Genetic Diversity Analysis in Natural Chinese Holly Using ISSR and SCoT Markers
Abstract
1. Introduction
2. Materials and Methods
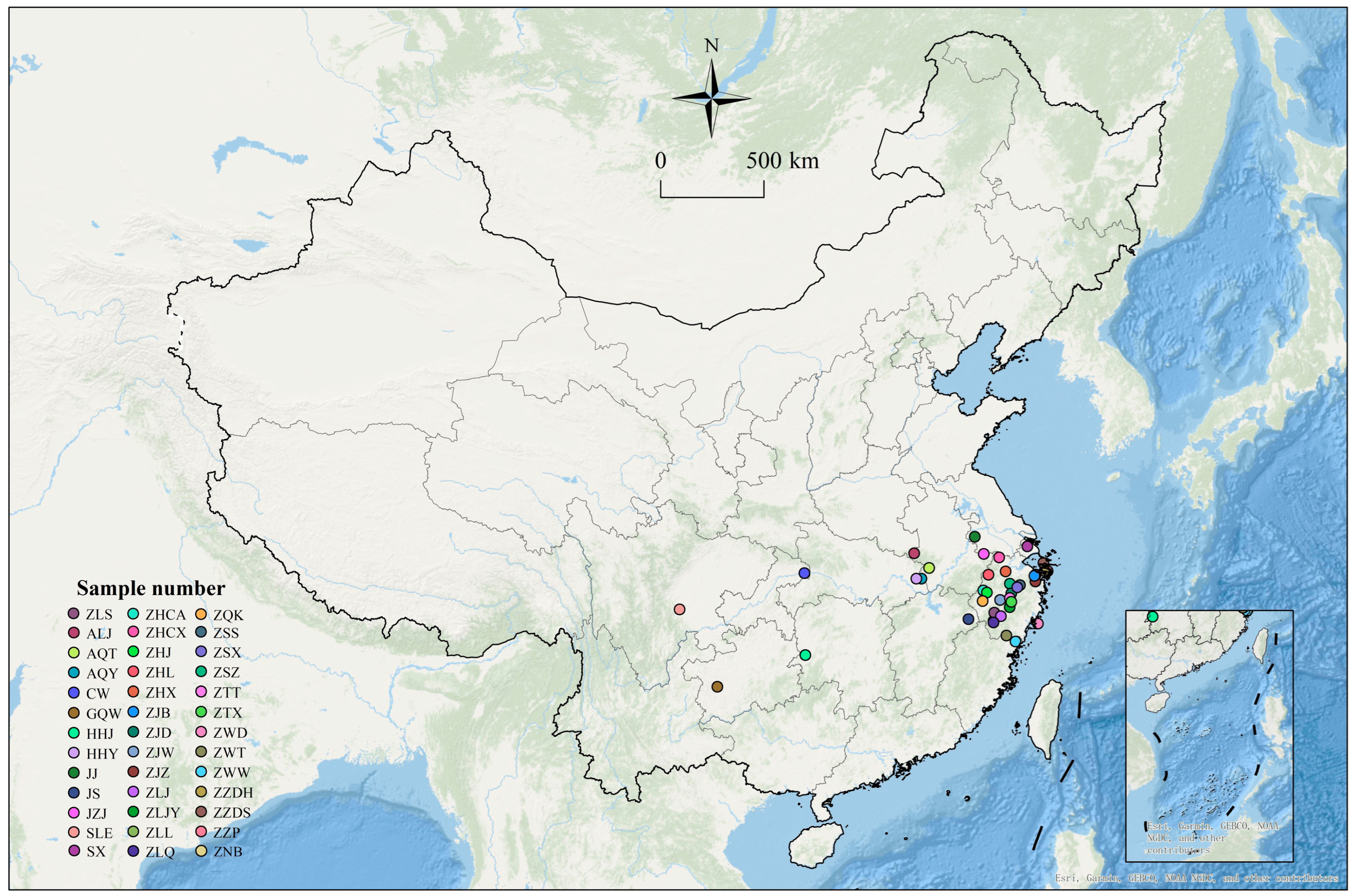
2.1. Plant Materials
2.2. DNA Extraction Procedure
2.3. ISSR and SCoT Amplification
2.4. Data Analysis
3. Results
3.1. Primer Amplification Efficiency and Polymorphism Performance
3.2. Genetic Diversity Parameters
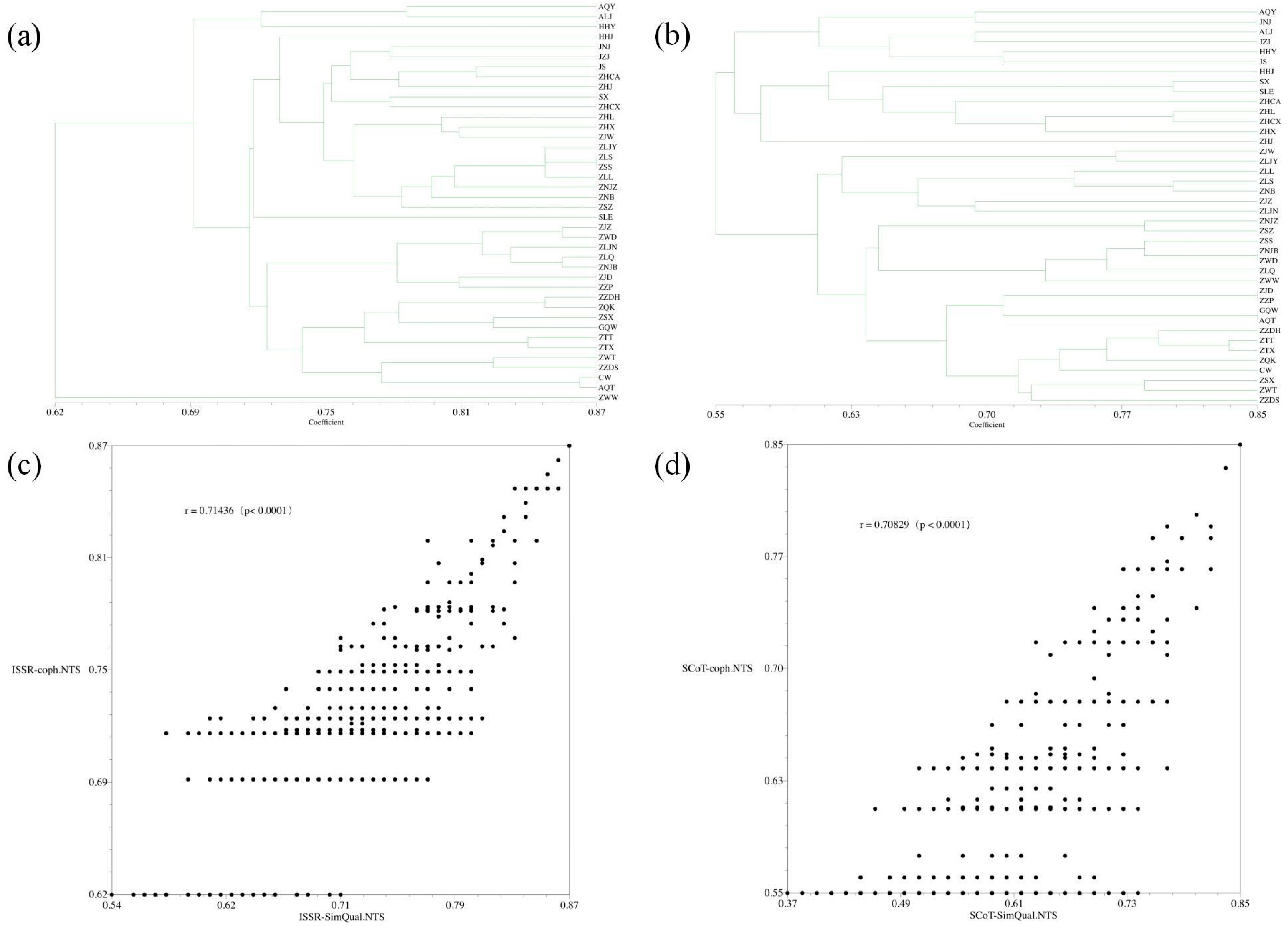
3.3. Genetic Similarity Coefficients
3.4. Cluster Analysis
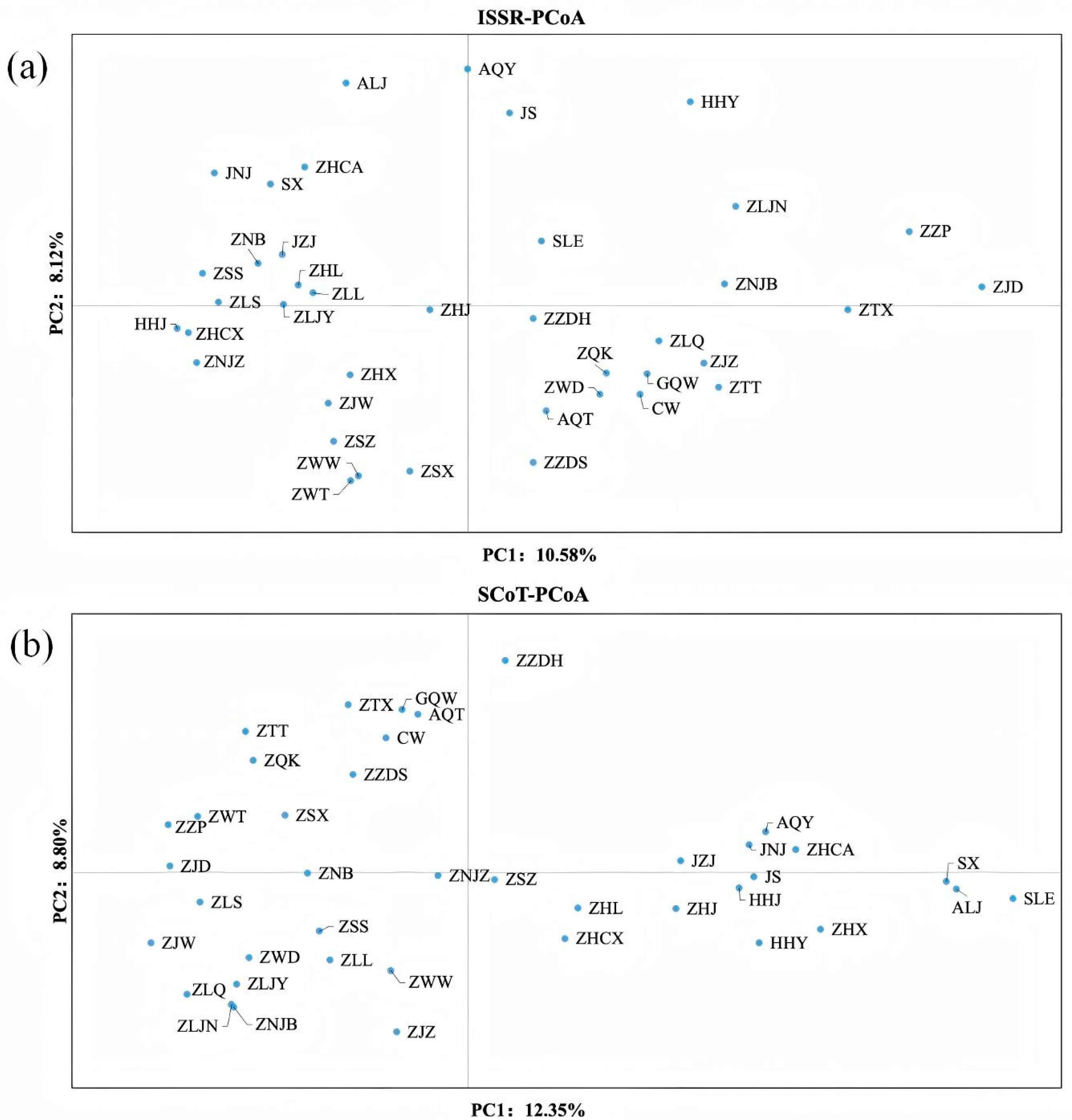
3.5. Principal Coordinates Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ISSR | Inter-Simple Sequence Repeat |
| SCoT | Start Codon Targeted Polymorphism |
| AFLP | Amplified Fragment Length Polymorphism |
| SSR | Simple Sequence Repeats |
| SRAP | Sequence-Related Amplified Polymorphism |
| SNP | Single Nucleotide Polymorphisms |
| TB | Total Number of Amplification Bits |
| PB | Number of Polymorphic Bits |
| PPB | Percentage of Polymorphic Bands |
| Na | Allele Number |
| Ne | Effective Allele Number |
| I | Shan-non’s information index |
| H | Nei’s gene diversity index |
| AQY | Yuexi County, Anqing City, Anhui Province |
| ALJ | Jinzhai County, Lu’an City, Anhui Province |
| HHY | Yingshan County, Huanggang City, Hubei Province |
| HHJ | Jingzhou County, Huaihua City, Hunan Province |
| JNJ | Jiangning District, Nanjing City, Jiangsu Province |
| JZJ | Jurong City, Zhenjiang City, Jiangsu Province |
| JS | Shangrao City, Jiangxi Province |
| SX | Xuhui District, Shanghai City |
| SLE | E’mei Mountain City, Leshan City, Sichuan Province |
| ZHCA | Chun’an County, Hangzhou City, Zhejiang Province |
| ZHJ | Jiande City, Hangzhou City, Zhejiang Province |
| ZHL | Lin’an District, Hangzhou City, Zhejiang Province |
| ZHX | Xihu District, Hangzhou City, Zhejiang Province |
| ZHCX | Changxing County, Huzhou City, Zhejiang Province |
| ZJW | Wucheng District, Jinhua City, Zhejiang Province |
| ZLJY | Jinyun County, Lishui City, Zhejiang Province |
| ZLL | Longquan City, Lishui City, Zhejiang Province |
| ZLS | Suichang County, Lishui City, Zhejiang Province |
