, G.; Thirugnanasampandan, R.; Gogulramnath, M. Effect of colchicine induced tetraploidy on morphology, cytology, essential oil composition, gene expression and antioxidant activity of Citrus limon (L.) Osbeck. Physiol. Mol. Biol. Plants 2020, 26, 271–279. [Google Scholar] [CrossRef]
- Jiang, J.; Yang, N.; Li, L.; Qin, G.; Ren, K.; Wang, H.; Deng, J.; Ding, D. Tetraploidy in Citrus wilsonii enhances drought tolerance via synergistic regulation of photosynthesis, phosphorylation, and hormonal changes. Front. Plant Sci. 2023, 14, 1269493. [Google Scholar] [CrossRef]
- Yin, H.; Wang, X.; Shi, X.; Chen, Y.; Qi, K.; Xie, Z.; Qin, C.; Zhang, S.; Xiao, W. Induction and characterization of tetraploid pear from the seeds of ‘Dangshansuli’ (Pyrus bretschneideri Rehd.). Fruit Res. 2023, 3, 14. [Google Scholar] [CrossRef]
- Wójcik, D.; Marat, M.; Marasek-Ciołakowska, A.; Klamkowski, K.; Buler, Z.; Podwyszyńska, M.; Tomczyk, P.P.; Wójcik, K.; Treder, W.; Filipczak, J. Apple autotetraploids—Phenotypic characterisation and response to drought stress. Agronomy 2022, 12, 161. [Google Scholar] [CrossRef]
