Abstract
Waterlogging is a critical abiotic stressor that significantly impacts plant growth. Plants under waterlogging stress release metabolic signals that recruit rhizosphere microorganisms and enhance stress resistance. However, the mechanisms through which the non-adaptive species R. delavayi responds to waterlogging stress via the synergistic interaction between root metabolites and rhizosphere microbiota remain poorly elucidated. Here, we employed pot experiments to characterize the responses of the root metabolite–microbiota complex in R. delavayi during waterlogging stress and subsequent recovery. Our results revealed that waterlogging altered the root morphology, the root metabolite profile, rhizosphere microbial diversity and network complexity, and these effects persisted during recovery. A significant correlation between root metabolites and the rhizosphere microbial community structure during waterlogging stress and recovery. Importantly, some differentially accumulated metabolites had significant effects on the assembly of rhizosphere microbes. Most of the core microbes in the rhizosphere microbial community under waterlogging and post–waterlogging recovery treatment were likely beneficial bacteria. Based on these findings, we propose a model for how root metabolites and rhizosphere microbes interact to help R. delavayi cope with waterlogging and recover. Based on these findings, we propose a possible response pattern of root metabolites and rhizosphere microbiota complex in R. delavayi under waterlogging stress and recovery. This work provides new insights into the synergistic mechanisms enhancing plant waterlogging tolerance and highlights the potential of harnessing rhizosphere microbiota to improve resilience in rhododendrons.
1. Introduction
Due to the marked increase in heavy precipitation and recurrent precipitation events, the response mechanisms of plants to waterlogging stress have become a key research focus. Rhododendrons, both evergreen and deciduous woody plants, have high horticultural value and are cultivated worldwide [1]. In the northwest of Guizhou Province, the Baili Azalea Nature Reserve (BANR) contains approximately 40 species of Rhododendron and serves as an important rhododendron germplasm bank. Rhododendron delavayi (R. delavayi) is a crucial member of the BANR [2]. With global climate change, the frequency and intensity of waterlogging events in August in Guizhou have increased significantly [2]. Unfortunately, waterlogging hinders rhododendron growth and development and can be fatal. However, research on their response patterns to waterlogging stress and post-waterlogging recovery remains limited.
The rhizosphere—the soil region within 2 mm of the root system—is a hotspot for microbial activity [3]. Rhizosphere microbiotas are highly sensitive to environmental fluctuations, particularly changes in soil moisture [4]. Growing evidence underscores their critical role in plant health and adaptation to waterlogging stress [5,6]. For instance, waterlogging alters spring wheat root microbiota, depleting beneficial bacteria [7], while enriching Geobacter in soybeans [8] and Pseudomonas in sugar beets [6]. Hypoxia during waterlogging imposed severe impacts on both plants and their microbiota, favoring anaerobic taxa (e.g., Firmicutes, Fusobacteria) [9]. Although the effects of waterlogging on rhizosphere communities have been documented [10], post–waterlogging recovery—particularly whether microbiota could restore functional communities to support plant resilience—remains poorly understood [11]. Waterlogging stress modulates rhizosphere microbiota assembly through root-mediated metabolites, altering plant–microbe interactions. Waterlogging-induced shifts in root metabolites, including mucilage composition, phytohormone fluxes such as strigolactones [12], and anaerobic byproducts (ethanol, aldehydes) [13,14], drive microbial community restructuring [15,16]. Following waterlogging, recovering plants face multiple challenges, including the detoxification of phytotoxins accumulated during inundation, nutrient deficiencies, and increased susceptibility to biotic stress [17,18]. To overcome these challenges, recovering plants rely on root metabolites to recruit specific rhizosphere microbial communities that aid in stress mitigation and recovery [9,15]. Consequently, understanding the responses of the root metabolite–rhizosphere microbiota complex to waterlogging stress and subsequent recovery is essential for elucidating the mechanisms by which waterlogging impacts rhododendrons.