| ZNB | Beilun District, Ningbo City, Zhejiang Province |
| ZNJZ | Yinzhou District, Ningbo City, Zhejiang Province |
| ZSS | Shengzhou City, Shaoxing City, Zhejiang Province |
| ZSZ | Zhuji City, Shaoxing City, Zhejiang Province |
| ZJZ | Zhenhai District, Ningbo City, Zhejiang Province |
| ZLJN | Jingning County, Lishui City, Zhejiang Province |
| ZJD | Dongyang City, Jinhua City, Zhejiang Province |
| ZZP | Putuo District, Zhoushan City, Zhejiang Province |
| ZLQ | Qingtian County, Lishui City, Zhejiang Province |
| ZNJB | Jiangbei District, Ningbo City, Zhejiang Province |
| ZWD | Dongtou County, Wenzhou City, Zhejiang Province |
| ZWW | Wencheng County, Wenzhou City, Zhejiang Province |
| ZZDH | Dinghai District, Zhoushan City, Zhejiang Province |
| ZQK | Kaihua County, Quzhou City, Zhejiang Province |
| ZSX | Xinchang County, Shaoxing City, Zhejiang Province |
| ZTT | Tiantai County, Taizhou City, Zhejiang Province |
| ZTX | Xianju County, Taizhou City, Zhejiang Province |
| CW | Wushan County, Chongqing Municipality |
| ZWT | Taishun County, Wenzhou City, Zhejiang Province |
| ZZDS | Daishan County, Zhoushan City, Zhejiang Province |
| GQW | Wangmo County, Qiannan Buyi and Miao Autonomous Prefecture, Guizhou Province |
| AQT | Tongcheng County, Anqing City, Anhui Province |
References
- Hu, S. The genus Ilex in China. J. Arnold Arbor. 1949, 30, 233–344. [Google Scholar] [CrossRef]
- Yao, X.; Zhang, F.; Corlett, R.T. Utilization of the hollies (Ilex L. spp.): A review. Forests 2022, 13, 94. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, D. Ornamental plant resources from China. In Proceedings of the XXVI International Horticultural Congress: Asian Plants with Unique Horticultural Potential: Genetic Resources, Cultural 620, Toronto, ON, Canada, 11–17 August 2002; pp. 365–375. [Google Scholar]
- Liu, J.; Coomes, D.A.; Gibson, L.; Hu, G.; Liu, J.; Luo, Y.; Wu, C.; Yu, M. Forest fragmentation in China and its effect on biodiversity. Biol. Rev. 2019, 94, 1636–1657. [Google Scholar] [CrossRef]
- Liu, J.; Zhan, J.; Deng, X. Spatio-temporal patterns and driving forces of urban land expansion in China during the economic reform era. AMBIO J. Hum. Environ. 2005, 34, 450–455. [Google Scholar] [CrossRef][Green Version]
