Genome-Wide Identification and Analysis of the CCT Gene Family Contributing to Photoperiodic Flowering in Chinese Cabbage (Brassica rapa L. ssp. pekinensis)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, Growth Conditions, and Stress Treatments
2.2. Identification of the CCT Gene Family in Chinese Cabbage
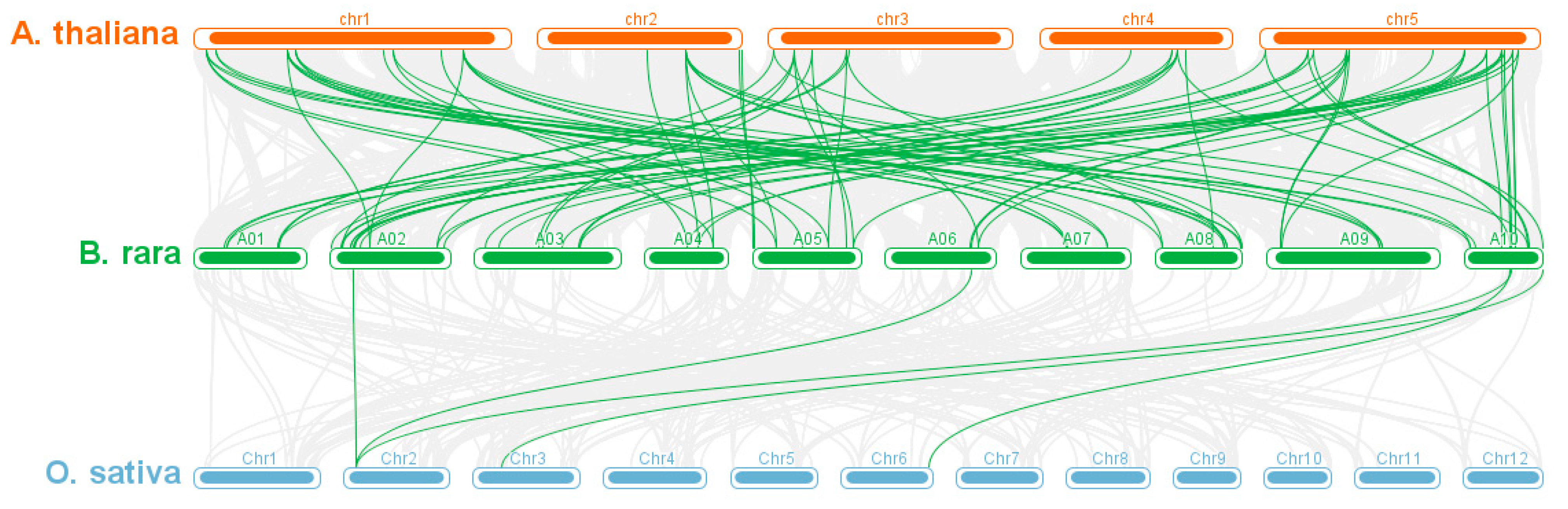
2.3. Analysis of the Gene Location, Duplication Relationship, and Collinearity
2.4. Analysis of Gene Structure and Motif Structure
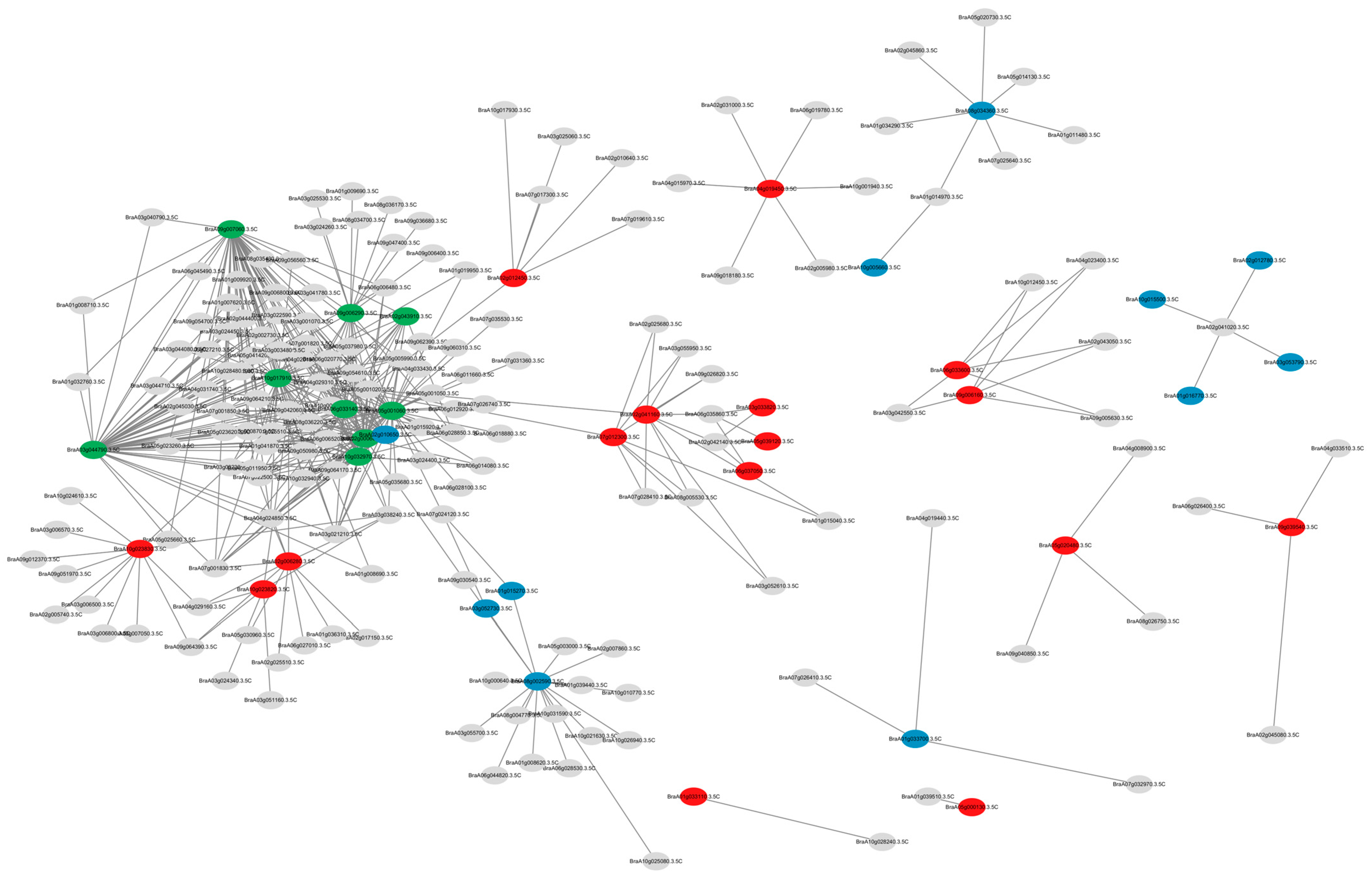
2.5. Co-Expression Network Analysis of the CCT Family Genes
2.6. Multiple Sequence Alignment and Phylogenetic Analysis
2.7. RNA Isolation, cDNA Synthesis, Transcriptome, and Quantitative Real-Time PCR Analysis
3. Results
3.1. Flowering Time of Chinese Cabbage Under LD and SD Conditions
3.2. Genome-Wide Identification and Physicochemical Property Analysis of the BrCCT Gene Family
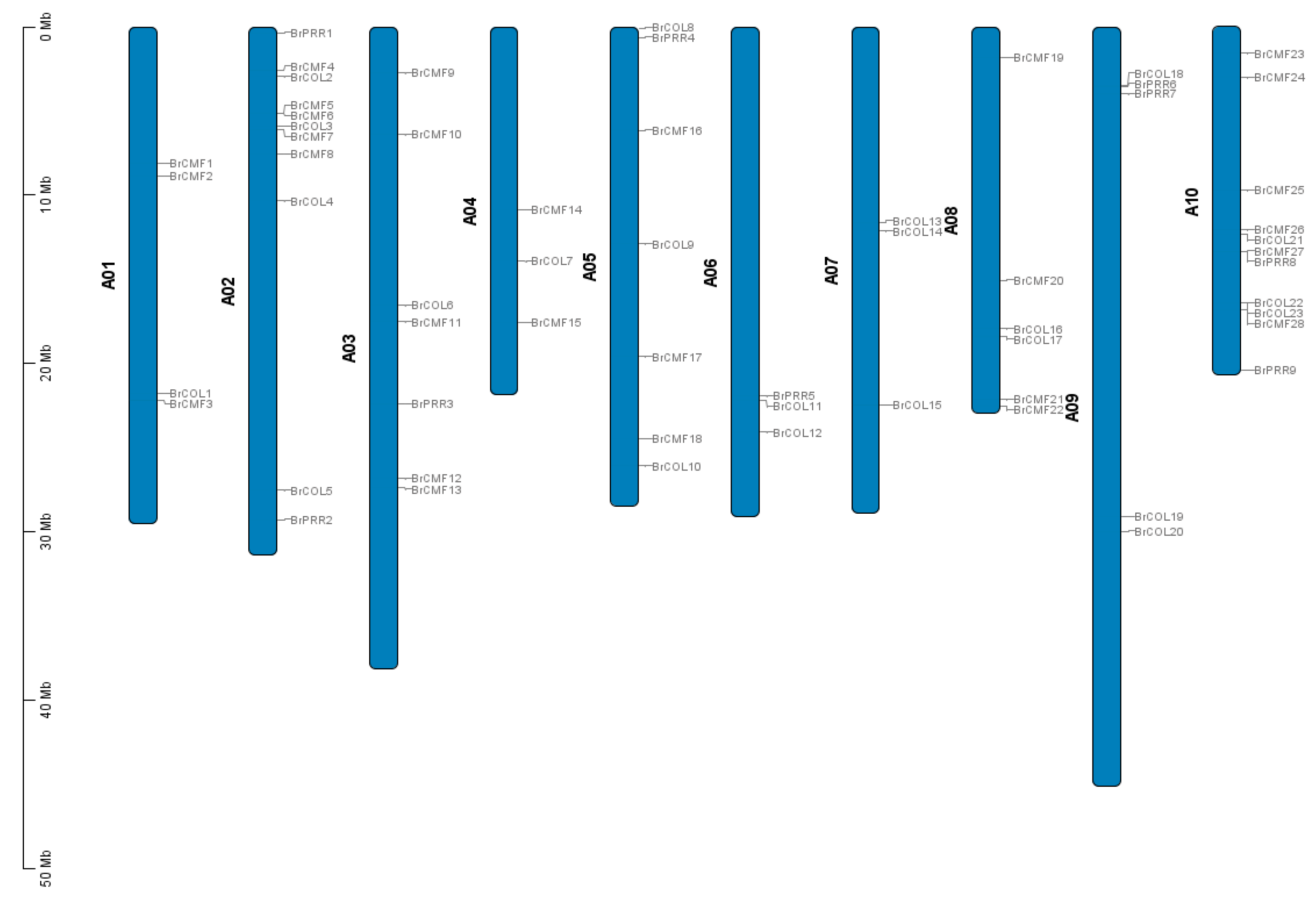
3.3. Chromosomal Localization Analysis of the BrCCT Gene Family
3.4. Gene Duplication Analysis of the BrCCT Gene Family
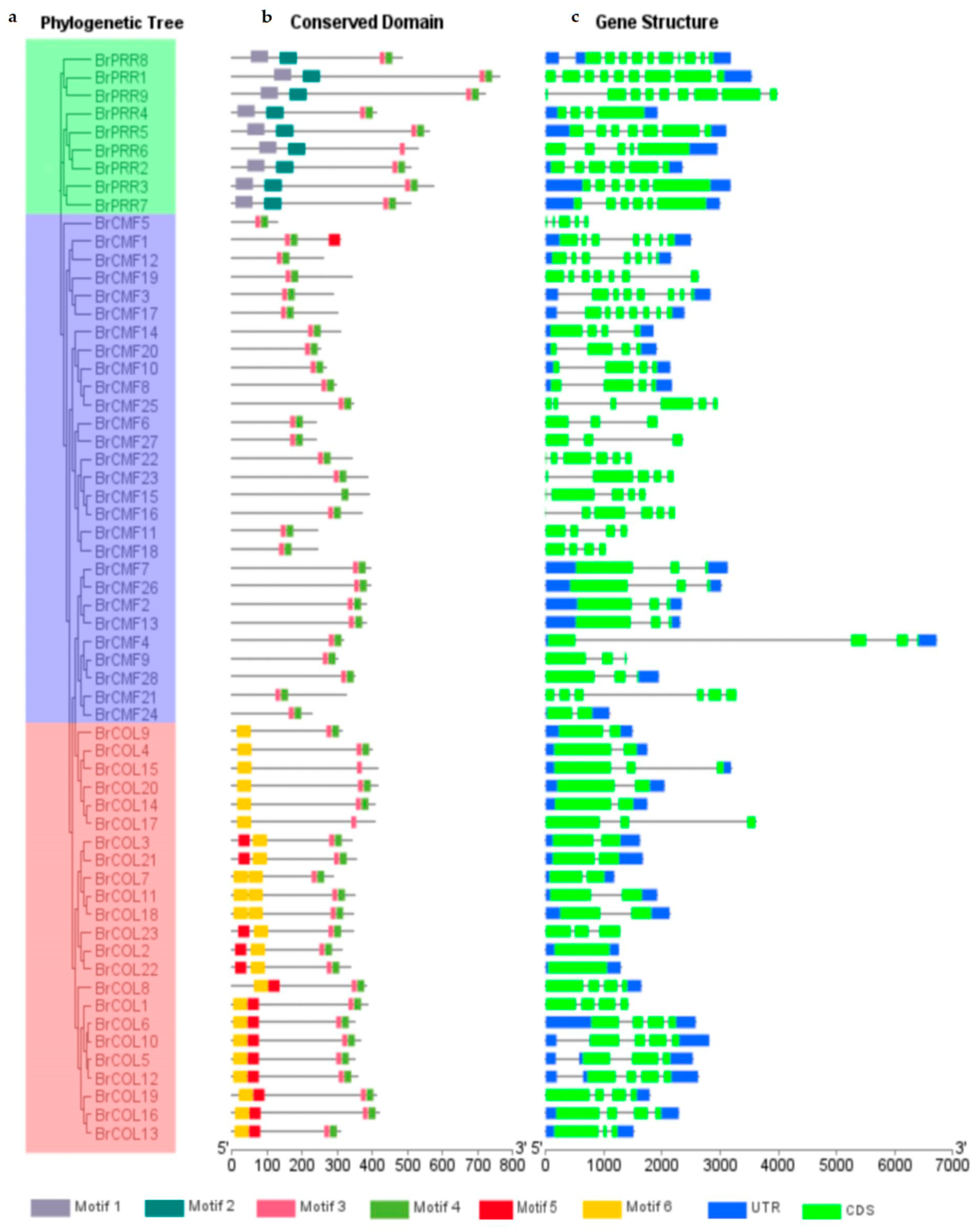
3.5. Gene Structure Analysis of the BrCCT Gene Family
3.6. Conserved Motif Analysis of the BrCCT Gene Family
3.7. Co-Expression Network Analysis of the BrCCT Gene Family
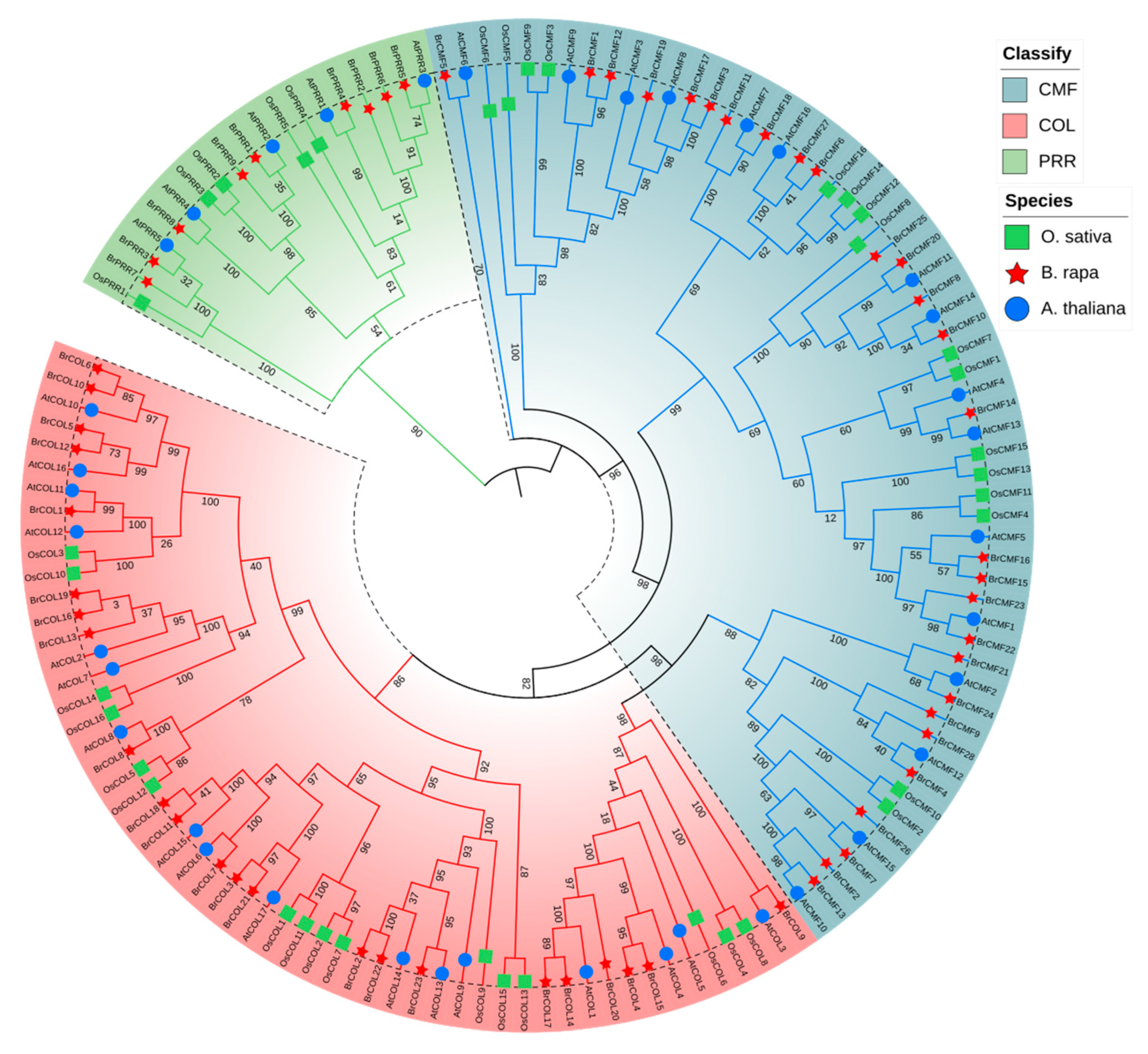
3.8. Multiple Sequence Alignment and Phylogenetic Tree Analysis of the BrCCT Gene Family
3.9. Transcriptome Analysis and Expression Analysis in Different Tissues of BrCMF Genes
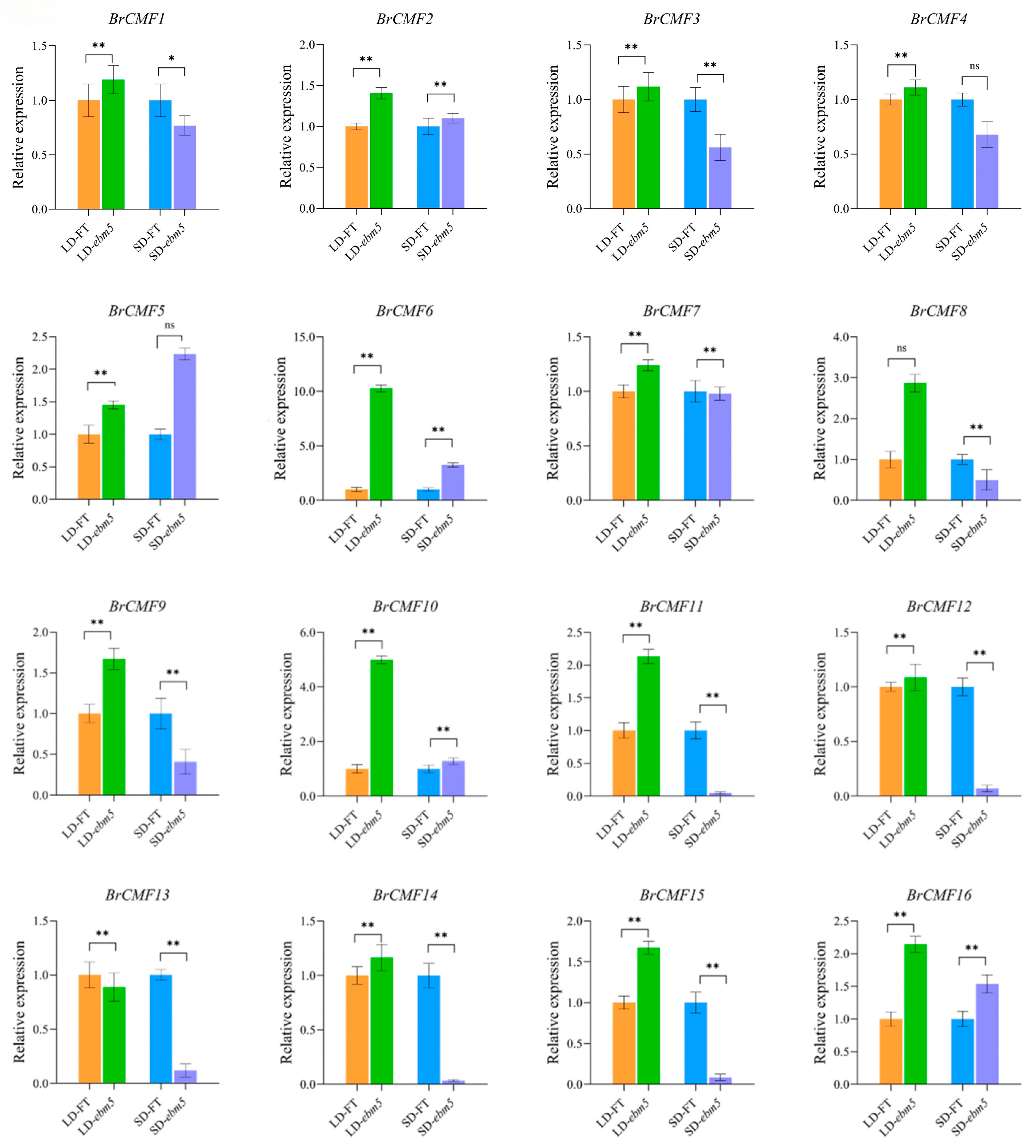
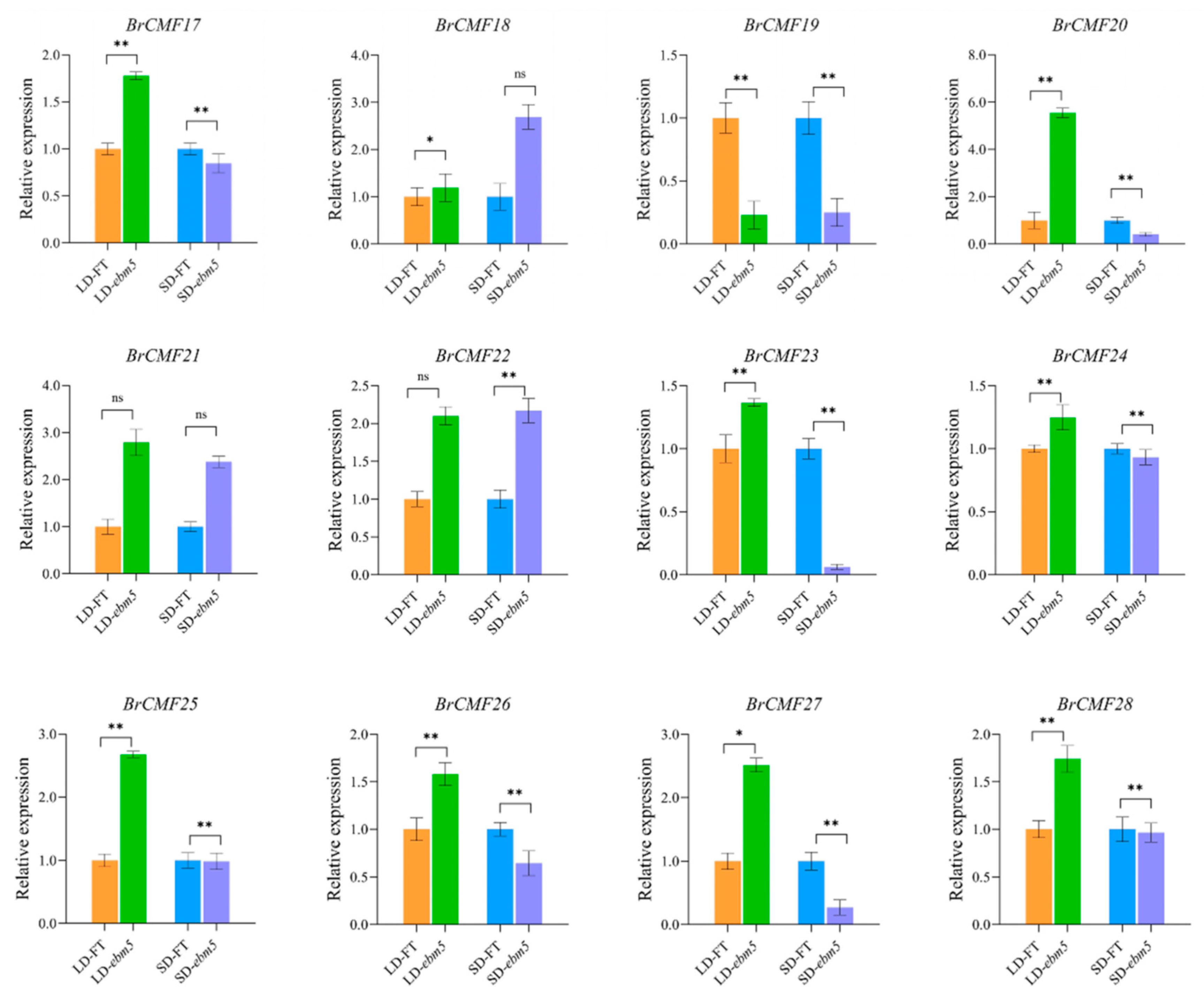
3.10. Expression Profiles Analysis of BrCMF Genes Under LD and SD Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| CCT | CONSTANS, CONSTANS-LIKE and TOC1 |
| LD | long-day |
| SD | short-day |
| CMF | CCT MOTIF FAMILY |
| COL | CONSTANS-like |
| PRR | PREUDORESPONSE REGULATOR |
| FT | FLOWERING LOCUS T |
| Hd1 | Heading Date 1 |
| Ehd1 | EH domain-containing protein 1 |
