Genome-Wide Identification of the Growth-Regulating Factor (GRF) Gene Family in Three Cymbidium Species and Expression Patterns in C. goeringii
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of the GRF Gene Family in Three Cymbidium Species
2.2. Physicochemical Property Analysis and Phylogenetic Analysis
2.3. Gene Structure Analysis and Conserved Domain of GRF
2.4. Chromosome Localization and Colinearity Analysis
2.5. Cis-Acting Element Prediction
2.6. qRT-PCR Analysis
3. Results
3.1. The Characterization of the GRF Genes in Three Cymbidium Species
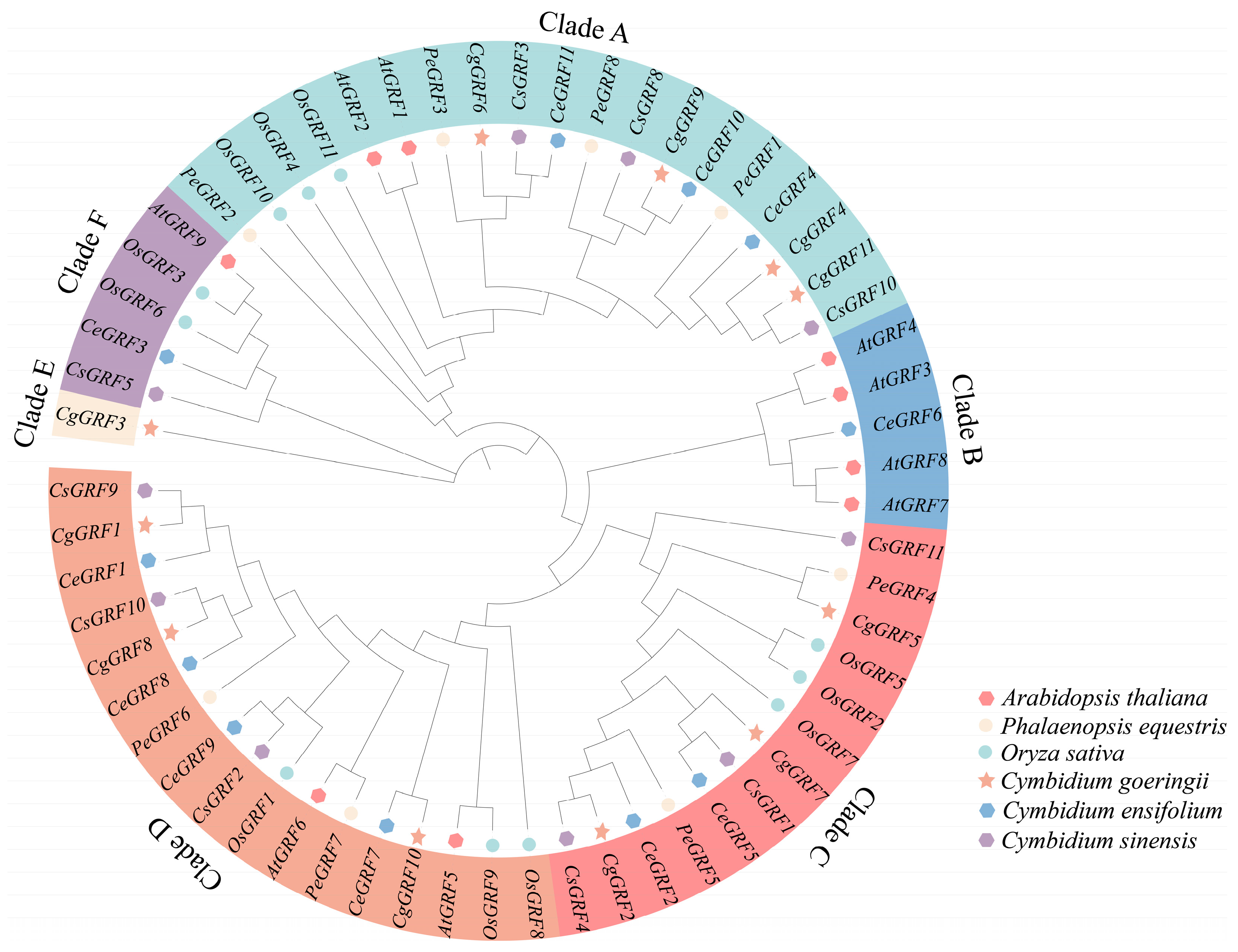
3.2. Phylogeny Analysis of the GRF Genes
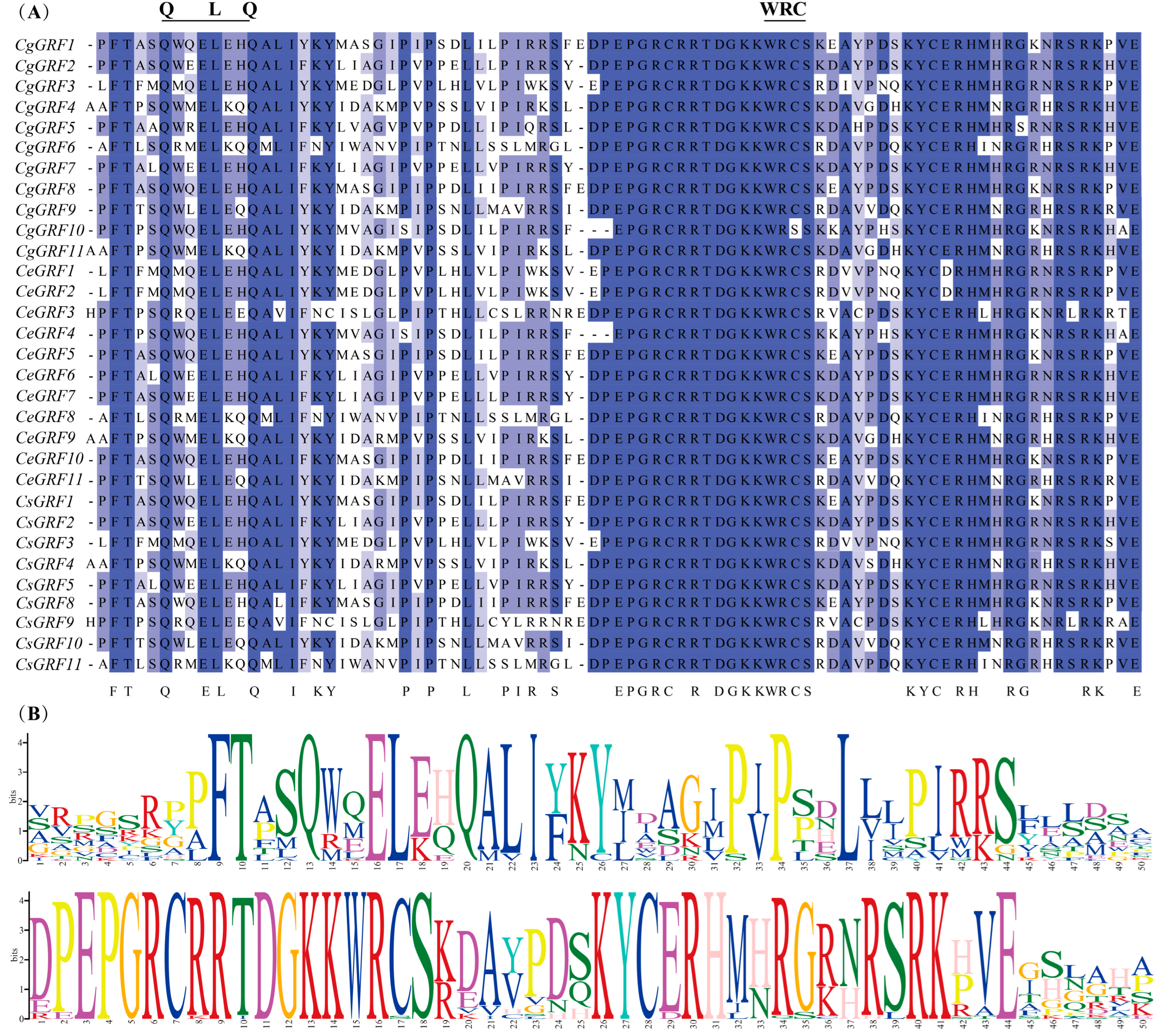
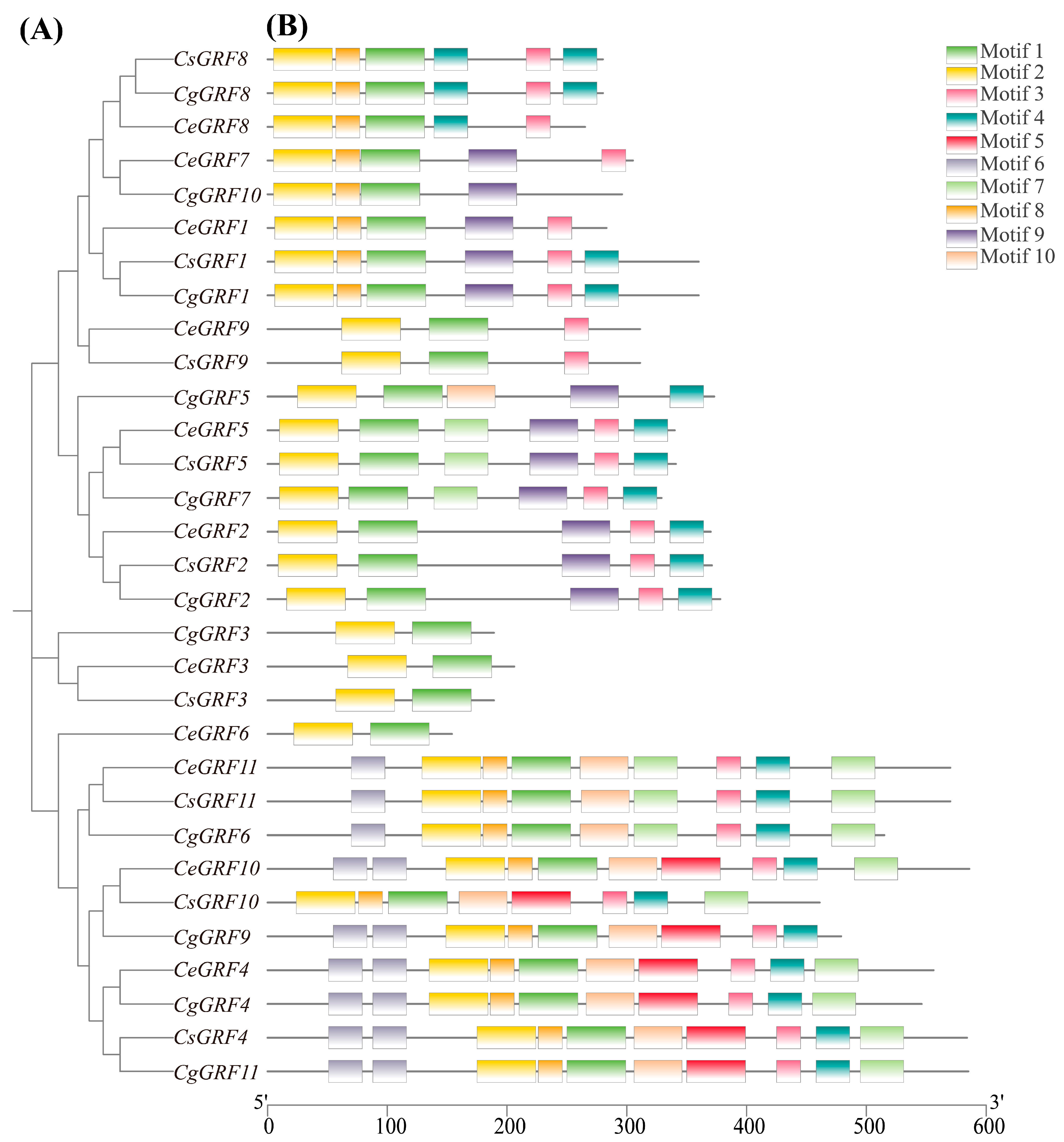
3.3. Conserved Domain and Gene Structure Analysis of GRF
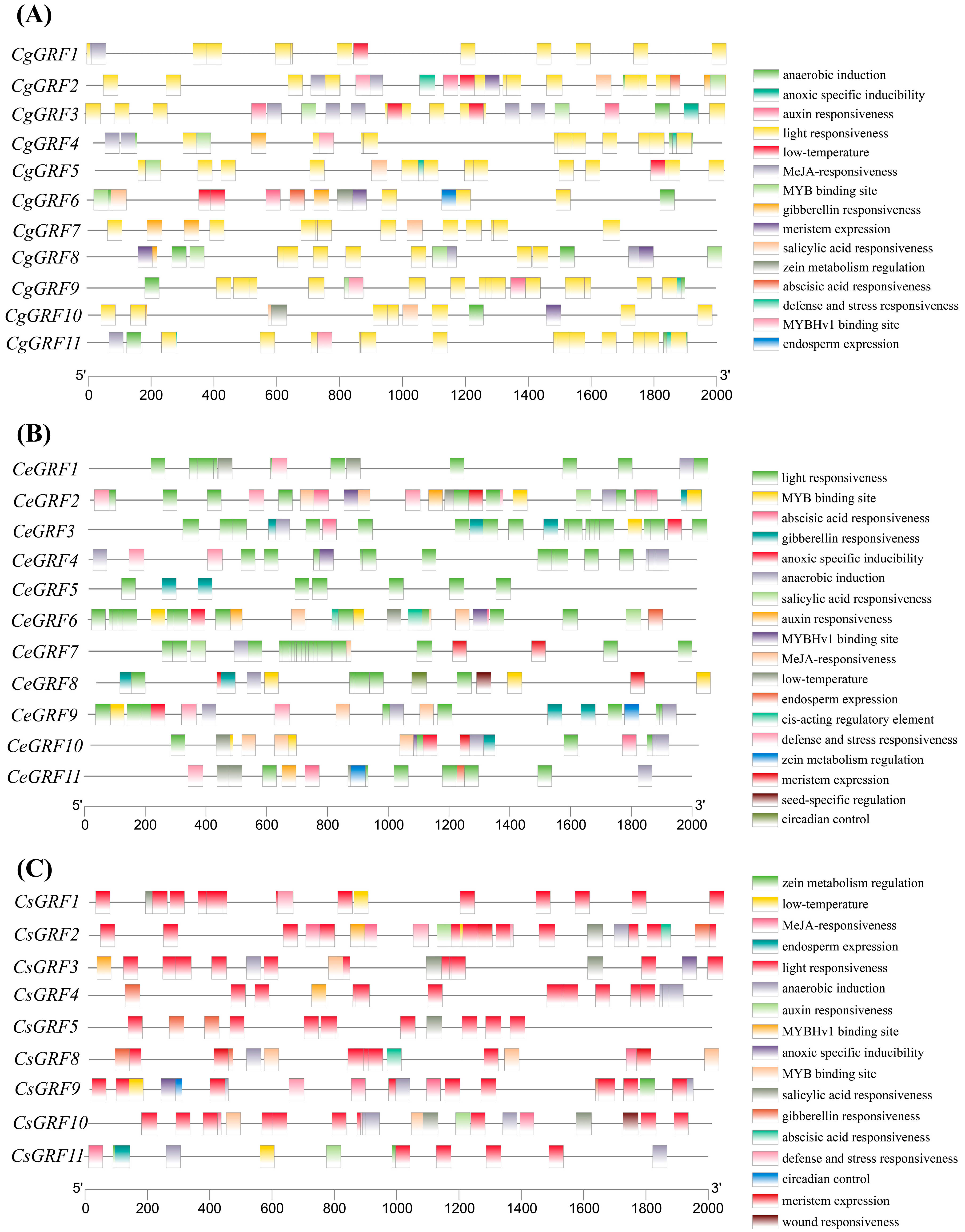
3.4. Cis-Acting Element Analysis of Three Cymbidium Species
3.5. Chromosome Location and Collinearity Analysis
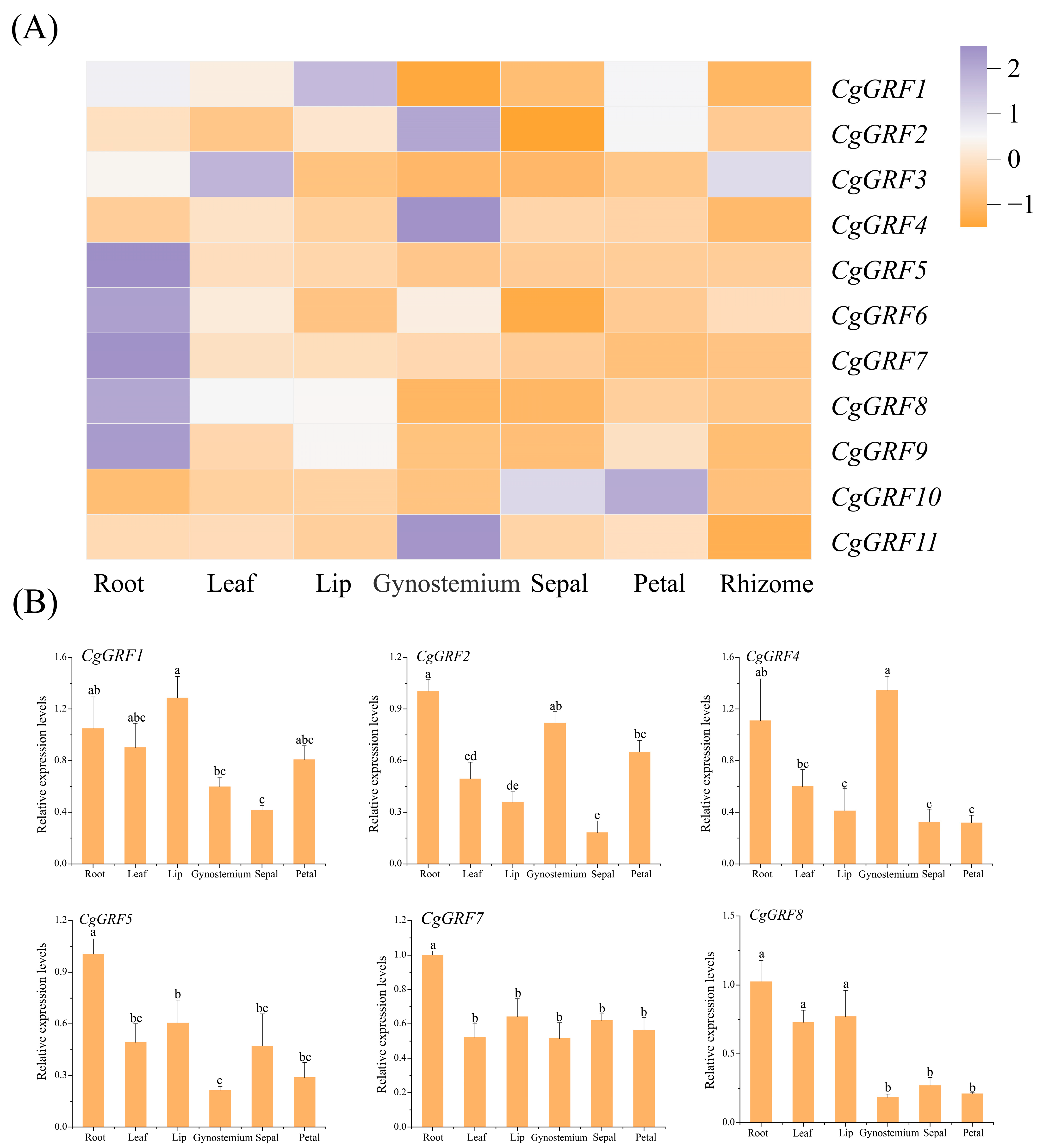
3.6. Expression Pattern of GRF Genes in Three Cymbidium Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Filiz, E.; Koc, I.; Tombuloğlu, H. Genome-wide identification and analysis of growth regulating factor genes in Brachypodium distachyon: In silico approaches. Turk. J. Biol. 2014, 38, 296–306. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; MuellerRoeber, B. Growth-regulating factors (GRFs): A small transcription factor family with important functions in plant biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef] [PubMed]
- Zan, T.; Zhang, L.; Xie, T.; Li, L. Genome-Wide Identification and Analysis of the Growth-Regulating Factor (GRF) Gene Family and GRF-Interacting Factor Family in Triticum aestivum L. Biochem. Genet. 2020, 58, 705–724. [Google Scholar] [CrossRef] [PubMed]