- Li, R.F.; Xia, W.Q.; Liu, J.B.; Cui, B.S.; Hou, Q.; Sun, H.; Li, S. New triterpenoid saponins from the leaves of Ilex chinensis. Fitoterapia 2018, 131, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Li, R.; Sun, H.; Li, S. New triterpenoid saponins from the leaves of Ilex chinensis and their hepatoprotective activity. Chin. J. Nat. Med. 2021, 19, 376–384. [Google Scholar] [CrossRef]
- College, J.N.M. Chinese Native Medicine Big Dictionary; Jiangsu Science and Technology Press: Nanjing, China, 1979. [Google Scholar]
- McLenachan, P.A.; Stöckler, K.; Winkworth, R.C.; McBreen, K.; Zauner, S.; Lockhart, P.J. Markers derived from amplified fragment length polymorphism gels for plant ecology and evolution studies. Mol. Ecol. 2000, 9, 1899–1903. [Google Scholar] [CrossRef]
- Ivandic, V.; Hackett, C.A.; Nevo, E.; Keith, R.; Thomas, W.T.; Forster, B.P. Analysis of simple sequence repeats (SSRs) in wild barley from the Fertile Crescent: Associations with ecology, geography and flowering time. Plant Mol. Biol. 2002, 48, 511–527. [Google Scholar] [CrossRef] [PubMed]
- Ayesiga, S.B.; Rubaihayo, P.; Oloka, B.M.; Dramadri, I.O.; Sserumaga, J.P. Genome-wide association study and pathway analysis to decipher loci associated with Fusarium ear rot resistance in tropical maize germplasm. Genet. Resour. Crop. Evol. 2024, 71, 2435–2448. [Google Scholar] [CrossRef]
- Joshi, P.; Dhawan, V. Assessment of genetic fidelity of micropropagated Swertia chirayita plantlets by ISSR marker assay. Biol. Plant. 2007, 51, 22–26. [Google Scholar] [CrossRef]
- Collard, B.C.; Mackill, D.J. Start codon targeted (SCoT) polymorphism: A simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol. Biol. Rep. 2009, 27, 86–93. [Google Scholar] [CrossRef]
- Kwok, P.; Chen, X. Detection of single nucleotide polymorphisms. Curr. Issues Mol. Biol. 2003, 5, 43–60. [Google Scholar]
- Daneva, V.; Beniwal, R.; Kajla, S.; Poonia, A.K.; Kumar, M.; Kajal. Assessment of genetic diversity in Tecomella undulata by using ISSR markers. Vegetos 2023, 36, 1526–1534. [Google Scholar] [CrossRef]