| TE | transposable element |
| DH | doubled haploid |
| TAIR | the Arabidopsis Information Resource |
| RGAP | Rice Genome Annotation Project |
| PPI | Protein-protein interaction |
| aa | Amino acids |
| pI | Isoelectric point |
| MW | Molecular weight |
| Da | Dalton |
| Mb | Megabases |
| UTRs | Untranslated regions |
| CDS | Coding DNA Sequence |
References
- Blümel, M.; Dally, N.; Jung, C. Flowering time regulation in crops—What did we learn from Arabidopsis? Curr. Opin. Biotechnol. 2015, 32, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Yi, D.; Yang, J.; Liu, X.; Pang, Y. Genome-wide identification, expression analysis and functional study of CCT gene family in Medicago truncatula. Plants 2020, 9, 513. [Google Scholar] [CrossRef] [PubMed]
- Pajoro, A.; Biewers, S.; Dougali, E.; Leal Valentim, F.; Mendes, M.A.; Porri, A.; Coupland, G.; Van de Peer, Y.; van Dijk, A.D.; Colombo, L.; et al. The (r)evolution of gene regulatory networks controlling Arabidopsis plant reproduction: A two-decade history. J. Exp. Bot. 2014, 65, 4731–4745. [Google Scholar] [CrossRef] [PubMed]
- Cockram, J.; Thiel, T.; Steuernagel, B.; Stein, N.; Taudien, S.; Bailey, P.C.; O’Sullivan, D.M. Genome dynamics explain the evolution of flowering time CCT domain gene families in the Poaceae. PLoS ONE 2012, 7, e45307. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Qu, X.; Zhou, Y.; Song, G.; Abiri, N.; Xiao, Y.; Liang, F.; Jiang, D.; Hu, Z.; Yang, D. Osprr37 confers an expanded regulation of the diurnal rhythms of the transcriptome and photoperiodic flowering pathways in rice. Plant Cell Environ. 2018, 41, 630–645. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Liu, H.; Zhou, X.; Li, Q.; Zhang, J.; Lu, L.; Liu, T.; Liu, H.; Zhang, C.; Zhang, Z.; et al. Natural variation in ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res. 2013, 23, 969–971. [Google Scholar] [CrossRef] [PubMed]