- Mohammadian, R.N. Phylogenetic, structure and expression analysis of growth regulatory factors (GRF) genes in wheat (Triticum aestivum L.) using in silico methods. Crop Biotechnol. 2020, 10, 45–60. [Google Scholar]
- Sahoo, R.K.; Jeughale, K.P.; Sarkar, S.; Selvaraj, S.; Singh, N.R.; Swain, N.; Samantaray, S. Growing Conditions and Varietal Ecologies Differently Regulates the Growth-regulating-factor (GRFs) Gene Family in Rice. Iran. J. Biotechnol. 2024, 22, e3697. [Google Scholar]
- Liu, Y.; Guo, P.; Wang, J.; Xu, Z.Y. Growth-regulating factors: Conserved and divergent roles in plant growth and development and potential value for crop improvement. Plant J. 2023, 113, 1122–1145. [Google Scholar] [CrossRef]
- van der Knaap, E.; Kim, J.H.; Kende, H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 2000, 122, 695–704. [Google Scholar] [CrossRef]
- Tian, Y.; Zhao, Y.; Sun, Y.; El-Kassaby, Y.A.; Song, G.; Mi, Y.; Han, J.; Li, Y. PagGRF11 overexpression promotes stem development and dwarfing in Populus. Int. J. Mol. Sci. 2022, 23, 7858. [Google Scholar] [CrossRef]
- Cui, D.; Song, Y.; Jiang, W.; Ye, H.; Wang, S.; Yuan, L.; Liu, B. Genome-wide characterization of the GRF transcription factors in potato (Solanum tuberosum L.) and expression analysis of StGRF genes during potato tuber dormancy and sprouting. Front. Plant Sci. 2024, 15, 1417204. [Google Scholar] [CrossRef]
- Nazir, N.; Iqbal, A.; Hussain, H.; Ali, F.; Almaary, K.S.; Aktar, M.N.; Sajid, M.; Bourhia, M.; Salamatullah, A.M. In silico genome-wide analysis of the growth-regulating factor gene family and their expression profiling in Vitis vinifera under biotic stress. Cell Biochem. Biophys. 2025, 83, 1207–1221. [Google Scholar] [CrossRef]
- Liu, X.; Guo, L.X.; Jin, L.F.; Liu, Y.Z.; Liu, T.; Fan, Y.H.; Peng, S.A. Identification and transcript profiles of citrus growth-regulating factor genes involved in the regulation of leaf and fruit development. Mol. Biol. Rep. 2016, 43, 1059–1067. [Google Scholar] [CrossRef]