- Wang, L.J.; Cao, Q.Z.; Zhang, X.Q.; Jia, G.X. Effects of polyploidization on photosynthetic characteristics in three Lilium species. Sci. Hortic. 2021, 284, 110098. [Google Scholar] [CrossRef]
- Oberprieler, C.; Talianova, M.; Griesenbeck, J. Effects of polyploidy on the coordination of gene expression between organellar and nuclear genomes in Leucanthemum Mill. (Compositae, Anthemideae). Ecol. Evol. 2019, 9, 9100–9110. [Google Scholar] [CrossRef]
- Warner, D.A.; Edwards, G.E. Effects of polyploidy on photosynthesis. Photosynth. Res. 1993, 35, 135–147. [Google Scholar] [CrossRef]
- Xue, H.; Zhang, B.; Tian, J.-R.; Chen, M.-M.; Zhang, Y.-Y.; Zhang, Z.-H.; Ma, Y. Comparison of the morphology, growth and development of diploid and autotetraploid ‘Hanfu’ apple trees. Sci. Hortic. 2017, 225, 277–285. [Google Scholar] [CrossRef]
- Mao, H.T.; Pang, X.; Li, T.; Qin, Y.; Zhang, Z.W.; Yuan, S.; Yuan, M.; Brestic, M.; Chen, Y.E. Chlorophyll b is essential for the growth, photoprotection, and photosystem I assembly in wheat. Plant J. 2025, 123, e70442. [Google Scholar] [CrossRef]