- Strayer, C.; Oyama, T.; Schultz, T.F.; Raman, R.; Somers, D.E.; Más, P.; Panda, S.; Kreps, J.A.; Kay, S.A. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 2000, 289, 768–771. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Q.; Dong, H.; He, Q.; Liang, L.; Tan, C.; Han, Z.; Yao, W.; Li, G.; Zhao, H.; et al. Three CCT domain-containing genes were identified to regulate heading date by candidate gene-based association mapping and transformation in rice. Sci. Rep. 2015, 5, 7663. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Roussot, C.; Suárez-López, P.; Corbesier, L.; Vincent, C.; Piñeiro, M.; Hepworth, S.; Mouradov, A.; Justin, S.; Turnbull, C.; et al. Constans acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 2004, 131, 3615–3626. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, J.L.; An, L.; Wu, T.; Yang, L.; Cheng, Y.S.; Nie, X.S.; Qin, Z.Q. Genome-wide analysis of the CCT gene family in chinese white pear (Pyrus bretschneideri rehd.) and characterization of pbprr2 in response to varying light signals. BMC Plant Biol. 2022, 22, 81. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Shen, Y.; Chang, H.C.; Hou, Y.; Harris, A.; Ma, S.F.; McPartland, M.; Hymus, G.J.; Adam, L.; Marion, C.; et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010, 187, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Suárez-López, P.; Wheatley, K.; Robson, F.; Onouchi, H.; Valverde, F.; Coupland, G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 2001, 410, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Robson, F.; Costa, M.M.; Hepworth, S.R.; Vizir, I.; Piñeiro, M.; Reeves, P.H.; Putterill, J.; Coupland, G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001, 28, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hu, Y.; Xu, L.-H.; He, Q.; Fan, X.-W.; Xing, Y.-Z. The CCT domain-containing gene family has large impacts on heading date, regional adaptation, and grain yield in rice. J. Integr. Agric. 