- Bolaños-Villegas, P.; Chen, F.C. Advances and perspectives for polyploidy breeding in orchids. Plants 2022, 11, 1421. [Google Scholar] [CrossRef]
- Debernardi, J.M.; Mecchia, M.A.; Vercruyssen, L.; Smaczniak, C.; Kaufmann, K.; Inze, D.; Rodriguez, R.E.; Palatnik, J.F. Post-transcriptional control of GRF transcription factors by micro RNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 2014, 79, 413–426. [Google Scholar] [CrossRef]
- Ercoli, M.F.; Ferela, A.; Debernardi, J.M.; Perrone, A.P.; Rodriguez, R.E.; Palatnik, J.F. GIF transcriptional coregulators control root meristem homeostasis. Plant Cell 2018, 30, 347–359. [Google Scholar] [CrossRef]
- Pajoro, A.; Madrigal, P.; Muiño, J.M.; Matus, J.T.; Jin, J.; Mecchia, M.A.; Debernardi, J.M.; Palatnik, J.F.; Balazadeh, S.; Arif, M.; et al. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 2014, 15, R41. [Google Scholar] [CrossRef]
- Cai, M.; Cheng, W.; Bai, Y.; Mu, C.; Zheng, H.; Cheng, Z.; Gao, J. PheGRF4e initiated auxin signaling during moso bamboo shoot development. Mol. Biol. Rep. 2022, 49, 8815–8825. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, Y.; Yuan, B.; Peng, M.; Zhao, Y.; Chen, T.; Lu, J.; Li, F.; Lu, X.; Yang, J. Identification of the citrus GRF gene family and its expression in fruit peel thickening mediated by gibberellin. BMC Plant Biol. 2025, 25, 216. [Google Scholar] [CrossRef]
- Bao, M.; Bian, H.; Zha, Y.; Li, F.; Sun, Y.; Bai, B.; Chen, Z.; Wang, J.; Zhu, M.; Han, N. miR396a-mediated basic helix–loop–helix transcription factor bHLH74 repression acts as a regulator for root growth in Arabidopsis seedlings. Plant Cell Physiol. 2014, 55, 1343–1353. [Google Scholar] [CrossRef]