In this study, a pot water control experiment was conducted to investigate the effects of waterlogging and post–waterlogging recovery on 3–year–old R. delavayi seedlings. The responses of root metabolites and rhizosphere microbial communities to waterlogging stress and recovery were analyzed using 16S rRNA gene sequencing (targeting the V4 region) and ultrahigh-performance liquid chromatography–mass spectrometry (UPLC–MS/MS). The objectives of this study were to (i) elucidate the responses of rhizosphere microbiotas and root metabolites in rhododendrons to waterlogging stress; (ii) assess whether rhizosphere microbiotas and root metabolites return to their pre-waterlogging state during the recovery phase; and (iii) explore how Rhododendron root metabolites unite specific rhizosphere microorganisms to enhance resistance to waterlogging stress. Insights into the roles of microorganisms and root metabolites under waterlogging stress may provide a valuable reference for understanding the mechanisms underlying the effects of waterlogging on rhododendrons from the perspective of plant–microbe interactions.
2. Result
2.1. Generating Metabolites and Rhizosphere Microbiotas for R. delavayi Root During Waterlogging and Recovery
To survey the metabolic and microbial characteristics of R. delavayi roots during waterlogging and recovery, we collected the root tissues and rhizosphere soil of 21 three-year-old R. delavayi seedlings (Figure 1a). The seedlings of R. delavayi were exposed to waterlogging stress for 0, 10, 20, or 30 days (W0, W10, W20, W30). In the post–waterlogging recovery treatments, waterlogging was performed for 10, 20, or 30 days, followed by an equal recovery period of 10, 20, or 30 days, designated as WR10, WR20, and WR30, respectively. In addition, we performed 16s rRNA sequencing on microbes in the rhizosphere soils (Figure 1b). We used UPLC–MS/MS to determine the contents of metabolites in each root sample (Figure 1c). Through the experimental design, we identified 693 metabolites in the root system of R. delavayi and classified a total of 6808 operational taxonomic units (OTUs) from its rhizosphere microbial community, covering 81 phyla and 687 genera.
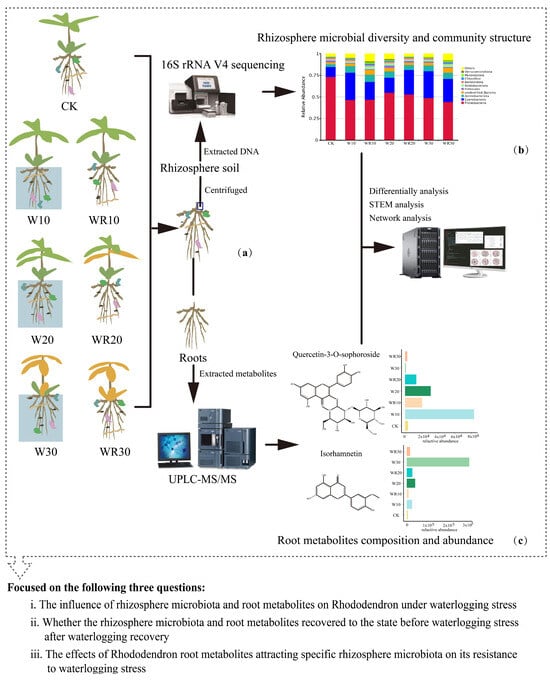
Figure 1.
Overview of the experimental design. (a) 21 samples were retrieved for normal (CK), waterlogging (W), and recovery (WR) on 3-year-old the R. delavayi seedlings. The root of R. delavayi seedling was collected and divided into two parts. (b) The root part was used to analyze the root metabolites by UPLC-MS/MS. (c) The rhizosphere soil was used to analyze the microbiota by sequencing.
2.2. Effects of Waterlogging Stress and Restoration on R. delavayi Seedling Roots
Waterlogging stress significantly reduced fresh root weight (Supplementary Figure S1). This decline persisted during recovery, exacerbated by prolonged prior stress durations. Adventitious roots emerged following 20 days of waterlogging but diminished during recovery beyond 20 days. By WR30, all adventitious roots were absent, with the remaining roots exhibiting darkening and necrosis alongside a pronounced reduction in root biomass. These results demonstrated persistent and irreversible root damage in R. delavayi under extended waterlogging stress and recovery.
2.3. Changes in Root Metabolites During Waterlogging Stress and Recovery Period
Root metabolomic profiling of R. delavayi via UPLC–MS/MS identified 693 metabolites (Supplementary Figure S2), predominantly lipids (17.5%), phenolic acids (17.0%), flavonoids (14.9%), and others (50.6%). Principal component analysis (PCA) revealed a marked divergence between waterlogging and recovery root metabolite profiles (Supplementary Figure S3A). Bray–Curtis dissimilarity was greatest between W0 and W30, exceeding all other group comparisons except W0 vs. WR30 (Supplementary Figure S3B). Both extended waterlogging (30 days) and prolonged recovery (30 days) induced significantly stronger root metabolite changes than shorter durations (10/20 days), demonstrating time-effect metabolic changes.
