Abstract
Red epicarp in pears is an important trait for breeding. Exploring the genes regulating pear anthocyanin synthesis and developing molecular markers associated with these traits are important for obtaining new varieties of red pears. We performed whole-genome resequencing (WGS) on 127 ‘Yuluxiang (Pyrus bretschneideri)’ × ‘Xianghongli (Pyrus communis)’ F1 populations and identified a total of 510,179 single-nucleotide polymorphism (SNP) sites in the population. In total, 1972 bins were screened to form a high-density genetic map with a total map length of 815.507 cM, covering 17 linkage groups with an average genetic distance of 0.414 cM between markers. Three red skin quantitative trait loci (QTLs), located on LG4 and LG5, that explained 18.7% of the phenotypic variance, were detected. The QTL intervals contained 1658 genes, including 94 transcription factors (TF), subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. Four key candidate genes (Pspp.Chr05.01969, Pspp.Chr05.01908, Pspp.Chr05.02419, and Pspp.Chr04.01087) that may play a role in promoting pear anthocyanin synthesis were screened and identified by a quantitative polymerase chain reaction (qPCR). Overall, our study deepens our understanding of the genetics of red peel traits in pears and accelerates pear breeding.
1. Introduction
Pears are one of the world’s leading fruits because of their unique taste and nutritional value [1]. They are classified into two categories: Western pears and Asian pears. These pears are different in appearance and taste [2].
The color of pears is a significant trait in determining the appearance and quality of pear fruits [3]. Red pears are likely to be favored by consumers because of their vibrant appearance and high anthocyanin content [4,5]. In recent years, many breeders have made red fruit a breeding goal, and considering that the red color is a dominant trait, when selecting parents for crossbreeding, at least one parent should be selected for a red variety.
The red color traits of fruits exhibit a variety of intensity phenotypes, including reddish blush on the peel, fully red fruits, and semi-red fruits. Some varieties are only red at maturity, whereas others are red at the young fruit stage [6,7]. Pear fruit anthocyanin synthesis mechanisms are very complex, and factors affecting pear fruit coloring can be mainly divided into inherited factors and environmental factors. Inherited factors are mainly caused by genetic differences, such as ‘Zaosu’ pear PyBBX24 on a section of the 14bp deletion, resulting in the color of its fruit spanning from green to red [8]. Environmental factors include temperature, light, and growth regulators. For example, low temperatures are more favorable for anthocyanin synthesis [9,10], blue light can promote anthocyanin accumulation in ‘Zaosu Red’ pears, while red light cannot prevent the color shift [11], and the jasmonic acid treatment of ripening Chinese red pear fruits promotes anthocyanin synthesis, while ethylene inhibits it [12]. Genetic factors are the most critical determinants of red color traits in fruits. Therefore, quantitative trait loci (QTLs) localization and mining of candidate genes for red skin are necessary.
Genetic mapping has been used to identify fruit color traits in several species. For example, Wang used a constructed genetic map of tetraploid kiwifruit to identify the master fruit color QTL on LG9, which explained 32.2% of phenotypic variance [13]. Nashima constructed a pineapple genetic linkage map using the F1 population of ‘Yugafu’ × ‘Yonekura’ and detected two QTLs for fruit shell color at LG2 and LG8, and the AcCCD4 gene was identified as the key gene controlling fruit shell color [14]. Wang used the red hawthorn variety ‘Shandong Da Mianqiu’ as the female and the yellow hawthorn variety ‘Da Huang Mianzha’ as the male to construct a segregating population and drew a 2297.75 cM genetic map. Five QTLs related to fruit color were localized on LG1, LG3, and LG5 [15]. QTLs for apple fruit color can also be used to predict the degree of fruit coloration in the progeny [16]. It has also been used in cucumber (Cucumis sativus L.) [17], pepper (Capsicum annuum L.) [18], sweet cherry (Prunus avium L.) [19], and strawberry (Fragaria x ananassa Duch.) [20]. Simple sequence repeat (SSR) and amplified fragment length polymorphism (AFLP) markers were previously used to construct genetic linkage maps in pears, which had a small total length, a small number of molecular markers, and a large average distance between markers. It could not be efficiently mined for the major QTLs of the trait or the molecular markers linked to them [21]. Terakami constructed a genetic linkage map of ‘Housui’ using SSR and AFLP markers. The total length of the map was 1174 cM, including 335 molecular markers, and the average distance between markers was 3.5 cM [22]. With the development of sequencing technology, the use of single-nucleotide polymorphism (SNP) markers to construct linkage maps has many advantages, including longer map distances, more molecular markers, and smaller average distances between markers. Gabay constructed a genetic map of ‘Spadona’ × ‘Harrow Sweet’ using the genotyping by sequencing (GBS) technique, which had a total map length of 1433 cM and contained 2036 markers with an average distance between markers of 0.7 cM [23]. The use of whole-genome resequencing (WGS) technology to construct bin maps can reduce the amount of data computation and provide the most representative markers, and this has been applied to several species in recent years, such as chili peppers [24], soybeans [25], rice [26], and apples [27].
Progress has recently been made in using genetic linkage mapping to locate QTLs for fruit color and identify key genes controlling fruit color. Yao used the ‘Bayuehong’ × ‘Dangshansuli’ F1 generation segregation population to construct a genetic linkage map, revealing stable pear red skin QTLs on LG5 and LG16 for several consecutive years, and verified the function of PyMYB114 in promoting anthocyanin synthesis [28]. Nashi pear (Pyrus pyrifolia Nakai) fruit color is mostly russet. Based on a study of F1 segregating populations of Japanese pear or sand pear varieties, the pear russet peel-related QTL was localized to LG8 [29,30]. Kim constructed a genetic map of the ‘Whangkeumbae’ × ‘Minibae’ population, identified pear russet-associated QTLs on LG8, and developed a CAPS marker that can be used to predict the degree of russet coloration in pear F1 generation [31]. Ntladi detected two major QTLs associated with fruit red color on LG5 and LG9 of the genetic map of ‘Flamingo’ × ‘Abate Fetel’ and identified two SSR markers, NB101a and SAmsCO865954, that were strongly associated with the fruit red trait [32].
High-density genetic linkage mapping is the basis for identifying the locations of QTLs for target traits. Therefore, in the present study, genetic maps were constructed using WGS technology (1) to detect SNP loci in each sample after screening high-quality bin markers. We also performed (2) QTL localization for red skin and (3) the mining of candidate genes to regulate red fruit color traits and develop bin markers related to red skin.