- Wei, Y.; Lin, Z.; Jin, J.; Zhu, W.; Gao, J.; Li, J.; Xie, Q.; Lu, C.; Zhu, G.; Yang, F. Genome-wide identification and functional characterization of LBD gene family in four Cymbidium species (Orchidaceae) and potential regulatory role of CsiLBD27 in floral development of Cymbidium sinense. BMC Genom. 2025, 26, 536. [Google Scholar] [CrossRef] [PubMed]
- Ai, Y.; Li, Z.; Sun, W.H.; Chen, J.; Zhang, D.Y.; Ma, L.; Zhang, Q.H.; Chen, M.K.; Zheng, Q.D.; Liu, J.F.; et al. The Cymbidium genome reveals the evolution of unique morphological traits. Hortic. Res. 2021, 8, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.X.; Gao, J.; Wei, Y.L.; Ren, R.; Zhang, G.Q.; Lu, C.Q.; Jin, J.P.; Ai, Y.; Wang, Y.Q.; Chen, L.J.; et al. The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 2021, 19, 2501–2516. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Rychlik, W. OLIGO 7 primer analysis software. In PCR Primer Design; Humana Press: Totowa, NJ, USA, 2007; pp. 35–59. [Google Scholar]
- Wang, F.; Chen, X.; Cheng, M.; Zhou, C.; Zheng, R.; Wu, X.; Duan, Y.; Ahmad, S.; Liu, Z.; Chen, J.; et al. Genome-Wide Identification of the WUSCHEL-Related Homeobox (WOX) Gene Family in Three Cymbidium Species and Expression Patterns in C. goeringii. Horticulturae 2024, 10, 645. [Google Scholar] [CrossRef]
- Chen, G.Z.; Huang, J.; Zhou, X.Q.; Hao, Y.; Chen, J.L.; Zhou, Y.Z.; Ahmad, S.; Lan, S.R.; Liu, Z.J.; Peng, D.H. Comprehensive analysis for GRF transcription factors in sacred lotus (Nelumbo nucifera). Int. J. Mol. Sci. 2022, 23, 6673. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Sun, Y.; Xiao, W.; Wang, Q.N.; Wang, J.; Kong, X.D.; Ma, W.H.; Zhang, Y.J. Multiple variation patterns of terpene synthases in 26 maize genomes. BMC Genom. 2023, 24, 46. [Google Scholar] [CrossRef]
- Kim, J.H.; Choi, D.; Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003, 36, 94–104. [Google Scholar] [CrossRef]
- Lu, Y.; Meng, Y.; Zeng, J.; Luo, Y.; Feng, Z.; Bian, L.; Gao, S. Coordination between GROWTH-REGULATING FACTOR1 and GRF-INTERACTING FACTOR1 plays a key role in regulating leaf growth in rice. BMC Plant Biol. 2020, 20, 200. [Google Scholar] [CrossRef]