Using orthogonal partial least squares discriminant analysis (OPLS–DA), a total of 470 differentially accumulated metabolites (DAMs) were identified across waterlogging–recovery phases (Supplementary Figure S4A). Venn analysis showed that less than 30% of the DAMs reversed their changes within the recovery period after experiencing waterlogging (Supplementary Figure S4B). Temporal profiling via Short Time series Expression Miner (STEM) revealed five significant expression patterns encompassing 97 DAMs (Supplementary Figure S4C), which were predominantly enriched in flavonoid biosynthesis, purine metabolism, linoleic acid metabolism, and alpha-linolenic acid metabolism. A total of 22 flavonoid metabolites were identified in the metabolome of R. delavayi roots under waterlogging stress and recovery, including 13 DAMs (Supplementary Figure S4D). Most of these differential flavonoid metabolites exhibited significant upregulation during the waterlogging stress period, such as Quercetin, Hesperetin, and Phloretin.
Through a comprehensive literature review of metabolites, we identified 48 out of the 97 DAMs that played a critical role in plant resistance to adverse conditions (Supplementary Table S2). Overall, the functions of these 48 DAMs were categorized into four main groups: (1) scavenging reactive oxygen species (ROS) and maintaining redox balance (e.g., L–Proline, α–Linolenic Acid, Quercetin), (2) antioxidation (e.g., Protocatechualdehyde, Phloretin, Dihydrokaempferol), (3) antifungal, antibacterial, and antiviral effects (e.g., Hesperetin, 9,10,13–Trihydroxy–11–Octadecenoic Acid), and (4) regulation of root growth (e.g., Isorhamnetin, Quercetin–3–O–sophoroside). These results suggested that the root metabolism of R. delavayi actively participated in both waterlogging stress and recovery.
2.4. Effects of Waterlogging and Post–Waterlogging Recovery on Rhizosphere Microbial Diversity and Community Structure
Utilizing sequencing of the V4 region of the 16S rRNA gene and subsequent analysis, a total of 6808 OTUs were classified in the rhizosphere microbiotas of R. delavayi under waterlogging stress and restoration. Only 17% (998/6068) constituted the consistently presenting microbiotas across all treatments (Supplementary Figure S5A). Waterlogging induced pronounced community turnover, retaining merely 40.5% of W0 OTUs (1265/3124) during stress and 40.5% (1267) post-recovery. Likelihood ratio testing identified 363 and 376 differentially abundant OTUs during waterlogging and recovery, respectively (Supplementary Table S3). These results revealed substantial rhizosphere microbiome restructuring in R. delavayi under waterlogging–recovery cycles. Notably, during waterlogging and the subsequent recovery period, a large number of OTUs in the rhizosphere exhibit decreased or increased abundance. Then, α-diversity analyses revealed a time-dependent shift in the microbial community of the R. delavayi rhizosphere under waterlogging–recovery cycles. The recovery phases (WR10, WR30) exhibited elevated Shannon diversity (p < 0.05) compared to W0, while waterlogging duration differentially impacted richness: PD whole tree and Chao1 indices peaked at W20 before declining at W30 (Supplementary Table S4). Non–Metric Multidimensional Scaling (NMDS) ordination of Bray–Curtis distances demonstrated a marked divergence between waterlogged and recovery communities (PERMANOVA R2 = 0.32, p = 0.001) (Supplementary Figure S5B). Unweighted UniFrac distances showed maximal structural shifts between the W0–W30 (waterlogging climax) and W20–WR20 (recovery transition) groups (Supplementary Figure S5C).