2. Materials and Methods
2.1. Plant Materials
The F1 generation population in this study was ‘Yuluxiang’ × ‘Xianghongli’, which was pollinated in 2015 and planted at the pear base of Hebei Agricultural University in 2017. Plants were cultivated with a spacing of 1 m × 4 m under conventional management. Female ‘Yuluxiang’ is a Chinese white pear (Pyrus bretschneideri) with reddish blush skin. The male ‘Xianghongli’ is an entirely red fruit that belongs to the Western pear (Pyrus communis) [33]. The hybrid progeny exhibited varying degrees of fruit coloration, and 127 hybrids were investigated for fruit coloration between 2023 and 2024. Individuals numbered 1-9 (red fruit) and 4-62 (green fruit) of the F1 generation at 120 days after flowering were used for the qPCR analysis.
2.2. DNA Extraction
In May 2023, young leaves from 127 hybrid progeny and parents were collected, placed on ice, and DNA was extracted using the CTAB method [34].
2.3. Sequencing Library Construction and SNP Genotyping
By applying NEBNext dsDNA fragmentase (NEB, Ipswich, MA, USA), we fragmented genomic DNA and performed DNA end repair. Further, through using an Illumina X-ten system (Illumina, San Diego, CA, USA), we amplified the captured sequences by 150bp paired-end sequencing.
After obtaining the clean reads from each sample of the F1 generation population, we used the 0.7.16a-r1181 Burrows–Wheeler Aligner (BWA) [35] to align those reads against the Yunhong NO.1 reference genome [36]. Next, we conducted multi-sample variant calling using the 3.8.1 Genome Analysis Toolkit [37] (GATK) Unified Genotyper with local realignment and base quality score recalibration. Further, we obtained SNPs and InDels filtered using GATK’s Variant Filtration tool with standard parameters (-Window 4, -filter “QD < 2.0 || FS > 60.0 || MQ < 40.0 “, -G_filter “GQ < 20”). Low-quality SNPs and InDels were subsequently discarded.
2.4. High-Density Genetic Bin Map Construction
The obtained SNP markers were filtered by (1) removing loci with a deletion percentage higher than 5%, (2) removing loci with a heterozygosity percentage greater than 50%, (3) removing loci with a chi-square test of p < 0.5, and (4) retaining markers with the segregation types lmxll, nnxnp, and hkxhk. After filtering, one of the markers with the highest completeness (lowest deletion rate) at each genetic position was taken as the bin marker. We used the OrderMarkers2 module of the 0.2 Lep-MAP3 software [38] to build the genetic map and the pseudo-crossover and CP modeling to calculate the genetic distances between the markers and map distances for individual chromosomes. For the set of parameters, refer to [39].
2.5. QTL Mapping of Red Skin
Red skin was investigated in 2023 and 2024. The results indicated that the phenotypes were stable and the data were consistent across both years (the data have been published; refer to [40]).
We used the composite interval mapping (CIM) functions in R/qtl to perform QTL mapping [41], including interval mapping and composition interval mapping. To further confirm the threshold of LOD (the logarithm of odds) values, we conducted 1000 permutations. When the LOD > 3, we identified the genetic groups with QTLs. The QTL mapping results were completed at a 5% significance level.
2.6. Expression Analysis
The pears were preserved and brought back to the laboratory, where the skin was cleaned with RNA-free water. Then, the skin was gently removed with a scalpel and quickly placed in liquid nitrogen for grinding. RNA was extracted using an RNA kit (Tiangen, Beijing, China), and reverse transcription was performed using a reverse transcription SuperMix kit (Tiangen, Beijing, China). qPCR primer design, reference gene, and reaction procedures were performed according to Zhang’s method [42]. Primer information is shown in Table S4.
Data statistics were assessed using Excel 2021 software, and significance tests and graphing were conducted with Prism 9 software.
3. Results
3.1. Specific-Locus Amplified Fragment (SLAF)-Seq and SNP Identification
We performed whole-genome resequencing on 127 hybrid progeny and parents, yielding 5319.8 million clean reads, with an average of 41,239.3 million clean reads per individual (Table S1). After filtering low-quality data, the sequencing base quality values of Q30 were all above 90%, meeting the quality requirements for subsequent analysis. (Table 1). After comparing the reads to the ‘Yunhong NO.1’ reference genome, the average mapping rate was 90.34% (Table S2). A total of 6,674,635 SNPs and 631,043 indels were identified using the GATK. These SNPs and inDels are mainly located in intergenic regions, followed by intronic regions (Figure S1). The functional annotation results of SNPs and inDels have shown that nonsynonymous SNVs (single-nucleotide variants) are the most numerous, followed by synonymous SNVs (Figure S2). After typing the markers, 227,034 markers of type <hkxhk>, 1567,195 markers of type <lmxll>, and 1,517,570 markers of type <nnxnp> were obtained. A total of 510,179 markers were obtained after filtering. Subsequently, bin markers were filtered for the construction of the genetic map.

Table 1.
Statistical results of base information.
3.2. Construction of the Bin Genetic Linkage Map
A total of 1972 bin markers from 127 progeny were used to construct genetic linkage maps covering 17 linkage groups. This resulted in a total map length of 815.507 cM, a total map length of 913.529 cM for males and a total map length of 717.484 cM for females, and an average distance between markers of 0.414 cM (Table 2). The length of the linkage groups ranged from 29.136 cM (LG4) to 63.393 cM (LG5), and the average distance between the markers was a minimum of 0.389 cM (LG6), containing 83 bin markers, and a maximum of 0.483 cM (LG12), containing 89 bin markers. In addition, there were no gaps > 5 cM in the linkage group (Figure 1).

Table 2.
Bin genetic map statistical information.
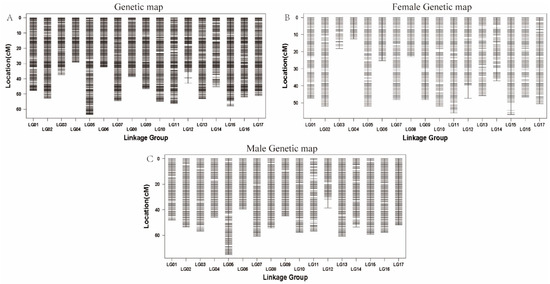
Figure 1.
Genetic linkage map constructed by Lep-MAP3 software using 1972 bin markers and 127 ‘Yuluxiang’ × ‘Xianghongli’ progeny. (A) The integrated common genetic map; (B) the female ‘Yuluxiang’ genetic map; (C) the male ‘Xianghongli’ genetic map.