- Sheikh, Z.N.; Sharma, V.; Shah, R.A.; Sharma, N.; Summuna, B.; Al-Misned, F.A.; El-Serehy, H.A.; Mir, J.I. Genetic diversity analysis and population structure in apricot (Prunus armeniaca L.) grown under north-western himalayas using ISSR markers. Saudi J. Biol. Sci. 2021, 28, 5986–5992. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Su, H.; Pu, S.; Chen, H.; El-Kassaby, Y.A.; Yang, Z.; Feng, J. High-yield hybrid breeding of Camellia oleifolia based on ISSR molecular markers. BMC Plant Biol. 2024, 24, 517. [Google Scholar] [CrossRef] [PubMed]
- Sidhiq, D.F.; Widiyastuti, Y.; Subositi, D.; Pujiasmanto, B.; Yunus, A. Morphological diversity, total phenolic and flavonoid content of Echinacea purpurea cultivated in Karangpandan, Central Java, Indonesia. Biodiversitas J. Biol. Divers. 2020, 21, 1265–1271. [Google Scholar] [CrossRef]
- Vishnu, B.; Siril, E. Start Codon Targeted (SCoT) markers unveiled the genetic diversity of Nelumbo nucifera Gaertn. from Kerala, South India. Gene Rep. 2025, 40, 102262. [Google Scholar] [CrossRef]
- Altaf, M.T.; Nadeem, M.A.; Ali, A.; Liaqat, W.; Bedir, M.; Baran, N.; Ilić, A.; Ilyas, M.K.; Ghafoor, A.; Dogan, H. Applicability of Start Codon Targeted (SCoT) markers for the assessment of genetic diversity in bread wheat germplasm. Genet. Resour. Crop Evol. 2025, 72, 1205–1218. [Google Scholar] [CrossRef]
- Al-Qurainy, F.; Khan, S.; Nadeem, M.; Tarroum, M. SCoT marker for the assessment of genetic diversity in Saudi Arabian date palm cultivars. Pak. J. Bot. 2015, 47, 637–643. [Google Scholar]
- Nouri, A.; Golabadi, M.; Etminan, A.; Rezaei, A.; Mehrabi, A.A. Comparative assessment of SCoT and ISSR markers for analysis of genetic diversity and population structure in some Aegilops tauschii Coss. accessions. Plant Genet. Resour. Charact. Util. 2021, 19, 375–383. [Google Scholar] [CrossRef]
- Hassan, F.A.S.; Ismail, I.A.; Mazrou, R.; Hassan, M. Applicability of inter-simple sequence repeat (ISSR), start codon targeted (SCoT) markers and ITS2 gene sequencing for genetic diversity assessment in Moringa oleifera Lam. J. Appl. Res. Med. Aromat. Plants 2020, 18, 100256. [Google Scholar] [CrossRef]