The 6808 OTUs were mostly divided into 81 Phyla (Supplementary Table S5). The highest relative abundances at the phylum level were for Proteobacteria (44.38~73.29%), followed by Cyanobacteria (11.29~31.50%) and Actinobacteriota (3.29~8.31%). Following the waterlogging and post–waterlogging recovery of the rhizosphere microbiota, there was a notable reduction in the relative abundances of Proteobacteria, while Cyanobacteria and Actinobacteriota exhibited marked increases. Interestingly, there was a substantial number of unidentified_bacteria (2.23~5.68%), and their abundance shows a significant increase following waterlogging and subsequent recovery. Moreover, to evaluate variations in the rhizosphere microbiota during waterlogging and post–waterlogging recovery, we evaluated differentially abundant genera in the waterlogging and recovery groups. A total of 687 genera were annotated in the R. delavayi rhizosphere microbiota, of which 187 genera were affected by waterlogging stress and restoration (Supplementary Figure S5D). Most genera (115) were significantly affected by waterlogging stress. Thirty-three genera only significantly changed during the post-waterlogging recovery period but experienced no significant changes during waterlogging. All of the results suggested that waterlogging and subsequent recovery exert substantial influence on rhizosphere microbiotas.
2.5. Effects of Waterlogging and Post–Waterlogging Recovery on Rhizosphere Microbial Co–Occurrence Network Complexity and Core Microbes
To evaluate the influence of waterlogging and the subsequent recovery period on the complexity of rhizosphere microbial co-occurrence networks, six distinct networks were constructed. We calculated various network topological parameters, including the total number of nodes, number of links, network diameter, relative modularity (RM), average clustering coefficient (average CC), and average degree. Analysis of network topological parameters indicated that the rhizosphere microbial co-occurrence networks manifested divergent trends in response to differing waterlogging durations and recovery periods (Figure 2a, Table 1). Prolonged waterlogging progressively increased the complexity and density of the overall structure of the co-occurrence network (nodes, links, average degree, and diameter increased from W10 to W30), with elevated modularity (RM increased) and reduced clustering (average CC reduced), indicating a transition toward compartmentalized communities. Intriguingly, the network structure within post–waterlogging recovery exhibited phase–dependent variation. During a shorter post-waterlogging recovery period (10 days), the network structure underwent simplification and loss (nodes, links, and average degree reduced, but diameter increased), concurrent with the decline in compartmentalization (RM and average CC reduced). Conversely, with prolonged waterlogging and subsequent restoration (20 or 30 days), the structure of the co-occurrence network increased again in complexity and density, with a higher degree of compartmentalization. Furthermore, we evaluated the robustness (resistance to node loss), cohesion, and negative co-occurrence ratio of rhizosphere microbial co-occurrence networks to examine how different durations of waterlogging and subsequent recovery periods affected network interaction and stability (Figure 2b). The robustness of waterlogging groups increased with stress duration, whereas recovery phases exhibited temporal heterogeneity. The 10 d and 30 d recovery groups showed reduced robustness compared to their corresponding waterlogging groups, while the 20 d recovery group displayed higher robustness than its waterlogging counterpart. In addition, we assessed the cohesion to measure the microbial interactions [19]. By analyzing the positive cohesion and negative cohesion, we found that the microbial interactions in the 10 d and 30 d waterlogging groups were higher than in their corresponding post–waterlogging recovery groups. Interestingly, the 20 d group exhibited the opposite trend. Notably, although robustness and cohesion exhibited fluctuations, these variations did not reach statistical significance. Under disturbances, the presence of negative interactions enhanced network stability [20]. Although the negative co-occurrence ratio was higher in the 10 d waterlogging group, it remained notably low across all networks following both waterlogging and recovery periods (ratio < 2.0%). The effects of waterlogging and subsequent recovery on microbial interactions and network stability were limited.
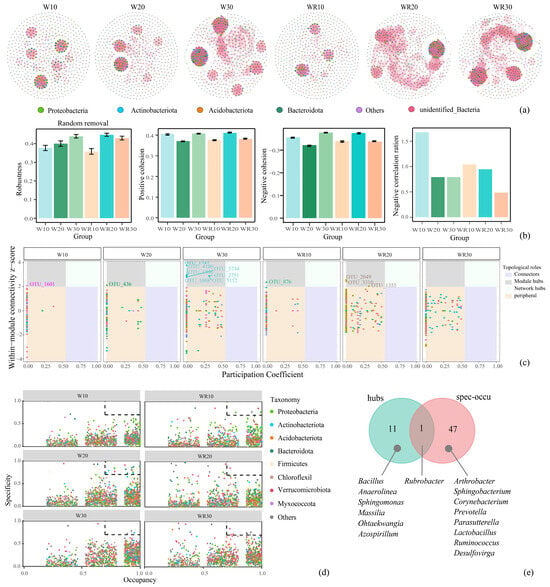
Figure 2.