3.3. Collinearity of the Genetic and Physical Maps
The better the covariance between the genetic and physical maps, the more accurate the platings. We generated a heatmap of 17 LGs based on the pairwise recombination values of 1972 recombinant bin markers to assess the accuracy of the genetic mapping in this study. The overall diagonal distribution from the heatmap of the linkage indicated that the genetic map was constructed accurately. The Pearson correlation coefficients of the genetic and physical locations on the genetic map can illustrate the quality of the map. The genetic and physical locations on the 17 LGs were highly correlated, with the highest Pearson correlation coefficient of 99.4% for LG5 and LG11, the lowest of 94.9% for LG13, and an average value of 98.2%, indicating that the 1972 bin markers are accurately localized on 17 chromosomes, which can cover the whole pear genome and meet the requirements of the QTL localization analysis (Table 3, Figure 2).

Table 3.
Pearson correlation coefficient statistics between genetic and physical maps.
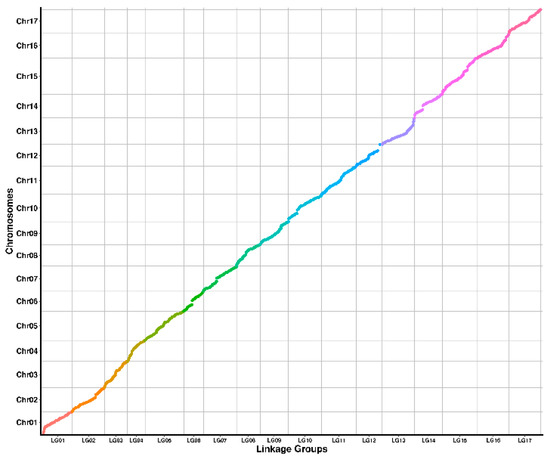
Figure 2.
Genetic map and physical map covariance results.
3.4. QTLs Associated with Red Skin
We detected QTLs in red fruit peels using composite interval mapping (CIM). When LOD > 3, it was considered a major QTL. The localization results of QTLs in this study showed that the QTLs associated with the red skin were localized on LG4 and LG5 of the ‘Yuluxiang’ × ‘Xianghongli’ genetic map (Figure 3). The length of the QTL interval on LG4 was 10.237 cM, and there were two QTLs on LG5 with interval lengths of 7.087 cM and 19.293 cM. The QTL for red skin on LG4 had a peak with an LOD value of 6.120, located at a physical position of 14.569 cM on LG4, and explained 5.519% of the phenotypic variance. The QTLs for red skin on LG5 had two peaks with LOD values of 6.826 and 4.711, respectively, and were located at physical positions of 28.000 cM and 51.189 cM on LG5. The two QTLs explained 7.806% and 5.398% of the phenotypic variance. Together, the three QTLs explained 18.723% of the phenotypic variance and contained 186 bin markers (Table 4 and Table S3).
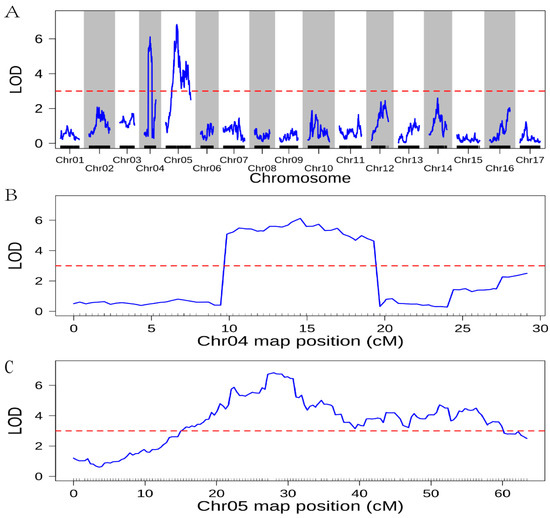
Figure 3.
Localization of QTLs for red skin on the genetic map of ‘Yuluxiang’ × ‘Xianghongli’. Note: The red dashed line indicates that the LOD value is 3. (A) results of localization of QTLs for red skin; (B) logarithm of odds (LOD) curve graph on LG4; (C) LOD curve graph on LG5.

Table 4.
QTL information related to red skin on LG4 and LG5 of ‘Yuluxiang’ × ‘Xianghongli’ genetic mapping.
3.5. Identification and Expression Analysis of Candidate Genes Associated with Red Skin
To better understand the functions of candidate genes within the QTLs, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of the candidate genes. Overall, the QTLs contained 1658 genes, including 94 transcription factors (TFs), some of which are related to the regulation of anthocyanosides in fruit (Table 5). We selected eight NAC family genes, four WRKY family genes, and seven MYB family genes from the candidate genes, totaling nineteen TFs, and performed quantitative qPCR analysis on progeny 1-9 (red fruit) and 4-62 (green fruit). Their results showed that the expression of four TFs was significantly higher in red fruits than in green fruits, namely Pspp.Chr05.01969 (MYB family), Pspp.Chr05.01908 (NAC family), Pspp.Chr05.02419 (WRKY family), and Pspp. Chr04.01087 (MYB family), which are four TFs that may promote anthocyanin synthesis in pears (Figure 4); the primer information is shown in Table S4. In addition, we categorized and annotated 1658 candidate genes using the GO database. The candidate genes were most enriched in biological process, followed by molecular function, and least enriched in the cellular component. The genes involved in biological processes were mainly related to cellular processes, metabolic processes, and biological regulation. The genes involved in molecular functions were mainly related to binding and catalytic activity. The genes involved in the cellular components were mainly related to cellular anatomical entities and a protein-containing complex (Figure S3). We also performed a more in-depth analysis of the GO-enriched biological process pathway, with 34 genes enriched in the GO:0006066 alcohol metabolic process pathway of biological process and 18 genes enriched in the GO:0009733 response to auxin pathway (Figure 5). To study the candidate genes at greater depths, the KEGG database was used to annotate the candidate genes, and the top 20 significantly enriched pathways were screened. Among these, the top three q-values were associated with flavone and flavonol biosynthesis, glycosphingolipid biosynthesis—lacto and neolacto series—and plant hormone signal transduction (Figure 6).

Table 5.
TFs within the interval of QTLs that are associated with anthocyanins.