- Xu, R.; An, J.; Song, J.; Yan, T.; Li, J.; Zhao, X.; Ding, C. OsGRF1 and OsGRF2 play unequal redundant roles in regulating leaf vascular bundle formation. J. Exp. Bot. 2025, eraf193. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Zhang, W.; Wang, Y.; He, Q.; Shu, F.; Liu, H.; Deng, H. OsGRF4 controls grain shape, panicle length and seed shattering in rice. J. Integr. Plant Biol. 2016, 58, 836–847. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Chang, C.; Guo, X.; Wang, B.; Wang, Y.; Zhang, M.; Yu, F.; Tang, Z. CrGRF1/4 mediating light signal to regulate monoterpenoid Indole alkaloid biosynthesis in Catharanthus roseus. Plant Cell Environ. 2025, 48, 4886–4901. [Google Scholar] [CrossRef]
- Huang, J.; Chen, G.Z.; Ahmad, S.; Hao, Y.; Chen, J.L.; Zhou, Y.Z.; Peng, D.H. Genome-wide identification and characterization of the GRF gene family in Melastoma dodecandrum. Int. J. Mol. Sci. 2023, 24, 1261. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Z.; Jin, J.; Xie, X.; Zhang, H.; Chen, Q.; Luo, Z.P.; Yang, J. Genome-wide identification and analysis of the growth-regulating factor family in tobacco (Nicotiana tabacum). Gene 2018, 639, 117–127. [Google Scholar] [CrossRef]
- Zhu, R.; Cao, B.; Sun, M.; Wu, J.; Li, J. Genome-wide identification and evolution of the GRF gene family and functional characterization of PbGRF18 in Pear. Int. J. Mol. Sci. 2023, 24, 14690. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Lu, C.; Wei, Y.; Wu, J.; Ren, R.; Gao, J.; Ahmad, S.; Jin, J.; Xv, Y.; Liang, G.; et al. Organ-Specific Gene Expression Reveals the Role of the Cymbidium ensifolium-miR 396/Growth-Regulating Factors Module in Flower Development of the Orchid Plant Cymbidium ensifolium. Front. Plant Sci. 2022, 12, 799778. [Google Scholar] [CrossRef]