- Yilmaz, A.; Ciftci, V. Genetic relationships and diversity analysis in Turkish laurel (Laurus nobilis L.) germplasm using ISSR and SCoT markers. Mol. Biol. Rep. 2021, 48, 4537–4547. [Google Scholar] [CrossRef]
- Igwe, D.O.; Ihearahu, O.C.; Osano, A.A.; Acquaah, G.; Ude, G.N. Assessment of genetic diversity of Musa species accessions with variable genomes using ISSR and SCoT markers. Genet. Resour. Crop Evol. 2022, 69, 49–70. [Google Scholar] [CrossRef]
- Tsaktsira, M.; Chavale, E.; Kostas, S.; Pipinis, E.; Tsoulpha, P.; Hatzilazarou, S.; Ziogou, F.-T.; Nianiou-Obeidat, I.; Iliev, I.; Economou, A.; et al. Vegetative Propagation and ISSR-Based Genetic Identification of Genotypes of Ilex aquifolium ‘Agrifoglio Commune’. Sustainability 2021, 13, 10345. [Google Scholar] [CrossRef]
- Leng, X.; Wang, Z.; An, S.; Feng, J.; Liu, Y.; Wang, G. ISSR analysis of genetic diversity of Ilex integra, an insular endemic plant. Biodiv. Sci. 2005, 13, 546–554. [Google Scholar] [CrossRef]
- Roldan-Ruiz, I.; Dendauw, J.; Van Bockstaele, E.; Depicker, A.; De Loose, M. AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol. Breed. 2000, 6, 125–134. [Google Scholar] [CrossRef]
- Yeh, F.; Yang, R.; Boyle, T. Microsoft Window-Based Freeware for Population Genetic Analysis, Quick User Guide, POPGENE version 1.31; Center for International Forestry Research, University of Alberta: Edmonton, AB, Canada, 1999; pp. 1–29. [Google Scholar]
- Rohlf, F.J. NTSYS-Pc: Numerical Taxonomy and Multivariate Analysis System, version 2.1, Exeter Software: New York, NY, USA, 2000.
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, S.; Wang, S.; Su, H.; De, J. Genetic Diversity and Metabolic Profile of Tibetan Medicinal Plant Saussurea obvallata. Genes 2025, 16, 593. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Ramírez, M.E.; Magaña-Álvarez, A.; Pérez-Brito, D.; Cortés-Velázquez, A.; Nexticapan-Garcéz, Á.; Tapia-Tussell, R.; Martín-Mex, R. Exploring the Genetic Variability of Gmelina arborea Roxb. in Mexico with Molecular Markers to Establish an Efficient Improvement Program. Plants 2025, 14, 1888. [Google Scholar] [CrossRef] [PubMed]