- Biswal, A.K.; Pattanayak, G.K.; Ruhil, K.; Kandoi, D.; Mohanty, S.S.; Leelavati, S.; Reddy, V.S.; Govindjee, G.; Tripathy, B.C. Reduced expression of chlorophyllide a oxygenase (CAO) decreases the metabolic flux for chlorophyll synthesis and downregulates photosynthesis in tobacco plants. Physiol. Mol. Biol. Plants 2024, 30, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Koziara-Ciupa, M.; Trojak-Goluch, A. The effect of polyploidization on the physiological parameters, biochemical profile, and tolerance to abiotic and biotic stresses of plants. Agronomy 2025, 15, 1918. [Google Scholar] [CrossRef]
- Chen, T.; Sheng, Y.; Hao, Z.; Long, X.; Fu, F.; Liu, Y.; Tang, Z.; Ali, A.; Peng, Y.; Liu, Y.; et al. Transcriptome and proteome analysis suggest enhanced photosynthesis in tetraploid Liriodendron sino-americanum. Tree Physiol. 2021, 41, 1953–1971. [Google Scholar] [CrossRef] [PubMed]
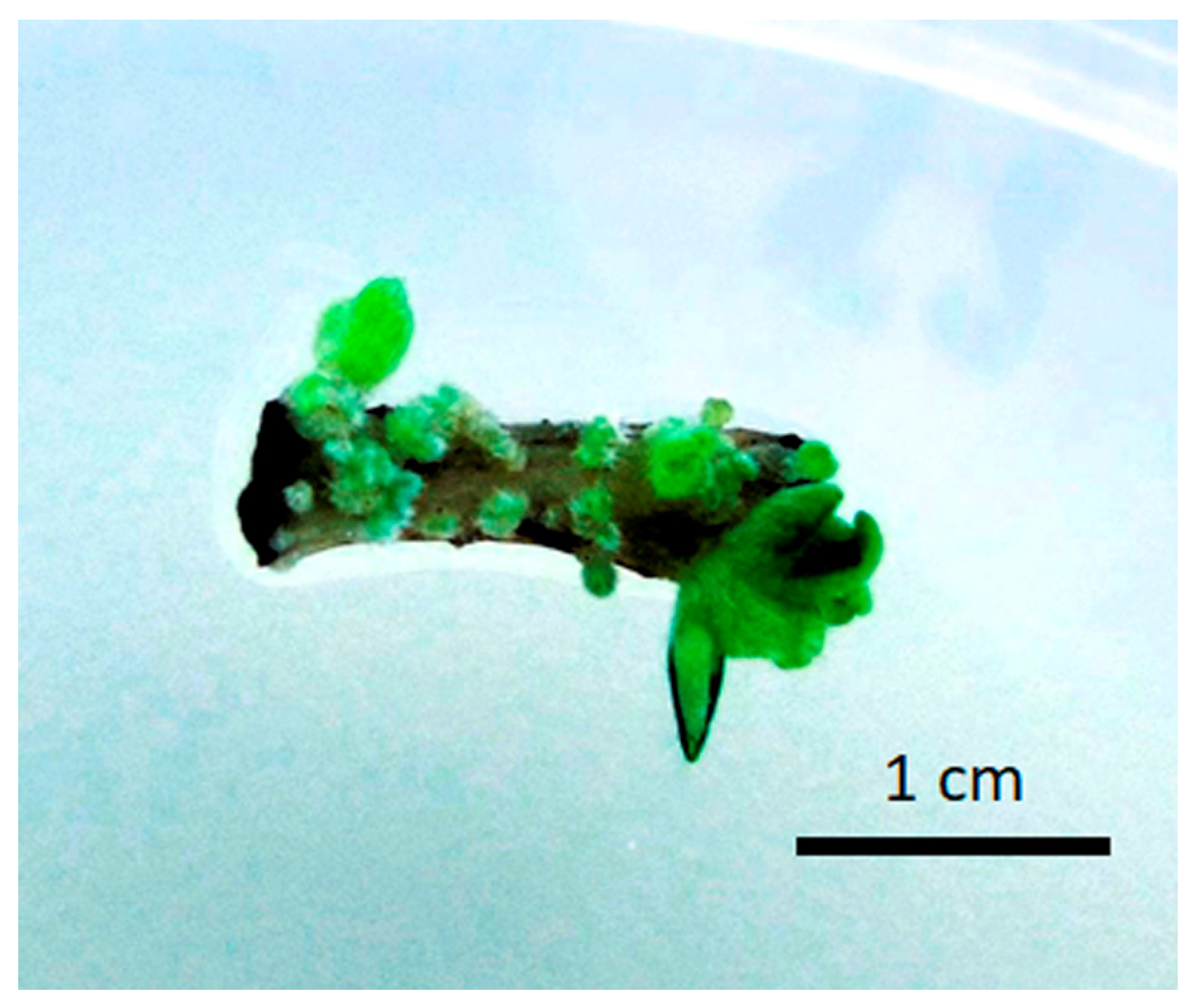
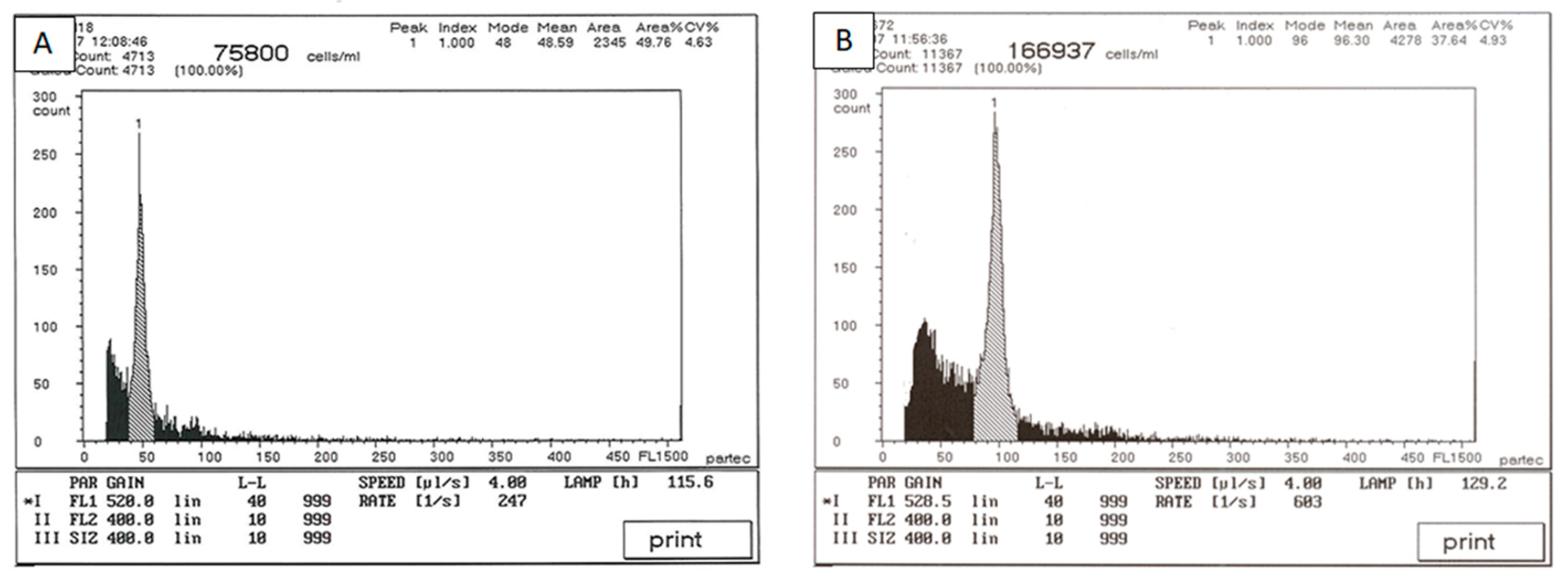
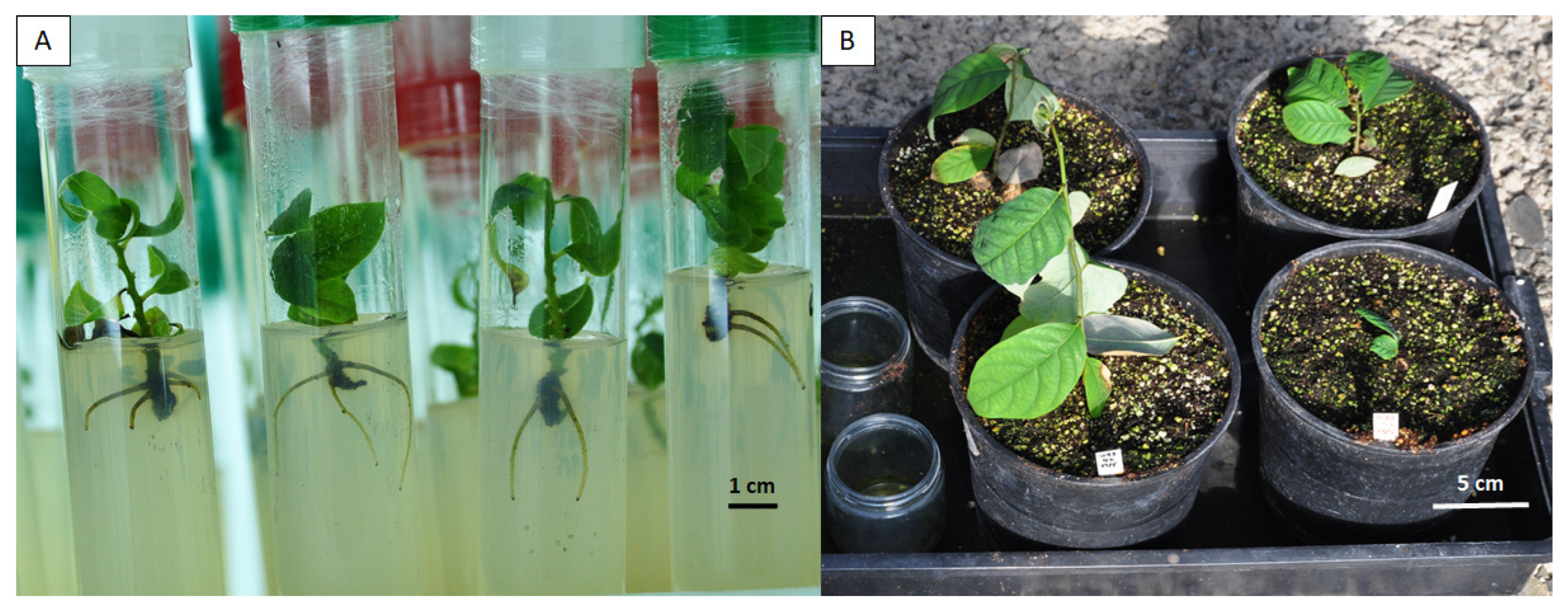
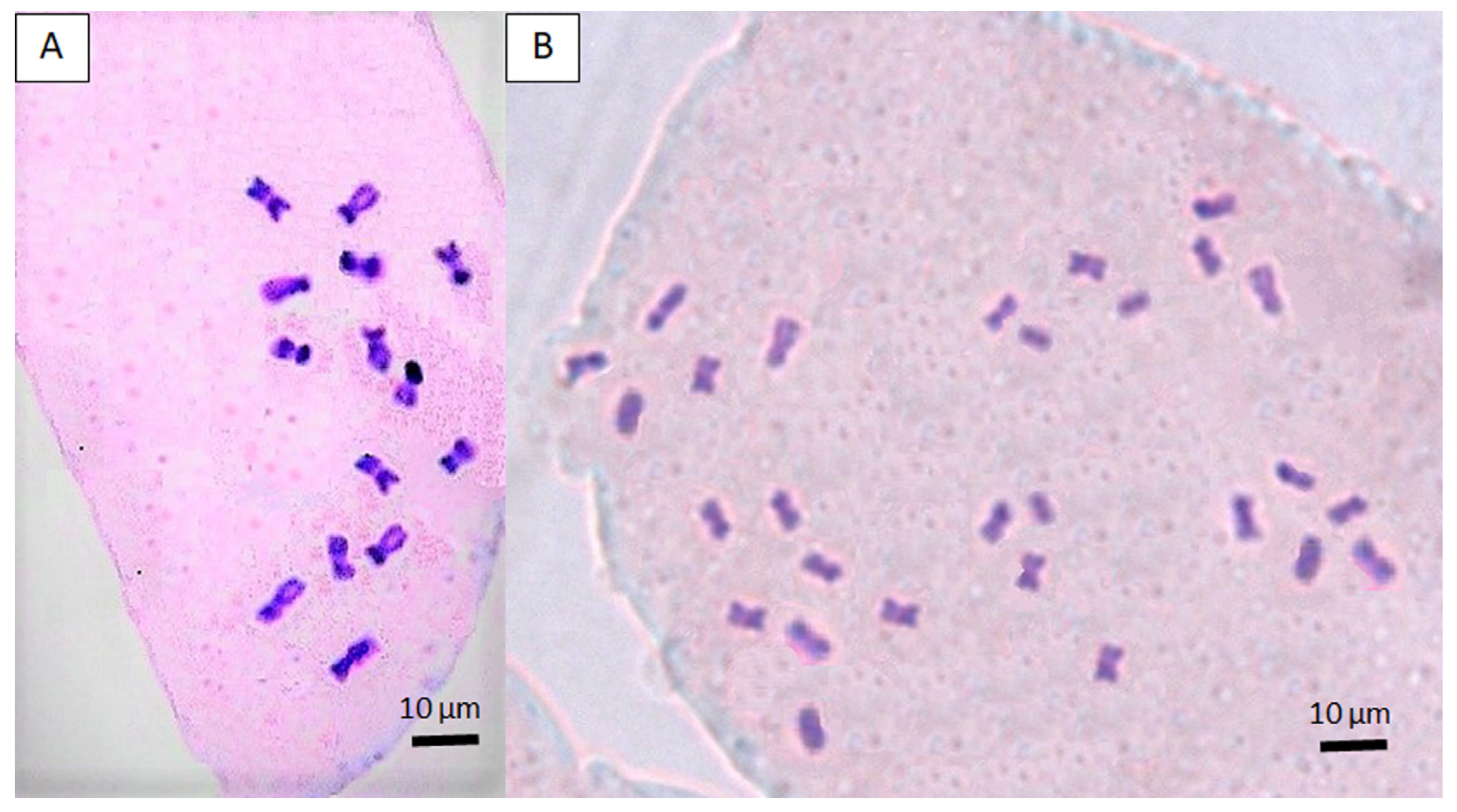
| % Colchicine | No. of Explants Initiated | % Regeneration 1 | Average Shoot No. 2 | % Shoot Survival 1 |
|---|---|---|---|---|
| 24 h colchicine treatment | ||||