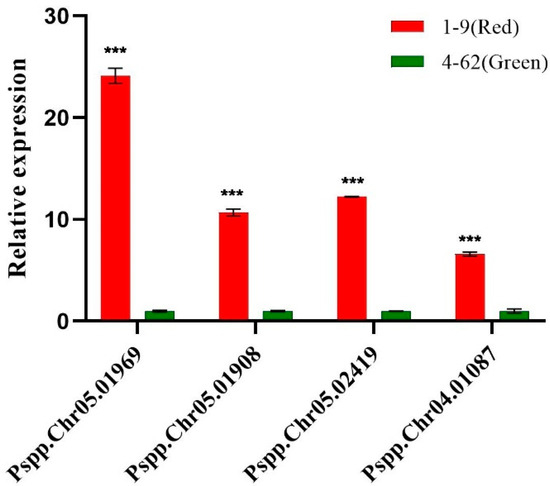
Figure 4.
q-PCR results of the four most critical candidate genes in 1-9 (red fruit) and 4-62 (green fruit). Note: *** indicates significance at the p < 0.05 level.
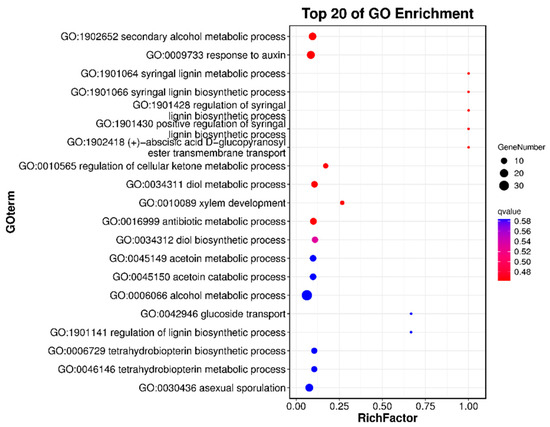
Figure 5.
Enrichment results of candidate genes involved in the biological process pathway using GO analysis.
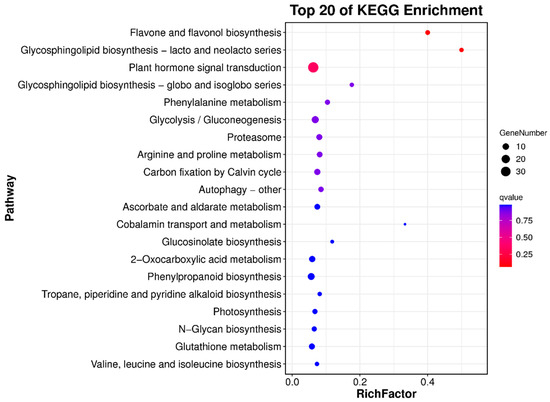
Figure 6.
Candidate gene enrichment results using KEGG analysis.
3.6. Bin Marker Screening Associated with Red Skin
We extracted 15 bin markers from the LOD peaks of three QTLs, and all 15 marker types were labeled <abxcd> (Table S5). Based on the extent of red coverage on the fruit peel, the hybrid progeny is classified into five categories: 1, no red coloration; 2, 1/4 red; 3, 1/2 red; 4, 3/4 red; and 5, completely red fruit peel. The female is 2, and the male is 5. After correlating the 15 bin markers with different genotypes and phenotypes in the F1 generation, we found that Chr05-29972420 was highly correlated with red skin (Figure 7). Their results showed that when the genotypes of the ‘Yuluxiang’ × ‘Xianghongli’ progeny population were BD and BC, their fruits were significantly redder than those of AD and AC. The Chr05-29972420 marker is located at position 29972420 on chromosome 5 within the gene Pspp.Chr05.02339, which is annotated as a J-domain-containing protein required for the chloroplast accumulation response 1-like factor.
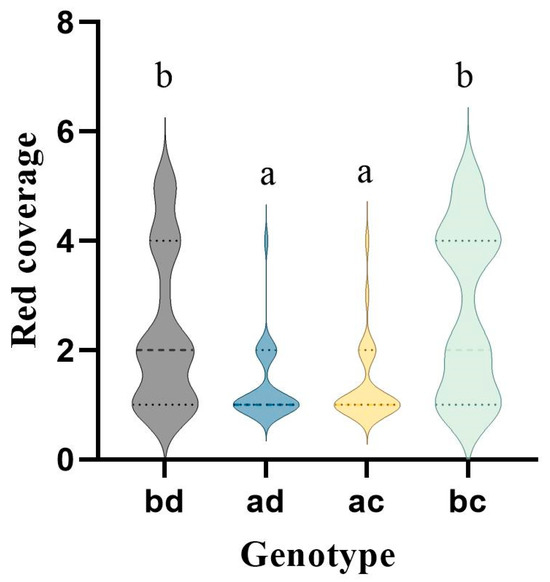
Figure 7.
Chr05-29972420 labeling different genotypes and F1 posterior phenotype association results. Note: The X-axis represents the genotype of the F1 generation, and the Y-axis represents the red coverage corresponding to different genotypes. a and b represent significance levels of p < 0.05.
4. Discussion
Genetic linkage mapping is a powerful tool for studying quantitative traits [43]. The construction of high-quality genetic linkage maps has been empowered by using high-throughput sequencing technology [44]. In this study, whole-genome resequencing (WGR) was used to construct an interspecific genetic map of ‘Yuluxiang’ × ‘Xianghongli’ (Pyrus bretschneideri × Pyrus communis), and a total of 510,179 single-nucleotide polymorphisms (SNPs) were obtained. Because many SNPs have similar genetic distances on the genetic map, we selected representative bin markers from SNPs with similar genetic distances, which can reduce the amount of computation and improve the accuracy of marker localization [45]. We used 1972 bin markers to build the genetic map, with a total map distance of 815.507 cM and a mean distance of 0.414 cM between markers. The pear bin genetic map was constructed through the WGR technique, which has been reported only once so far [46]. Qin used 176 ‘Niitaka’ × ‘Hongxiangsu’ populations to construct the genetic map, whose length was 1358.5 cM, containing 3190 bins, and the mean distance of the markers was 0.43 cM. The number of populations we used (127) was smaller than that of Qin’s map (176); therefore, the length and number of markers in our map were smaller. Also, the average distance between markers in our map was smaller (0.414 cM). Compared to the pear genetic map constructed using the GBS method, our map is more advantageous in terms of marker density [23]. In addition, the 17 linkage groups were highly covariant with the pear reference genome, which proved that the map was of good quality and provided a key basis for subsequent QTL localization analysis.