Rhizosphere microbial correlation networks and core microbiotas with different waterlogging and recovery periods. (a) Visualization of constructed networks in 6 different waterlogging and recovery period treatments. (b) The robustness, positive and negative cohesion, and negative correlation ratio of the networks. (c) Hub microbiotas with 6 rhizosphere microbial correlation networks. (We identified module hubs (Zi ≥ 2.0, Pi < 0.55), connectors (Zi < 2.0, Pi ≥ 0.55), and network hubs (Zi ≥ 2.0, Pi ≥ 0.55), which are referred to as keystone nodes). (d) The SPEC-OCCU plots of 6 different waterlogging and recovery period treatment groups. (e) Venn analysis between the hub microbiotas and specialist microbiotas.

Table 1.
Topological properties of rhizosphere bacterial networks.
Native core microbiomes associated with the plant rhizoplane hold the key to harnessing the full potential of soil microbiomes for supporting plant growth [21]. Here, two methods were used to jointly define the core microbes. Firstly, by calculating the within-module connectivity (Zi) and among-module connectivity (Pi), network hub microbiotas were obtained. The W10 network contained one module hub microbiotas, while W20, W30, WR10, and WR20 harbored one, seven, one, and three hub microbiota, respectively (Figure 2c). Secondly, to identify specialist microbiotas associated with each treatment, we selected OTUs with specificity and occupancy greater than or equal to 0.7 in the specificity–occupancy (SPEC–OCCU) plots. Here, a total of 49 specialist OTUs were found (Figure 2d, Supplementary Table S6). A total of 59 core microbes were found using the two methods, of which only Rubrobacter fulfilled the criteria for hub microbiotas and specialist microbiotas. (Figure 2e).
2.6. Co-Occurrence Between Root Metabolites and Rhizosphere Microbial Communities and Microbial Assembly Processes Under Waterlogging and Post-Waterlogging Recovery
Plants selectively recruit specific rhizosphere microbial communities through root metabolites, thereby establishing plant–rhizosphere microbial complexes that synergistically enhance plant resistance to stressors [22,23]. Root metabolites were significantly correlated with the composition of the rhizosphere microbial community under waterlogging and post–waterlogging recovery, based on a Mantel test (p = 0.021). We propose that root metabolites during waterlogging and recovery correlate with dynamic shifts in the rhizosphere microbiota. Hence, we investigated the potential driving role of metabolites in the microbial community assembly process under these conditions. By calculating the β–nearest taxon index (βNTI) between sample pairs, we found that deterministic and stochastic assembly processes were observed across all the rhizosphere bacterial communities of waterlogging and post–waterlogging recovery (W10, W30, WR10 of βNTI > 2, W20, WR20, WR30 of |βNTI| < 2) (Figure 3a). Furthermore, dispersal limitation (RCbray > 0.95) was found to be dominant among the stochastic processes, and deterministic processes were dominated by homogeneous selection. Finally, correlation analyses integrated βNTI with 48 DAMs identified through OPLS–DA, STEM clustering, and literature validation. A total of 16 metabolites (including L–valine, L–citrulline, N–glycyl–L–leucine, etc.) demonstrated significant associations with microbial community assembly processes (Figure 3b). To explore the root metabolite–rhizomicrobiota response patterns under waterlogging stress and post–waterlogging recovery, cross–domain network pipeline analysis was conducted between 48 different metabolites screened above and the rhizosphere microbiota of the network. It was found that 24 metabolites were significantly correlated with 57 microorganisms (Figure 3c). Quercetin–3–O–sophoroside, L–Citrulline, L–Valine, N–Glycyl–L–leucine, Guanine, and L–Glycyl–L–phenylalanine exhibited significant co–occurrences with a greater number of rhizosphere microbiotas, most of which were positive.