| 0 | 50 | 100 ± 0 a | 4.5 ± 1.0 a | 100 ± 0 a |
| 0.01 | 50 | 74 ± 6 b | 4.3 ± 1.0 a | 70 ± 4 b |
| 0.05 | 50 | 48 ± 7 c | 4.2 ± 1.0 a | 45 ± 5 c |
| 0.1 | 50 | 42 ± 7 c | 4.1 ± 0.9 a | 40 ± 5 c |
| 0.2 | 50 | 18 ± 5 de | 1.0 ± 0.0 b | 22 ± 14 cd |
| 48 h colchicine treatment | ||||
| 0 | 50 | 100 ± 0 a | 4.3 ±1.0 a | 100 ± 0 a |
| 0.01 | 50 | 32 ± 7 cde | 1.2 ± 0.4 b | 21 ± 9 d |
| 0.05 | 50 | 16 ± 5 de | 1.0 ± 0.0 b | 13 ± 12 d |
| 0.1 | 50 | 12 ± 5 e | 1.0 ± 0.0 b | 17 ± 15 cd |
| 0.2 | 50 | 14 ± 5 e | 1.0 ± 0.0 b | 0 ± 0 d |
| % Colchicine | No. of Explants Analyzed | Polyploidization (%) | Triploid (3x) No. (%) | Tetraploid (4x) No. (%) | Mixoploid (2x–4x) No. (%) |
|---|---|---|---|---|---|
| 0 | 20 | 0 ± 0 c | 0 (0 ± 0 a) | 0 (0 ± 0 b) | 0 (0 ± 0 a) |
| 0.01 | 155 | 1.3 ± 0.9 bc | 2 (1.3 ± 0.9 a) | 0 (0 ± 0 b) | 0 (0 ± 0 a) |
| 0.05 | 99 | 4.0 ± 2.0 ab | 2 (2.0 ± 1.4 a) | 2 (2.0 ±1.4 ab) | 0 (0 ± 0 a) |
| 0.1 | 86 | 10.5 ± 3.3 a | 1 (1.2 ± 1.2 a) | 5 (5.8 ± 2.5 a) | 3 (3.5 ± 2.0 a) |
| Ploidy Level | Code of Polyploid Shoots | Survival Rate: (No. of Survivors/Total) | Code of Surviving Shoots |
|---|---|---|---|