Many QTL localizations for fruit color have been reported in fruit trees. For example, in apples, fruit color QTLs are localized in LG9, LG10, and LG13 [47]. Frett detected four quantitative trait loci in peach pericarps in LG3 and LG4 [48]. The major QTL associated with grape berry color is located in LG2 [49]. In this study, we identified red skin QTLs using the interspecific population ‘Yuluxiang’ × ‘Xianghongli’, and three QTLs were located on LG4 and LG5. To verify whether the QTL on LG5 in this study was consistent with the QTL on LG5 in Yao’s study, we compared PyMYB114 in the QTL interval with the reference genome of this study and found that it was located at 34907938 on Chr05, which was far away from the location of the QTLs in this study, suggesting that the QTLs reported in this study were different from those previously reported. This may have been caused by the different genetic backgrounds [28].
TFs are crucial for turning on genes associated with flavonoid biosynthesis, and related studies have shown that TFs regulate anthocyanin synthesis by regulating the MBW complex [50]. In our study, after annotating the candidate genes within the QTL interval using the KEGG database, the top three q-values were flavone and flavonol biosynthesis, glycosphingolipid biosynthesis (lacto and neolacto series), and plant hormone signal transduction, of which flavone and flavonol biosynthesis and plant hormone signal transduction are inextricably linked to anthocyanin biosynthesis [51,52]. Some of the 94 TFs contained in the candidate genes are associated with anthocyanin synthesis. For example, the overexpression of PpWRKY44 promotes the accumulation of pear anthocyanins [53]; MYB4 is involved in pear anthocyanin biosynthesis [54], and MYB44 negatively regulates anthocyanin accumulation in strawberries [55]. These results also prove the accuracy of the QTLs. According to previous reports, the MYB [56], NAC [57], and WRKY gene families [58] play important roles in regulating pear flavonoid biosynthesis. We obtained four key candidate genes by q-PCR; the four genes belonged to the MYB family, NAC family, and WRKY family, and their expression levels were significantly higher in red fruits than in green fruits. We hypothesized that these four genes might promote the synthesis of pear anthocyanosides, and we will follow up with gene silencing or overexpression to further confirm their functions.
Although some SSR and RAPD molecular markers have been developed for pears, both SSR and RAPD use DNA fragment length changes as a means of detection and need to be verified by gel electrophoresis, which has the disadvantages of being time-consuming and genetically unstable [59,60]. Currently, the development of SNP molecular markers has become mainstream and has been applied in several species, with the advantages of a high marker number and genetic stability [61,62,63]. In this study, a bin marker (Chr05-29972420) associated with red skin was developed based on the results of the QTL localization. We can apply the Chr05-29972420 marker to the early filtering of red fruits at the sprouting stage, improving breeding efficiency.
5. Conclusions
In the present study, we constructed an interspecific genetic linkage map of pear F1 generation using whole-genome resequencing. Three QTLs for red skin were detected and distributed in LG4 and LG5, and four key candidate genes were screened using q-PCR. In addition, we developed a molecular marker for red skin. In the future, we will continue to validate the function of the candidate genes using various molecular biology techniques and verify the generalizability of these molecular markers to other populations.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae11080994/s1, Table S1: Reads statistical information sheet; Table S2: Comparison of reference genome statistical results for each sample; Table S3: Information on bin markers within the red skin QTL interval; Table S4: q-PCR primer information for 4 key candidate genes and reference gene; Figure S1: Location information of SNP-InDel in the genome; Figure S2: Functional annotation of SNP-InDel in the genome; Figure S3: Functional GO enrichment analysis of candidate genes; Table S5: A total of 15 bin markers at peak LOD for 3 QTLs.
Author Contributions
Data curation, M.T.; funding acquisition, H.M. and Y.Z.; investigation, X.Z. and M.T.; project administration, J.P.; supervision, J.P., H.M., and Y.Z.; writing—original draft, X.Z. and M.T.; writing—review and editing, X.Z. and H.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Pear Industrial Technology Engineering Research Center of the Ministry of Education and the Pear Technology Innovation Center of Hebei Province.
Data Availability Statement
The original contributions presented in this study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The manuscript has been approved by all authors for publication, and no conflicts of interest exist in the submission. We declare that the work carried out by all the authors listed is original and has not been previously published in any form. All the authors listed have contributed significantly to the work and agree to be included in the author list.
References