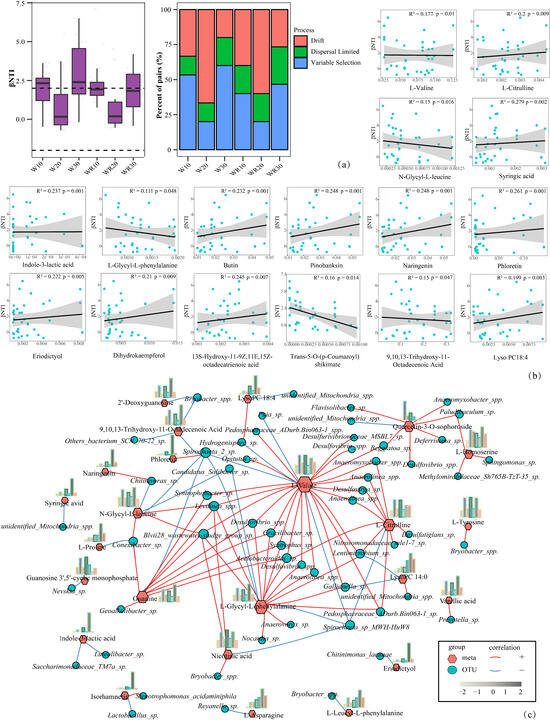
Figure 3.
Correlation analysis of root metabolites with rhizosphere microbial community and microbial assembly processes under waterlogging and post-waterlogging recovery. (a) Contributions of deterministic and stochastic processes to the aggregation of rhizosphere microbial communities were analyzed by βNTI, and each community assembly process was defined by the percentage of site pairs. (b) Scatter plots illustrating the correlation between βNTI and the relative abundance of the 16 root differential metabolites. Adjusted R2 and p values from linear regressions are shown. (c) Correlation network of metabolite production and microbial community of R. delavayi root under waterlogging and post-waterlogging recovery. (The bar chart illustrated the variation of expression for corresponding metabolites). From left to right, each bar represented the following groups: CK, W10, WR10, W20, WR20, W30, and WR30).
3. Discussion
3.1. Effects of Waterlogging and Post–Waterlogging Recovery on Root Metabolites and Rhizosphere Microbes
Global climate change has increased waterlogging frequency over six decades, with partial/complete inundation damaging most terrestrial plants. While plant acclimation to waterlogging is well studied, post-stress recovery mechanisms remain underexplored [24]. Although rhizosphere microbiome involvement in waterlogging resistance is established [9], its role during recovery remains unclear. Prolonged waterlogging caused progressive root damage in R. delavayi, with incomplete restoration observed post-recovery. Root metabolite alterations intensified with treatment duration, peaking at 30 days of waterlogging/recovery. Consistent with reported waterlogging-induced microbial shifts [6], our data revealed persistent community restructuring during recovery. Rhizosphere microbial diversity and phylogenetic complexity increased during waterlogging and persisted through recovery. These findings provide further evidence of continuous waterlogging–hypoxia impacts during plant recovery [25,26], emphasizing the critical need to investigate post-stress microbial and metabolic dynamics for enhancing plant resilience.
Waterlogging and subsequent recovery exert substantial influence on the rhizosphere microbiota across taxonomic ranks. Previous studies of rhizosphere microorganisms in sugar beet and maize found that Proteobacteria and Firmicutes were significantly enriched under waterlogging treatment [6,27]. However, another research on spring wheat (Triticum aestivum) showed that the proportion of Actinobacteria and Proteobacteria decreased [7]. We also found a notable reduction in Proteobacteria abundance in the rhizosphere of R. delavayi. At the same time, Cyanobacteria and Actinobacteriota exhibited a marked increase. We think that this large variation may be related to differences in the soil in which plants grow. Soil type could also significantly influence microbial community structure [8]. Compared with the neutral soil of sugar beet and maize, the planting soil of R. delavayi has higher acidity (pH 4.8 ± 0.3). Some studies have suggested that waterlogging is less harmful to neutral soils than acidic soils, at least on the level of microbial functions [8]. The variability in rhizosphere microbial responses, attributed to the dissimilarities of soil and plants, suggests that the current research on the interaction between plants and rhizosphere microorganisms for resisting stress is still profoundly inadequate. Moreover, there was a substantial number of unidentified bacteria in the rhizosphere of R. delavayi, and their abundance shows a significant increase following waterlogging and subsequent recovery. These indicated that there are still many unknown bacteria in the rhizosphere microbes of R. delavayi that we need to isolate and study, which may bring us more challenges. Many studies have reported that host plants recruit rhizosphere beneficial bacteria to promote plant growth under various stresses, such as waterlogging, high salt, etc. [28,29]. Through database comparison and identification, 44 potentially rhizosphere-beneficial bacteria were identified from the 187 genera of rhizosphere microbiota affected by waterlogging stress and subsequent recovery (Supplementary Table S7) [30]. Nearly all potentially rhizosphere-beneficial bacteria demonstrated varying degrees of enrichment under distinct waterlogging and recovery periods.