2017, 16, 2686–2697. [Google Scholar] [CrossRef]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12, 2473–2484. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Jeong, D.H.; Lee, D.Y.; Yi, J.; Ryu, C.H.; Kim, S.L.; Jeong, H.J.; Choi, S.C.; Jin, P.; Yang, J.; et al. Oscol4 is a constitutive flowering repressor upstream of Ehd1 and downstream of osphyB. Plant J. 2010, 63, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Liu, X.; Jia, W.; Liu, H.; Li, W.; Peng, Y.; Du, Y.; Wang, Y.; Yin, Y.; Zhang, X.; et al. Zmcol3, a CCT gene represses flowering in maize by interfering with the circadian clock and activating expression of ZmCCT. J. Integr. Plant Biol. 2018, 60, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Li, Z.; Li, W.; Ku, L.; Wang, C.; Ye, J.; Li, K.; Yang, N.; Li, Y.; Zhong, T.; et al. CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA 2013, 110, 16969–16974. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jiao, B.; Dong, F.; Liang, X.; Zhou, S.; Wang, H. Genome-wide identification of CCT genes in wheat (Triticum aestivum L.) and their expression analysis during vernalization. PLoS ONE 2022, 17, e0262147. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, X.; Wu, L.; Zhang, L.; Liu, G.; Xia, C.; Liu, X.; Kong, X. Transcriptome profiling of developing leaf and shoot apices to reveal the molecular mechanism and co-expression genes responsible for the wheat heading date. BMC Genom. 2021, 22, 468. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Xiang, R.; Jiang, Y.; Li, W.; Yang, Q.; Gan, G.; Li, W.; Yu, C.; Wang, Y. Genome-wide identification and expression profiling analysis of the CCT gene family in Solanum lycopersicum and Solanum melongena. Genes 2024, 15, 1385. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Xia, J.; Jiang, R.; Wang, J.; Yuan, X.; Dong, X.; Chen, Z.; Zhao, Z.; Wu, B.; Zhan, L.; et al. Genome-wide identification and characterization of the CCT gene family in rapeseed (Brassica napus L.). Int. J. Mol. Sci. 2024, 25, 5301. [Google Scholar] [CrossRef] [PubMed]