- Li, J.M.; Zhang, M.Y.; Li, X.L.; Khan, A.; Kumar, S.; Allan, A.C.; Kui, L.W.; Espley, R.V.; Wang, C.H.; Wang, R.Z.; et al. Pear genetics: Recent advances, new prospects, and a roadmap for the future. Hortic. Res. 2022, 9, uhab040. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Y.T.; Xu, J.B.; Korban, S.S.; Fei, Z.J.; Tao, S.T.; Ming, R.; Tai, S.S.; Khan, A.M.; Postman, J.D.; et al. Diversification and independent domestication of Asian and European pears. Genome Biol. 2018, 19, 77. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.N.; Yao, G.F.; Zheng, D.M.; Zhang, S.L.; Wang, C.; Zhang, M.Y.; Wu, J. Expression differences of anthocyanin biosynthesis genes reveal regulation patterns for red pear coloration. Plant Cell Rep. 2015, 34, 189–198. [Google Scholar] [CrossRef]
- Steyn, W.J.; Holcroft, D.M.; Wand, S.J.E.; Jacobs, G. Anthocyanin degradation in detached pome fruit with reference to preharvest red color loss and pigmentation patterns of blushed and fully red pears. J. Am. Soc. Hortic. Sci. 2004, 129, 13–19. [Google Scholar] [CrossRef]
- Wang, Z.G.; Du, H.; Zhai, R.; Song, L.Y.; Ma, F.W.; Xu, L.F. Transcriptome Analysis Reveals Candidate Genes Related to Color Fading of ‘Red Bartlett’ (Pyrus communis L.). Front. Plant Sci. 2017, 8, 455. [Google Scholar] [CrossRef]
- Wu, M.; Liu, J.L.; Song, L.Y.; Li, X.Y.; Cong, L.; Yue, R.R.; Yang, C.Q.; Liu, Z.; Xu, L.F.; Wang, Z.G. Differences among the Anthocyanin Accumulation Patterns and Related Gene Expression Levels in Red Pears. Plants 2019, 8, 100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.T.; Li, S.L.; Gu, X.J.; Lei, D.Y.; Zhao, B.; Tang, H.L.; Lin, Y.X.; Wang, Y.; Li, M.Y.; Chen, Q.; et al. Anthocyanin Accumulation and Related Gene Expression Profile in ‘Red Zaosu’ Pear and Its Green Mutant. Agriculture 2021, 11, 898. [Google Scholar] [CrossRef]
- Ou, C.Q.; Zhang, X.L.; Wang, F.; Zhang, L.Y.; Zhang, Y.J.; Fang, M.; Wang, J.H.; Wang, J.X.; Jiang, S.L.; Zhang, Z.H. A 14 nucleotide deletion mutation in the coding region of the PpBBX24 gene is associated with the red skin of “Zaosu Red” pear (Pyrus pyrifolia White Pear Group): A deletion in the PpBBX24 gene is associated with the red skin of pear. Hortic. Res. 2020, 7, 39. [Google Scholar] [CrossRef]
- Sun, Y.; Qian, M.; Wu, R.; Niu, Q.; Teng, Y.; Zhang, D. Postharvest pigmentation in red Chinese sand pears (Pyrus pyrifolia Nakai) in response to optimum light and temperature. Postharvest Biol. Technol. 2014, 91, 64–71. [Google Scholar] [CrossRef]
- Yao, W.; Lei, D.; Zhou, X.; Wang, H.; Lu, J.; Lin, Y.; Zhang, Y.; Wang, Y.; He, W.; Li, M.; et al. Effect of Different Culture Conditions on Anthocyanins and Related Genes in Red Pear Callus. Agronomy 2023, 13, 2032. [Google Scholar] [CrossRef]
- Tao, R.Y.; Bai, S.L.; Ni, J.B.; Yang, Q.S.; Zhao, Y.; Teng, Y.W. The blue light signal transduction pathway is involved in anthocyanin accumulation in ‘Red Zaosu’ pear. Planta 2018, 248, 37–48. [Google Scholar] [CrossRef]
- Ni, J.B.; Zhao, Y.; Tao, R.Y.; Yin, L.; Gao, L.; Strid, Å.; Qian, M.J.; Li, J.C.; Li, Y.J.; Shen, J.Q.; et al. Ethylene mediates the branching of the jasmonate-induced flavonoid biosynthesis pathway by suppressing anthocyanin biosynthesis in red Chinese pear fruits. Plant Biotechnol. J. 2020, 18, 1223–1240. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Bourke, P.M.; Li, S.K.; Lin, M.M.; Sun, L.M.; Gu, H.; Li, Y.K.; Visser, R.G.F.; Qi, X.J.; Maliepaard, C.; et al. QTL mapping of fruit quality traits in tetraploid kiwiberry (Actinidia arguta). Hortic. Plant J. 2025, 11, 1090–1102. [Google Scholar] [CrossRef]
- Nashima, K.; Takeuchi, M.; Moromizato, C.; Omine, Y.; Shoda, M.; Urasaki, N.; Tarora, K.; Irei, A.; Shirasawa, K.; Yamada, M.; et al. Identification of Quantitative Trait Loci of Fruit Quality and Color in Pineapples. Hortic. J. 2023, 92, 375–383. [Google Scholar] [CrossRef]
- Wang, D.S.; Cheng, B.B.; Zhang, J.J. High-density genetic map and quantitative trait loci map of skin color in hawthorn (Crataegus pinnatifida bge. Var. major NEBr.). Front. Genet. 2024, 15, 1405604. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.Y.; Shen, F.; Wang, W.Q.; Wu, B.; Wang, X.; Xiao, C.; Tian, Z.D.; Yang, X.L.; Yang, J.; Wang, Y.; et al. Quantitative trait loci-based genomics-assisted prediction for the degree of apple fruit cover color. Plant Genome 2020, 13, e20047. [Google Scholar] [CrossRef]
- Kishor, D.S.; Alavilli, H.; Lee, S.C.; Kim, J.G.; Song, K. Development of SNP Markers for White Immature Fruit Skin Color in Cucumber (Cucumis sativus L.) Using QTL-seq and Marker Analyses. Plants 2021, 10, 2341. [Google Scholar] [CrossRef]
- Brand, A.; Borovsky, Y.; Meir, S.; Rogachev, I.; Aharoni, A.; Paran, I. pc8.1, a major QTL for pigment content in pepper fruit, is associated with variation in plastid compartment size. Planta 2012, 235, 579–588. [Google Scholar] [CrossRef]
- Sooriyapathirana, S.S.; Khan, A.; Sebolt, A.M.; Wang, D.C.; Bushakra, J.M.; Lin-Wang, K.; Allan, A.C.; Gardiner, S.E.; Chagné, D.; Iezzoni, A.F. QTL analysis and candidate gene mapping for skin and flesh color in sweet cherry fruit (Prunus avium L.). Tree Genet. Genomes 2010, 6, 821–832. [Google Scholar] [CrossRef]
- Ürün, I.; Karci, H.; Attar, S.H.; Gölcü, A.E.; Sönmez, D.A.; Topçu, H.; Özgören, B.; Kafkas, S.; Kafkas, E. Identification of quantitative trait loci (QTL) and development of markers associated with fruit quality traits in strawberry (Fragaria × ananassa Duch.). Euphytica 2025, 221, 48. [Google Scholar] [CrossRef]