- Liu, H.; Guo, S.; Xu, Y.; Li, C.; Zhang, Z.; Zhang, D.; Xu, S.; Zhang, C.; Chong, K. OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol. 2014, 165, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ge, L.; Li, G.; Jiang, L.; Yang, Y. Characterization and expression analysis of growth regulating factor (GRF) family genes in cucumber. Arch. Biol. Sci. 2018, 70, 629–637. [Google Scholar] [CrossRef]
- Baloglu, M.C. Genome-wide in silico identification and comparison of Growth Regulating Factor (GRF) genes in Cucurbitaceae family. Plant Omics 2014, 7, 260–270. [Google Scholar]
- Qin, L.; Chen, H.; Wu, Q.; Wang, X. Identification and exploration of the GRF and GIF families in maize and foxtail millet. Physiol. Mol. Biol. Plants 2022, 28, 1717–1735. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, J.; Li, J.; Li, Y.; Zeng, X. Genome-wide identification and analysis of the growth-regulating factor family in Zanthoxylum armatum DC and functional analysis of ZaGRF6 in leaf size and longevity regulation. Int. J. Mol. Sci. 2022, 23, 9043. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H. Biological roles and an evolutionary sketch of the GRF-GIF transcriptional complex in plants. BMB Rep. 2019, 52, 227. [Google Scholar] [CrossRef] [PubMed]
- Baucher, M.; Moussawi, J.; Vandeputte, O.M.; Monteyne, D.; Mol, A.; Pérez-Morga, D.; El Jaziri, M. A role for the miR396/GRF network in specification of organ type during flower development, as supported by ectopic expression of Populus trichocarpa miR396c in transgenic tobacco. Plant Biol. 2013, 15, 892–898. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Gene ID | Number of Amino Acids (AA) | Theoretical PI | Molecular Weight (Mw) | Instability Index (II) | Aliphatic Index (AI) | Grand Average of Hydropathicity (GRAVY) | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| CgGRF1 | GL00240 | 359 | 7.2 | 40.92 | 61.65 | 57.10 | −0.669 | Nucleus |