- Mengarelli, D.A.; Zanor, M.I. Genome-wide characterization and analysis of the CCT motif family genes in soybean (Glycine max). Planta 2021, 253, 15. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhou, X.; Li, Q.; Wang, L.; Xing, Y. CCT domain-containing genes in cereal crops: Flowering time and beyond. Theor. Appl. Genet. 2020, 133, 1385–1396. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Y.; Yan, W.Y.; Chang, H.Y.; Sun, C.W. Arabidopsis CIA2 and CIL have distinct and overlapping functions in regulating chloroplast and flower development. Plant Direct 2022, 6, e380. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Huang, S.; Gao, Y.; Zhang, M.; Qu, G.; Wang, N.; Liu, Z.; Feng, H. Role of BrSDG8 on bolting in Chinese cabbage (Brassica rapa). Theor. Appl. Genet. 2020, 133, 2937–2948. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, T.; He, X.; Cai, X.; Lin, R.; Liang, J.; Wu, J.; King, G.; Wang, X. Brad v3.0: An upgraded Brassicaceae database. Nucleic Acids Res. 2022, 50, D1432–D1441. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProscan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Nie, S.; Zhang, M.; Zhang, L. Genome-wide identification and expression analysis of calmodulin-like (CML) genes in Chinese cabbage (Brassica rapa L. Ssp. Pekinensis). BMC Genom. 2017, 18, 842. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Tun, W.; Dai, S.F.; Li, J.Y.; Zhang, Q.J.; Yin, G.Y.; Yoon, J.; Cho, L.H.; An, G.; Gao, L.Z. Genome-wide analysis of CCT transcript factors to identify genes contributing to photoperiodic flowering in Oryza rufipogon. Front. Plant Sci. 2021, 12, 736419. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ma, J.; Wang, G.; Li, H.; Wang, H.; Wang, G.; Jiang, Y.; Liu, Y.; Liu, G.; Liu, G.; et al. Exploring the SiCCT gene family and its role in heading date in foxtail millet. Front. Plant Sci. 2022, 13, 863298. [Google Scholar] [CrossRef] [PubMed]