- Yamamoto, T.; Kimura, T.; Terakami, S.; Nishitani, C.; Sawamura, Y.; Saito, T.; Kotobuki, K.; Hayashi, T. Integrated reference genetic linkage maps of pear based on SSR and AFLP markers. Breed. Sci. 2007, 57, 321–329. [Google Scholar] [CrossRef]
- Terakami, S.; Kimura, T.; Nishitani, C.; Sawamura, Y.; Saito, T.; Hirabayashi, T.; Yamamoto, T. Genetic Linkage Map of the Japanese Pear ‘Housui’ Identifying Three Homozygous Genomic Regions. J. Jpn. Soc. Hortic. Sci. 2009, 78, 417–424. [Google Scholar] [CrossRef][Green Version]
- Gabay, G.; Dahan, Y.; Izhaki, Y.; Faigenboim, A.; Ben-Ari, G.; Elkind, Y.; Flaishman, M.A. High-resolution genetic linkage map of European pear (Pyrus communis) and QTL fine-mapping of vegetative budbreak time. BMC Plant Biol. 2018, 18, 175. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Jeong, H.J.; Yang, H.B.; Kang, S.M.; Kwon, J.K.; Kim, S.; Choi, D.; Kang, B.C. An ultra-high-density bin map facilitates high-throughput QTL mapping of horticultural traits in pepper (Capsicum annuum). DNA Res. 2016, 23, 81–91. [Google Scholar] [CrossRef]
- Patil, G.; Vuong, T.D.; Kale, S.; Valliyodan, B.; Deshmukh, R.; Zhu, C.S.; Wu, X.L.; Bai, Y.H.; Yungbluth, D.; Lu, F.; et al. Dissecting genomic hotspots underlying seed protein, oil, and sucrose content in an interspecific mapping population of soybean using high-density linkage mapping. Plant Biotechnol. J. 2018, 16, 1939–1953. [Google Scholar] [CrossRef]
- Jiang, N.F.; Shi, S.L.; Shi, H.; Khanzada, H.; Wassan, G.M.; Zhu, C.L.; Peng, X.S.; Yu, Q.Y.; Chen, X.R.; He, X.P.; et al. Mapping QTL for Seed Germinability under Low Temperature Using a New High-Density Genetic Map of Rice. Front. Plant Sci. 2017, 8, 1223. [Google Scholar] [CrossRef]
- Fu, W.H.; Zhao, L.; Qiu, W.J.; Xu, X.; Ding, M.; Lan, L.M.; Qu, S.C.; Wang, S.H. Whole-genome resequencing identifies candidate genes and allelic variation in the MdNADP-ME promoter that regulate fruit malate and fructose contents in apple. Plant Commun. 2024, 5, 100973. [Google Scholar] [CrossRef]
- Yao, G.F.; Ming, M.L.; Allan, A.C.; Gu, C.; Li, L.T.; Wu, X.; Wang, R.Z.; Chang, Y.J.; Qi, K.J.; Zhang, S.L.; et al. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017, 92, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Inoue, E.; Kasumi, M.; Sakuma, F.; Anzai, H.; Amano, K.; Hara, H. Identification of RAPD marker linked to fruit skin color in Japanese pear (Pyrus pyrifolia Nakai). Sci. Hortic. 2006, 107, 254–258. [Google Scholar] [CrossRef]
- Yamamoto, T.; Terakami, S.; Takada, N.; Nishio, S.; Onoue, N.; Nishitani, C.; Kunihisa, M.; Inoue, E.; Iwata, H.; Hayashi, T.; et al. Identification of QTLs controlling harvest time and fruit skin color in Japanese pear (Pyrus pyrifolia Nakai). Breed. Sci. 2014, 64, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Oh, S.; Han, H.Y.D.; Kim, D. QTL Analysis and CAPS Marker Development Linked with Russet in Pear (Pyrus spp.). Plants 2022, 11, 3196. [Google Scholar] [CrossRef] [PubMed]
- Ntladi, S.M.; Human, J.P.; Bester, C.; Vervalle, J.; Roodt-Wilding, R.; Tobutt, K.R. Quantitative trait loci (QTL) mapping of blush skin and flowering time in a European pear (Pyrus communis) progeny of ‘Flamingo’ × ‘Abate Fetel’. Tree Genet. Genomes 2018, 14, 70. [Google Scholar] [CrossRef]
- Yue WenQuan, Y.W.; Zhang Hai, Z.H.; Liu JinLi, L.J.; Xu JinTao, X.J.; Hao BaoFeng, H.B.; Gao LiJuan, G.L.; Li LongFei, L.L. Breeding Report of a New Red Pear Cultivar ‘Xianghongli’. Agric. Sci. Technol. 2016, 17, 2569. [Google Scholar]
- Schenk, J.J.; Becklund, L.E.; Carey, S.J.; Fabre, P.P. What is the “modified” CTAB protocol? Characterizing modifications to the CTAB DNA extraction protocol. Appl. Plant Sci. 2023, 11, e11517. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Rastas, P. Lep-MAP3: Robust linkage mapping even for low-coverage whole genome sequencing data. Bioinformatics 2017, 33, 3726–3732. [Google Scholar] [CrossRef]
- Chen, S.; Tian, Y.S.; Li, Z.T.; Li, Z.Q.; Liu, Y.; Wang, L.N.; Li, L.L.; Pang, Z.F.; Yang, C.J.; Wang, Q.B.; et al. Construction of a high-density genetic linkage map and QTL mapping for growth traits in hybrid Epinephelus fuscoguttatus (♀) and Epinephelus tukula (♂) progeny. Aquaculture 2022, 551, 737921. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, M.; Li, J.; Chi, Y.; Wang, K.; Peng, J.; Zhang, Y. Phenotypic Characters and Inheritance Tendency of Agronomic Traits in F1 Progeny of Pear. Plants 2025, 14, 1491. [Google Scholar] [CrossRef]
- Arends, D.; Prins, P.; Jansen, R.C.; Broman, K.W. R/qtl: High-throughput multiple QTL mapping. Bioinformatics 2010, 26, 2990–2992. [Google Scholar] [CrossRef]
- Zhang, X.J.; Wu, Y.J.; Wang, X.M.; Wang, W.F.; Huang, M.X.; Ma, Z.T.; Peng, J.Y. Ring Stripping, Ring Cutting, and Growth Regulators Promote Phase Change and Early Flowering in Pear Seedlings. Plants 2023, 12, 2933. [Google Scholar] [CrossRef]