- Igwe, D.O.; Afiukwa, C.A.; Ubi, B.E.; Ogbu, K.I.; Ojuederie, O.B.; Ude, G.N. Assessment of genetic diversity in Vigna unguiculata L. (Walp) accessions using inter-simple sequence repeat (ISSR) and start codon targeted (SCoT) polymorphic markers. BMC Genom. Data 2017, 18, 98. [Google Scholar] [CrossRef] [PubMed]
- Pakseresht, F.; Talebi, R.; Karami, E. Comparative assessment of ISSR, DAMD and SCoT markers for evaluation of genetic diversity and conservation of landrace chickpea (Cicer arietinum L.) genotypes collected from north-west of Iran. Physiol. Mol. Biol. Plants 2013, 19, 563–574. [Google Scholar] [CrossRef]
- Browne, L.; Karubian, J. Habitat loss and fragmentation reduce effective gene flow by disrupting seed dispersal in a neotropical palm. Mol. Ecol. 2018, 27, 3055–3069. [Google Scholar] [CrossRef] [PubMed]
- Toczydlowski, R.H.; Waller, D.M. Drift happens: Molecular genetic diversity and differentiation among populations of jewelweed (Impatiens capensis Meerb.) reflect fragmentation of floodplain forests. Mol. Ecol. 2019, 28, 2459–2475. [Google Scholar] [CrossRef] [PubMed]
- da Silva Carvalho, C.; Ribeiro, M.C.; Côrtes, M.C.; Galetti, M.; Collevatti, R.G. Contemporary and historic factors influence differently genetic differentiation and diversity in a tropical palm. Heredity 2015, 115, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Ålund, M.; Cenzer, M.; Bierne, N.; Boughman, J.W.; Cerca, J.; Comerford, M.S.; Culicchi, A.; Langerhans, B.; McFarlane, S.E.; Möst, M.H.; et al. Anthropogenic Change and the Process of Speciation. Cold Spring Harb. Perspect. Biol. 2023, 15, a041455. [Google Scholar] [CrossRef] [PubMed]
- Luz, G.C.; Strioto, D.K.; Mangolin, C.A.; Machado, M. ISSR markers to assess genetic diversity of cultivated populations from artificial selection of Stevia rebaudiana (Bert.) Bertoni. Breed Sci. 2020, 70, 508–514. [Google Scholar] [CrossRef]
- Slatkin, M. Gene flow and the geographic structure of natural populations. Science 1987, 236, 787–792. [Google Scholar] [CrossRef]