- Tribhuvan, K.U.; Kaila, T.; Srivastava, H.; Das, A.; Kumar, K.; Durgesh, K.; Joshi, R.; Singh, B.K.; Singh, N.K.; Gaikwad, K. Structural and functional analysis of CCT family genes in pigeonpea. Mol. Biol. Rep. 2022, 49, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Chapman, B.A.; Bowers, J.E.; Schulze, S.R.; Paterson, A.H. A comparative phylogenetic approach for dating whole genome duplication events. Bioinformatics 2004, 20, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Pirovano, W.; Heringa, J. Multiple sequence alignment. Methods Mol. Biol. 2008, 452, 143–161. [Google Scholar] [PubMed]



| Gene Name | Gene ID | Chromosome | Protein (aa) | pI | MW (Da) |
|---|---|---|---|---|---|
| BrCMF1 | BraA01g015270.3.5C | A01 | 310 | 6.05 | 33,558.27 |
| BrCMF2 | BraA01g016770.3.5C | A01 | 382 | 6.37 | 42,602.83 |
| BrCOL1 | BraA01g033110.3.5C | A01 | 386 | 5.54 | 43,028.5 |
| BrCMF3 | BraA01g033700.3.5C | A01 | 290 | 6.26 | 31,727.19 |
| BrPRR1 | BraA02g000660.3.5C | A02 | 760 | 8.23 | 82,882.73 |
| BrCMF4 | BraA02g005440.3.5C | A02 | 318 | 8.49 | 35,569.73 |
| BrCOL2 | BraA02g006280.3.5C | A02 | 315 | 7.14 | 35,249.63 |
| BrCMF5 | BraA02g010650.3.5C | A02 | 128 | 10.5 | 14,454.51 |
| BrCMF6 | BraA02g010730.3.5C | A02 | 241 | 6.41 | 26,671.66 |
| BrCOL3 | BraA02g012450.3.5C | A02 | 342 | 5.49 | 37,592.09 |
| BrCMF7 | BraA02g012780.3.5C | A02 | 394 | 7.73 | 43,853.06 |
| BrCMF8 | BraA02g015010.3.5C | A02 | 298 | 5.35 | 33,624.66 |
| BrCOL4 | BraA02g019020.3.5C | A02 | 401 | 5.39 | 45,585.3 |
| BrCOL5 | BraA02g041160.3.5C | A02 | 351 | 5.48 | 38,961.19 |
| BrPRR2 | BraA02g043910.3.5C | A02 | 509 | 7.63 | 57,088.59 |
| BrCMF9 | BraA03g006340.3.5C | A03 | 301 | 9.34 | 33,962.85 |
| BrCMF10 | BraA03g014440.3.5C | A03 | 269 | 4.77 | 30,419.14 |
| BrCOL6 | BraA03g033820.3.5C | A03 | 351 | 5.47 | 38,638.54 |
| BrCMF11 | BraA03g035990.3.5C | A03 | 244 | 5.9 | 28,167.87 |