3.2. Effects of Waterlogging and Post–Waterlogging Recovery on Core Microbes
Core microbes constitute an essential and interrelated group within the rhizosphere microbiome community, where changes in their presence and abundance can induce significant variations in microbiome structure and function [31,32]. A total of 59 core microbes were found using two methods, in which only Rubrobacter fulfilled the criteria for hub microbiotas and specialist microbiotas. (Figure 2e). Rubrobacter was present during the plant developmental processes and played a crucial role in promoting plant growth and nutrient cycling [33,34]. Interestingly, we found that the hub microbiotas were almost all potentially beneficial to plant growth or nutrient cycling. The genus Bacillus has long been recognized for its beneficial interactions with plants, enhancing growth, nutrient uptake, and stress resistance [35]. The genera Anaerolinea and Ohtaekwangia played a significant role in soil carbon and nitrogen cycles and promoted plant growth [36,37,38]. The genus Sphingomonas produced highly beneficial phytohormones and improved plant growth under stress conditions [39]. Massilia genera had rock phosphate solubilizing ability, improved plant phosphorus nutrition status, and driven plant growth [40,41]. Azospirillum genera was a nitrogen-fixing bacterium, which promoted plant growth and increased plant tolerance to water stress [42,43]. Some specialist microbiotas have also been shown to promote plant growth or nutrient cycling. The genus Arthrobacter is a nitrogen-fixing bacterium, which promotes plant growth. Sphingobacterium, Rubrobacter, and Lactobacillus are plant growth-promoting rhizobacteria [44]. In addition, we found that many of the specialist microbiotas were anaerobic bacteria or facultative anaerobes, which can adapt to anoxic conditions. Examples include the genera Corynebacterium, Sphingobacterium, Prevotella, Parasutterella, Lactobacillus, Ruminococcus, and Desulfovirga [45,46,47,48,49]. The results indicated that the core microbes in the rhizosphere microbial community under waterlogging and post–waterlogging recovery treatment may exhibit two key characteristics: (i) they are important for promoting plant growth and nutrition cycling; (ii) they are adapted to anoxic conditions. Nevertheless, the way in which the core potentially rhizosphere beneficial bacteria we discovered establish a defense mechanism against waterlogging damage in association with R. delavayi is still unclear. In the future, to address these issues, we can use synthetic communities to explore the extent to which they contribute to R. delavayi resistance to waterlogging stress [50,51].
3.3. Root Metabolites Assembled Rhizosphere Microbial Community to Resist Stress for Plant
Current research indicates that root metabolites are correlated with rhizosphere microbiota succession and might be particularly influential under stress conditions [52,53]. Plant roots under stress release metabolic signals to recruit rhizosphere microorganisms, which promote plant growth and contribute to stress resistance [29,54]. In our studies, the differentially accumulated metabolites were significantly correlated with the composition and assembly process of R. delavayi rhizosphere microbial communities under waterlogging and post–waterlogging recovery. Meanwhile, based on the existing literature on root metabolite function and rhizosphere beneficial bacteria, we found several interesting co–occurrences between metabolites and rhizosphere microbiota. L–Homoserine exhibited a significant positive co-occurrence with the beneficial bacterium Sphingomonas sp., which synthesizes highly advantageous plant hormones under adverse conditions, thereby promoting plant growth. The plant growth–promoting bacterium Bryobacter sp. correlated with Nicotinic acid, 9,10,13–Trihydroxy–11–Octadecenoic Acid, 2′–Deoxyguanosine, L–Leucyl–L–phenylalanine and L–Tyrosine. Nocardia sp. secreted Indoleacetic acid (IAA) and siderophores to promote plant growth, which correlated with L–Glycyl–L–phenylalanine. Lactobacillus sp. and Stenotrophomonas acidaminiphila demonstrated the ability to fix nitrogen, increase phosphorus, and increase potassium. Isorhamnetin was likely to concentrate Lactobacillus sp. and Stenotrophomonas acidaminiphila together to regulate the growth of both primary and lateral roots. Additionally, Quercetin–3–O–sophoroside, which similarly regulated the growth of both primary and lateral roots, enhanced the abundance of the plant growth–promoting bacterium Flavisolibacter sp. [30]. Hence, we suggested that enriching these metabolites may play a significant role in rhizosphere microbial community assembly and plant resistance to stress.