| CgGRF2 | GL00004 | 377 | 8.45 | 41.09 | 54.56 | 57.48 | −0.609 | Nucleus |
| CgGRF3 | GL13410 | 188 | 8.97 | 20.59 | 57.16 | 68.56 | −0.556 | Nucleus |
| CgGRF4 | GL18194 | 545 | 7.61 | 60.17 | 69.52 | 64.99 | −0.602 | Nucleus |
| CgGRF5 | GL18797 | 578 | 8.56 | 41.17 | 61.37 | 52.23 | −0.690 | Nucleus |
| CgGRF6 | GL13302 | 514 | 9.19 | 55.40 | 52.59 | 67.98 | −0.470 | Nucleus |
| CgGRF7 | GL16466 | 329 | 9.04 | 36.43 | 61.44 | 63.22 | −0.516 | Nucleus |
| CgGRF8 | GL16921 | 279 | 8.76 | 32.29 | 75.25 | 45.84 | −0.977 | Nucleus |
| CgGRF9 | GL08626 | 478 | 8.82 | 52.39 | 56.50 | 60.42 | −0.642 | Nucleus |
| CgGRF10 | GL14385 | 295 | 10.06 | 34.03 | 52.08 | 53.66 | −0.880 | Nucleus |
| CgGRF11 | GL21410 | 584 | 8.7 | 65.07 | 65.79 | 67.83 | −0.582 | Nucleus |
| CeGRF1 | JL019854 | 282 | 8.41 | 32.06 | 65.97 | 49.86 | −0.833 | Nucleus |
| CeGRF2 | JL000034 | 369 | 7.72 | 40.30 | 56.58 | 56.61 | −0.632 | Nucleus |
| CeGRF3 | JL009285 | 205 | 8.96 | 22.42 | 56.73 | 63.37 | −0.533 | Nucleus |
| CeGRF4 | JL020731 | 555 | 7.63 | 61.35 | 68.79 | 68.79 | −0.564 | Nucleus |
| CeGRF5 | JL011992 | 339 | 8.66 | 37.79 | 59.96 | 59.32 | −0.585 | Nucleus |
| CeGRF6 | JL009284 | 153 | 9.43 | 16.70 | 64.81 | 69.54 | −0.518 | Nucleus |
| CeGRF7 | JL023725 | 304 | 9.88 | 35.56 | 58.84 | 52.37 | −1.057 | Nucleus |
| CeGRF8 | JL007573 | 264 | 8.4 | 30.59 | 75.47 | 41.06 | −1.045 | Nucleus |
| CeGRF9 | JL018239 | 310 | 9.61 | 35.79 | 72.76 | 62.90 | −0.903 | Nucleus |
| CeGRF10 | JL006667 | 585 | 7.33 | 63.72 | 52.22 | 62.36 | −0.611 | Nucleus |
| CeGRF11 | JL001734 | 569 | 9.03 | 61.05 | 52.63 | 69.44 | −0.442 | Nucleus |
| CsGRF1 | cymsin_Mol016305 | 359 | 7.18 | 40.90 | 61.23 | 57.10 | −0.669 | Nucleus |
| CsGRF2 | cymsin_Mol005754 | 370 | 8.43 | 40.34 | 54.25 | 58.57 | −0.584 | Nucleus |