| Tetraploid (4x) | 2022, 2025, 2943, 2050, 2069, 2083, 2084 | 43 ± 19% (3/7) | 2069, 2083, 2084 |
| Triploid (3x) | 522, 615, 1010, 1015, 2019 | 40 ± 22% (2/5) | 522, 1015 |
| Mixoploid (2x–4x) | 2073, 2084, 2088 | 67 ± 27% (2/3) | 2073, 2084 |
| Cherimoya foliar size (in vitro) | 2n = 2x = 14 | 2n = 4x = 28 |
| Average Length (cm) | 1.6 ± 0.3 b | 2.1 ± 0.2 a |
| Average Width (cm) | 0.7 ±0.1 b | 1.2 ±0.1 a |
| Leaf Index (L/W) | 2.3 ± 0.1 a | 1.8 ± 0.1 b |
| Cherimoya foliar size (ex vitro) | 2n = 2x = 14 | 2n = 4x = 28 |
| Average Length (cm) | 18.4 ± 1.5 b | 22.2 ± 1.6 a |
| Average Width (cm) | 10.2 ± 1.2 b | 13.7 ± 1.1 a |
| Leaf Index (L/W) | 1.8 ± 0.2 a | 1.6 ± 0.1 b |
| Chlorophyll content in cherimoya leaves in vitro | 2n = 2x = 14 | 2n = 4x = 28 |
| Chlorophyll A (mg g−1 of FW) | 42.5 ± 0.4 b | 47.4 ± 0.4 a |
| Chlorophyll B (mg g−1 of FW) | 38.2 ± 0.3 b | 39.1 ± 0.2 a |
| Chlorophyll total (mg g−1 of FW) | 68.5 ± 0.5 b | 69.2 ± 0.2 a |
| Ratio Chl a/Chl b | 1.11 ± 0.01 b | 1.21 ± 0.01 a |
| Chlorophyll content in cherimoya leaves ex vitro | 2n = 2x = 14 | 2n = 4x = 28 |
| Chlorophyll A (mg g−1 of FW) | 23.4 ± 0.2 b | 27.9 ± 0.2 a |