- Chen, W.W.; Xiao, Z.Z.; Tong, X.; Liu, Y.P.; Li, Y.Y. Development and characterization of 25 microsatellite primers for Ilex chinensis (Aquifoliaceae). Appl. Plant Sci. 2015, 3, 1500057. [Google Scholar] [CrossRef]
- Hou, S.; Zhou, P.; Fang, Y.; Wang, X.; Zhang, M.; Zhang, Q. The Genetic Diversity of Natural Ilex chinensis Sims (Aquifoliaceae) Populations as Revealed by SSR Markers. Forests 2024, 15, 763. [Google Scholar] [CrossRef]
- Zhou, P.; Li, J.; Huang, J.; Li, F.; Zhang, Q.; Zhang, M. Genome Survey Sequencing and Genetic Background Characterization of Ilex chinensis Sims (Aquifoliaceae) Based on Next-Generation Sequencing. Plants 2022, 11, 3322. [Google Scholar] [CrossRef]
| Primer Name | Sequence of Primer (5′-3′) | Total No. of Amplification (B) | No. of Polymorphic (A) | Polymorphism (A/B) % |
|---|---|---|---|---|
| UBC811 | GAGAGAGAGAGAGAGAC | 12 | 10 | 83% |
| UBC815 | CTCTCTCTCTCTCTCTCTG | 11 | 10 | 91% |
| UBC824 | TCTCTCTCTCTCTCTCG | 7 | 7 | 100% |
| UBC834 | AGAGAGAGAGAGAGAGYT | 8 | 6 | 75% |
| UBC835 | AGAGAGAGAGAGAGAGYC | 9 | 6 | 67% |
| UBC836 | AGAGAGAGAGAGAGAGYA | 9 | 7 | 78% |
| UBC840 | GAGAGAGAGAGAGAGAYT | 11 | 11 | 100% |
| UBC841 | GAGAGAGAGAGAGAGAYC | 8 | 7 | 88% |
| UBC842 | CTCTCTCTCTCTCTCTRA | 9 | 7 | 78% |
| UBC843 | CTCTCTCTCTCTCTCTRC | 7 | 7 | 100% |
| UBC844 | CTCTCTCTCTCTCTCTRG | 9 | 9 | 100% |
| UBC873 | GACAGACAGACAGACA | 16 | 16 | 100% |
| UBC874 | CCCTCCCTCCCTCCCT | 10 | 8 | 80% |
| Total bands scored | 126 | 111 | 88% | |
| SCoT11 | AAGCAATGGCTACACCAA | 12 | 12 | 100% |
| SCoT13 | ACGACATGGGACACATCG | 16 | 16 | 100% |
| SCoT19 | ACCATGGCTACACCCGGC | 9 | 9 | 100% |
| SCoT20 | ACCATGGCTACACCCGGG | 10 | 10 | 100% |
| SCoT34 | ACCATGGCTACCCCGGA | 9 | 9 | 100% |
| SCoT35 | CATGGCTACCCCGGGCC | 9 | 9 | 100% |
| Total bands scored | 65 | 65 | 100% |
| Primer Type | Primer Name | PB | TB | PPB | Na | Ne | I | H | PIC |
|---|---|---|---|---|---|---|---|---|---|
| UBC811 | 10 | 12 | 83% | 1.833 | 1.524 | 0.296 | 0.436 | 0.30 | |
| UBC815 | 10 | 11 | 91% | 1.909 | 1.509 | 0.286 | 0.429 | 0.29 | |
| UBC824 | 7 | 7 | 100% | 2.000 | 1.461 | 0.279 | 0.432 | 0.28 | |
| UBC834 | 6 | 8 | 75% | 1.750 | 1.512 | 0.282 | 0.412 | 0.28 | |