| CsGRF3 | cymsin_Mol008035 | 188 | 8.97 | 20.58 | 56.99 | 68.03 | −0.558 | Nucleus |
| CsGRF4 | cymsin_Mol011492 | 583 | 8.79 | 65.11 | 63.89 | 67.77 | −0.587 | Nucleus |
| CsGRF5 | cymsin_Mol012650 | 340 | 9.07 | 37.84 | 61.42 | 59.15 | −0.550 | Nucleus |
| CsGRF8 | cymsin_Mol001074 | 279 | 8.76 | 32.28 | 74.81 | 45.84 | −0.975 | Nucleus |
| CsGRF9 | cymsin_Mol001920 | 310 | 9.49 | 35.82 | 69.21 | 63.87 | −0.886 | Nucleus |
| CsGRF10 | cymsin_Mol011781 | 460 | 8.54 | 49.96 | 50.25 | 64.89 | −0.603 | Nucleus |
| CsGRF11 | cymsin_Mol000179 | 569 | 9.12 | 61.12 | 53.02 | 70.30 | −0.433 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, Y.; Pan, Y.; Wang, F.; Chen, F.; Wu, X.; Chen, J.; Zhu, J.; Peng, D. Genome-Wide Identification of the Growth-Regulating Factor (GRF) Gene Family in Three Cymbidium Species and Expression Patterns in C. goeringii. Horticulturae 2025, 11, 1015. https://doi.org/10.3390/horticulturae11091015
Deng Y, Pan Y, Wang F, Chen F, Wu X, Chen J, Zhu J, Peng D. Genome-Wide Identification of the Growth-Regulating Factor (GRF) Gene Family in Three Cymbidium Species and Expression Patterns in C. goeringii. Horticulturae. 2025; 11(9):1015. https://doi.org/10.3390/horticulturae11091015
Chicago/Turabian StyleDeng, Yan, Yun Pan, Fei Wang, Feng Chen, Xiaopei Wu, Jinliao Chen, Jin Zhu, and Donghui Peng. 2025. "Genome-Wide Identification of the Growth-Regulating Factor (GRF) Gene Family in Three Cymbidium Species and Expression Patterns in C. goeringii" Horticulturae 11, no. 9: 1015. https://doi.org/10.3390/horticulturae11091015
APA StyleDeng, Y., Pan, Y., Wang, F., Chen, F., Wu, X., Chen, J., Zhu, J., & Peng, D. (2025). Genome-Wide Identification of the Growth-Regulating Factor (GRF) Gene Family in Three Cymbidium Species and Expression Patterns in C. goeringii. Horticulturae, 11(9), 1015. https://doi.org/10.3390/horticulturae11091015