- Ali, I.; Teng, Z.H.; Bai, Y.T.; Yang, Q.; Hao, Y.S.; Hou, J.; Jia, Y.B.; Tian, L.X.; Liu, X.Y.; Tan, Z.Y.; et al. A high density SLAF-SNP genetic map and QTL detection for fibre quality traits in Gossypium hirsutum. BMC Genom. 2018, 19, 879. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.F.; Zhang, Q.S.; Zhang, X.Q.; Tan, C.; Li, C.D. Construction of High-Density Genetic Map in Barley through Restriction-Site Associated DNA Sequencing. PLoS ONE 2015, 10, e0133161. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.X.; Ke, C.J.; Tang, W.Q.; Jiang, J.L.; Xia, J.; Xie, X.F.; Yang, M.; Duan, C.F.; Wu, W.R.; Zheng, Y. Construction of a High-Density Recombination Bin-Based Genetic Map Facilitates High-Resolution Mapping of a Major QTL Underlying Anthocyanin Pigmentation in Eggplant. Int. J. Mol. Sci. 2022, 23, 10258. [Google Scholar] [CrossRef] [PubMed]
- Qin, M.F.; Li, L.T.; Singh, J.; Sun, M.Y.; Bai, B.; Li, S.W.; Ni, J.P.; Zhang, J.Y.; Zhang, X.; Wei, W.L.; et al. Construction of a high-density bin-map and identification of fruit quality-related quantitative trait loci and functional genes in pear. Hortic. Res. 2022, 9, uhac141. [Google Scholar] [CrossRef]
- Oh, S.; Ahn, S.; Han, H.Y.D.; Kim, K.; Kim, S.A.; Kim, D. Genetic linkage maps and QTLs associated with fruit skin color and acidity in apple (Malus × domestica). Hortic. Environ. Biotechnol. 2023, 64, 299–310. [Google Scholar] [CrossRef]
- Frett, T.J.; Reighard, G.L.; Okie, W.R.; Gasic, K. Mapping quantitative trait loci associated with blush in peach Prunus persica (L.) Batsch. Tree Genet. Genomes 2014, 10, 367–381. [Google Scholar] [CrossRef]
- Sun, L.; Li, S.C.; Jiang, J.F.; Tang, X.P.; Fan, X.C.; Zhang, Y.; Liu, J.H.; Liu, C.H. New quantitative trait locus (QTLs) and candidate genes associated with the grape berry color trait identified based on a high-density genetic map. BMC Plant Biol. 2020, 20, 302. [Google Scholar] [CrossRef]
- Li, Y.Q.; Shan, X.T.; Gao, R.F.; Han, T.T.; Zhang, J.; Wang, Y.N.; Kimani, S.; Wang, L.; Gao, X. MYB repressors and MBW activation complex collaborate to fine-tune flower coloration in Freesia hybrida. Commun. Biol. 2020, 3, 396. [Google Scholar] [CrossRef]
- Gao, H.N.; Jiang, H.; Cui, J.Y.; You, C.X.; Li, Y.Y. Review: The effects of hormones and environmental factors on anthocyanin biosynthesis in apple. Plant Sci. 2021, 312, 111024. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.J.; Geng, Z.Q.; Wang, Y.X.; Wang, Y.G.; Liu, S.H.; Chen, C.W.; Song, A.P.; Jiang, J.F.; Chen, S.M.; Chen, F.D. A novel transcription factor CmMYB012 inhibits flavone and anthocyanin biosynthesis in response to high temperatures in chrysanthemum. Hortic. Res. 2021, 8, 248. [Google Scholar] [CrossRef]
- Alabd, A.; Ahmad, M.; Zhang, X.; Gao, Y.H.; Peng, L.; Zhang, L.; Ni, J.B.; Bai, S.L.; Teng, Y.W. Light-responsive transcription factor PpWRKY44 induces anthocyanin accumulation by regulating PpMYB10 expression in pear. Hortic. Res. 2022, 9, uhac199. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.Y.; Wang, L.; Wang, S.M.; Li, W.J.; Liu, D.; Guo, X.F.; Qu, B.H. Transcriptomic analysis of bagging-treated ‘Pingguo’ pear shows that MYB4-like1, MYB4-like2, MYB1R1 and WDR involved in anthocyanin biosynthesis are up-regulated in fruit peels in response to light. Sci. Hortic. 2019, 244, 428–434. [Google Scholar] [CrossRef]
- Yin, Y. Functional Study of FaMYB44.1 Transcription Factor in Regulating the Accumulation of Anthocyanins in Strawberry Fruits. Master’s Thesis, Yangzhou University, Yangzhou, China, 2023. [Google Scholar]
- Liu, X.Y.; Huang, Q.; Liang, Y.Q.; Lu, Z.; Liu, W.T.; Yuan, H.; Li, H.J. Genome-Wide Identification and Expression Analysis of ‘NanGuo’ Pear Revealed Key MYB Transcription Factor Family Genes Involved in Anthocyanin Accumulation. Horticulturae 2024, 10, 989. [Google Scholar] [CrossRef]
- Zhang, J.H.; Song, B.B.; Chen, G.S.; Yang, G.Y.; Ming, M.L.; Zhang, S.Q.; Xue, Z.L.; Han, C.H.; Li, J.M.; Wu, J. Transcriptome Analysis Identified PyNAC42 as a Positive Regulator of Anthocyanin Biosynthesis Induced by Nitrogen Deficiency in Pear (Pyrus spp.). Horticulturae 2024, 10, 980. [Google Scholar] [CrossRef]
- Cong, L.; Qu, Y.Y.; Sha, G.Y.; Zhang, S.C.; Ma, Y.F.; Chen, M.; Zhai, R.; Yang, C.Q.; Xu, L.F.; Wang, Z.G. PbWRKY75 promotes anthocyanin synthesis by activating PbDFR, PbUFGT, and PbMYB10b in pear. Physiol. Plant. 2021, 173, 1841–1849. [Google Scholar] [CrossRef]
- Hoy-Taek, K.I.M.; Robin, A.H.K.; Nou, I.-S. Parentage Confirmation of Korean Bred Pear Cultivars by Simple Sequence Repeat-Genotyping and S-Genotypes Analysis. Plant Breed. Biotechnol. 2016, 4, 198–211. [Google Scholar] [CrossRef]
- Teng, Y.W.; Tanabe, K.; Tamura, F.; Itai, A. Genetic relationships of Pyrus species and cultivars native to East Asia revealed by randomly amplified polymorphic DNA markers. J. Am. Soc. Hortic. Sci. 2002, 127, 262–270. [Google Scholar] [CrossRef]
- Nowak, B.; Tomkowiak, A.; Sobiech, A.; Bocianowski, J.; Kowalczewski, P.L.; Spychala, J.; Jamruszka, T. Identification and Analysis of Candidate Genes Associated with Yield Structure Traits and Maize Yield Using Next-Generation Sequencing Technology. Genes 2024, 15, 56. [Google Scholar] [CrossRef] [PubMed]
- Rameneni, J.J.; Islam, A.S.M.F.; Avila, C.A.; Shi, A. Improving genomic prediction of vitamin C content in spinach using GWAS-derived markers. BMC Genom. 2025, 26, 171. [Google Scholar] [CrossRef]
- Yang, F.; Lang, T.; Wu, J.; Zhang, C.; Qu, H.; Pu, Z.; Yang, F.; Yu, M.; Feng, J. SNP loci identification and KASP marker development system for genetic diversity, population structure, and fingerprinting in sweetpotato (Ipomoea batatas L.). BMC Genom. 2024, 25, 1245. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).