| Chlorophyll B (mg g−1 of FW) | 13.5 ± 0.3 b | 14.7 ± 0.3 a |
| Chlorophyll total (mg g−1 of FW) | 32.2 ± 0.8 b | 36.5 ± 0.3 a |
| Ratio Chl a/Chl b | 1.73 ± 0.02 b | 1.90 ± 0.04 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Encina, C.L.; Regalado, J.J. In Vitro Induction of Autotetraploids in the Subtropical Fruit Tree Cherimoya (Annona cherimola Mill.). Horticulturae 2026, 12, 25. https://doi.org/10.3390/horticulturae12010025
Encina CL, Regalado JJ. In Vitro Induction of Autotetraploids in the Subtropical Fruit Tree Cherimoya (Annona cherimola Mill.). Horticulturae. 2026; 12(1):25. https://doi.org/10.3390/horticulturae12010025
Chicago/Turabian StyleEncina, Carlos Lopez, and José Javier Regalado. 2026. "In Vitro Induction of Autotetraploids in the Subtropical Fruit Tree Cherimoya (Annona cherimola Mill.)" Horticulturae 12, no. 1: 25. https://doi.org/10.3390/horticulturae12010025
APA StyleEncina, C. L., & Regalado, J. J. (2026). In Vitro Induction of Autotetraploids in the Subtropical Fruit Tree Cherimoya (Annona cherimola Mill.). Horticulturae, 12(1), 25. https://doi.org/10.3390/horticulturae12010025