| BrPRR3 | BraA03g044790.3.5C | A03 | 576 | 6.67 | 64,811.77 |
| BrCMF12 | BraA03g052730.3.5C | A03 | 261 | 5.97 | 28,333.46 |
| BrCMF13 | BraA03g053790.3.5C | A03 | 383 | 6.22 | 42,959.09 |
| BrCMF14 | BraA04g014840.3.5C | A04 | 308 | 5.33 | 34,785.22 |
| BrCOL7 | BraA04g019450.3.5C | A04 | 289 | 8.27 | 31,501.53 |
| BrCMF15 | BraA04g025130.3.5C | A04 | 390 | 4.41 | 43,197.44 |
| BrCOL8 | BraA05g000130.3.5C | A05 | 382 | 8.7 | 43,594.07 |
| BrPRR4 | BraA05g001060.3.5C | A05 | 412 | 6.09 | 45,915.79 |
| BrCMF16 | BraA05g011500.3.5C | A05 | 370 | 4.7 | 41,413.53 |
| BrCOL9 | BraA05g020480.3.5C | A05 | 314 | 6.07 | 37,089.29 |
| BrCMF17 | BraA05g026820.3.5C | A05 | 301 | 6.31 | 33,252.02 |
| BrCMF18 | BraA05g035260.3.5C | A05 | 244 | 5.09 | 28,037.53 |
| BrCOL10 | BraA05g039120.3.5C | A05 | 368 | 5.58 | 40,383.3 |
| BrPRR5 | BraA06g033140.3.5C | A06 | 563 | 6.98 | 62,530.66 |
| BrCOL11 | BraA06g033600.3.5C | A06 | 351 | 5.56 | 37,943.14 |
| BrCOL12 | BraA06g037050.3.5C | A06 | 358 | 5.85 | 39,397.8 |
| BrCOL13 | BraA07g012300.3.5C | A07 | 308 | 5.8 | 34,026.11 |
| BrCOL14 | BraA07g013220.3.5C | A07 | 408 | 5.51 | 46,106.73 |
| BrCOL15 | BraA07g031770.3.5C | A07 | 415 | 5.48 | 47,062.04 |
| BrCMF19 | BraA08g002590.3.5C | A08 | 343 | 5.49 | 37,283.24 |
| BrCMF20 | BraA08g019720.3.5C | A08 | 252 | 5.06 | 28,599.34 |
| BrCOL16 | BraA08g025070.3.5C | A08 | 418 | 5.67 | 46,245.53 |
| BrCOL17 | BraA08g025990.3.5C | A08 | 407 | 5.58 | 45,398.2 |
| BrCMF21 | BraA08g034360.3.5C | A08 | 325 | 4.82 | 37,077.56 |
| BrCMF22 | BraA08g035370.3.5C | A08 | 340 | 4.87 | 37,905.93 |
| BrCOL18 | BraA09g006160.3.5C | A09 | 345 | 5.89 | 37,321.68 |
| BrPRR6 | BraA09g006290.3.5C | A09 | 528 | 8.4 | 59,627.47 |
| BrPRR7 | BraA09g007060.3.5C | A09 | 510 | 7.69 | 57,852.13 |
| BrCOL19 | BraA09g038260.3.5C | A09 | 412 | 8.45 | 45,914.97 |
| BrCOL20 | BraA09g039540.3.5C | A09 | 414 | 5.08 | 46,495.03 |
| BrCMF23 | BraA10g002920.3.5C | A10 | 386 | 4.48 | 43,148.13 |