| UBC835 | 6 | 9 | 67% | 1.667 | 1.275 | 0.181 | 0.286 | 0.18 | |
| ISSR | UBC836 | 7 | 9 | 78% | 1.778 | 1.539 | 0.299 | 0.438 | 0.30 |
| UBC840 | 11 | 11 | 100% | 2.000 | 1.477 | 0.281 | 0.433 | 0.28 | |
| UBC841 | 7 | 8 | 88% | 1.875 | 1.347 | 0.211 | 0.329 | 0.21 | |
| UBC842 | 7 | 9 | 78% | 1.778 | 1.529 | 0.294 | 0.429 | 0.29 | |
| UBC843 | 7 | 7 | 100% | 2.000 | 1.594 | 0.340 | 0.504 | 0.34 | |
| UBC844 | 9 | 9 | 100% | 2.000 | 1.309 | 0.209 | 0.348 | 0.21 | |
| UBC873 | 16 | 16 | 100% | 2.000 | 1.492 | 0.309 | 0.479 | 0.31 | |
| UBC874 | 8 | 10 | 80% | 1.800 | 1.424 | 0.253 | 0.384 | 0.25 | |
| AV. | 1.876 | 1.461 | 0.271 | 0.411 | 0.27 | ||||
| SCoT11 | 12 | 12 | 100% | 2.000 | 1.860 | 0.458 | 0.649 | 0.46 | |
| SCoT13 | 16 | 16 | 100% | 2.000 | 1.666 | 0.378 | 0.557 | 0.38 | |
| SCoT19 | 9 | 9 | 100% | 2.000 | 1.695 | 0.402 | 0.589 | 0.40 | |
| SCoT20 | 10 | 10 | 100% | 2.000 | 1.678 | 0.391 | 0.576 | 0.39 | |
| SCoT34 | 9 | 9 | 100% | 2.000 | 1.645 | 0.369 | 0.546 | 0.37 | |
| SCoT35 | 9 | 9 | 100% | 2.000 | 1.623 | 0.359 | 0.536 | 0.36 | |
| AV. | 2.000 | 1.695 | 0.393 | 0.575 | 0,39 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.; He, H.; Zhang, B.; Zuo, J.; Ding, W.; Zheng, B.; Jiao, J.; Wang, X. Genetic Diversity Analysis in Natural Chinese Holly Using ISSR and SCoT Markers. Horticulturae 2025, 11, 1078. https://doi.org/10.3390/horticulturae11091078
Liu M, He H, Zhang B, Zuo J, Ding W, Zheng B, Jiao J, Wang X. Genetic Diversity Analysis in Natural Chinese Holly Using ISSR and SCoT Markers. Horticulturae. 2025; 11(9):1078. https://doi.org/10.3390/horticulturae11091078
Chicago/Turabian StyleLiu, Meng, Huixue He, Baoxin Zhang, Jianfang Zuo, Wona Ding, Bingsong Zheng, Jiejie Jiao, and Xiaofei Wang. 2025. "Genetic Diversity Analysis in Natural Chinese Holly Using ISSR and SCoT Markers" Horticulturae 11, no. 9: 1078. https://doi.org/10.3390/horticulturae11091078
APA StyleLiu, M., He, H., Zhang, B., Zuo, J., Ding, W., Zheng, B., Jiao, J., & Wang, X. (2025). Genetic Diversity Analysis in Natural Chinese Holly Using ISSR and SCoT Markers. Horticulturae, 11(9), 1078. https://doi.org/10.3390/horticulturae11091078