| BrCMF24 | BraA10g005660.3.5C | A10 | 227 | 4.97 | 26,904.79 |
| BrCMF25 | BraA10g012030.3.5C | A10 | 347 | 5.08 | 38,537.5 |
| BrCMF26 | BraA10g015500.3.5C | A10 | 397 | 9.14 | 44,406.02 |
| BrCOL21 | BraA10g015920.3.5C | A10 | 356 | 6.08 | 39,055.67 |
| BrCMF27 | BraA10g017860.3.5C | A10 | 240 | 6.3 | 26,425.3 |
| BrPRR8 | BraA10g017910.3.5C | A10 | 484 | 5.81 | 53,046.54 |
| BrCOL22 | BraA10g023820.3.5C | A10 | 337 | 6.23 | 37,950.39 |
| BrCOL23 | BraA10g023830.3.5C | A10 | 345 | 7.19 | 39,033.48 |
| BrCMF28 | BraA10g024780.3.5C | A10 | 352 | 9.72 | 40,132.2 |
| BrPRR9 | BraA10g032970.3.5C | A10 | 722 | 7.62 | 78,528.65 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, W.; Jia, X.; Li, S.; Zhou, Y.; Zhang, X.; Jiang, L.; Hao, L. Genome-Wide Identification and Analysis of the CCT Gene Family Contributing to Photoperiodic Flowering in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Horticulturae 2025, 11, 848. https://doi.org/10.3390/horticulturae11070848
Fu W, Jia X, Li S, Zhou Y, Zhang X, Jiang L, Hao L. Genome-Wide Identification and Analysis of the CCT Gene Family Contributing to Photoperiodic Flowering in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Horticulturae. 2025; 11(7):848. https://doi.org/10.3390/horticulturae11070848
Chicago/Turabian StyleFu, Wei, Xinyu Jia, Shanyu Li, Yang Zhou, Xinjie Zhang, Lisi Jiang, and Lin Hao. 2025. "Genome-Wide Identification and Analysis of the CCT Gene Family Contributing to Photoperiodic Flowering in Chinese Cabbage (Brassica rapa L. ssp. pekinensis)" Horticulturae 11, no. 7: 848. https://doi.org/10.3390/horticulturae11070848
APA StyleFu, W., Jia, X., Li, S., Zhou, Y., Zhang, X., Jiang, L., & Hao, L. (2025). Genome-Wide Identification and Analysis of the CCT Gene Family Contributing to Photoperiodic Flowering in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Horticulturae, 11(7), 848. https://doi.org/10.3390/horticulturae11070848