3.4. The Response Patterns of Root Metabolite–Rhizomicrobiota in R. delavayi Under Waterlogging Stress and Post-Waterlogging Recovery
This study described the responses of root metabolites and rhizosphere microorganisms to waterlogging stress and post–waterlogging recovery. By combining our results, we attempt to describe the possible response patterns of root metabolites and rhizosphere microbiota complexes in R. delavayi under waterlogging stress and recovery (Figure 4). Three response patterns were identified: (1) The effects of waterlogging stress on R. delavayi persisted throughout the corresponding recovery period, as evidenced by the root morphology and the composition of root metabolites and rhizosphere microorganisms. (2) When R. delavayi was subjected to waterlogging stress, the abundances of numerous root metabolites and rhizosphere microorganisms changed significantly. Some metabolites associated with both biotic and abiotic stress responses were enriched. The enrichment of these metabolites may be a common response of R. delavayi to waterlogging stresses. There are many potentially beneficial bacteria in the rhizosphere microbiota of R. delavayi, which may contribute to waterlogging stress resistance. However, the enrichment degree of potentially beneficial bacteria differed across different waterlogging and recovery periods. Core microbes play a pivotal role in organizing the assembly and function of plant-associated microbial communities [31]. Notably, most of the core microbiotas of R. delavayi responding to waterlogging stress were potentially beneficial bacteria. (3) The waterlogged R. delavayi assembled specific rhizosphere microbiotas by means of root metabolites resistant to the biological or abiotic stresses of host plants, within which some beneficial bacteria in the rhizosphere were recruited.
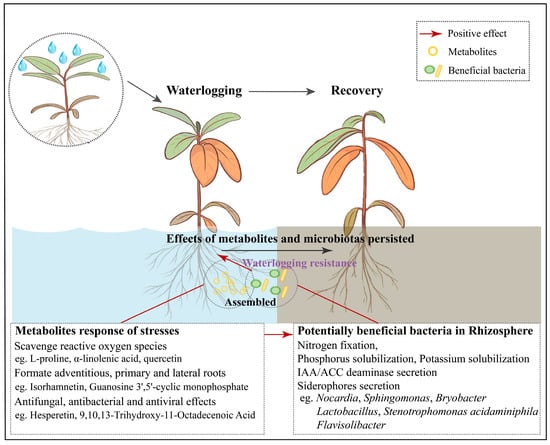
Figure 4.
The response patterns of root metabolites and rhizosphere microbiota complex in R. delavayi under waterlogging stress and post-waterlogging recovery.
Nonetheless, to propose more detailed mechanisms underlying rhizosphere responses to waterlogging stress in the future, it is important to recognize that the patterns identified in this study require further experimental validation and a more comprehensive explanation of several key aspects. First, the enrichment of synergistic beneficial microbiota by specific root metabolites needs to be experimentally confirmed. Future studies should also focus on elucidating the succession dynamics of these metabolites and microorganisms during the post–waterlogging recovery phase. Finally, to deepen our understanding of the roles of root metabolites and rhizosphere microorganisms in the waterlogging resistance of R. delavayi, further investigations should incorporate more detailed time-series analyses.
4. Materials and Methods
4.1. Plant Material and Pot Water Control Treatment
Three-year-old R. delavayi seedlings were selected for the pot waterlogging experiment, which included two treatments: waterlogging and recovery. These seedlings were cultured in plastic flowerpots (cylinders of diameter 8 cm and height 15 cm) with wild, healthy rhododendron soil from the Baili Rhododendron area. The soil pH was 4.8 ± 0.3, characteristic of acidic conditions. The culture conditions in a greenhouse were a photoperiod of 16 h/8 h, a temperature of 22 °C, a light intensity of 400 µmol m−2s−1, and a relative humidity of 60–70%. The plastic flowerpots were placed in trays (length × width × height, 55 cm × 30 cm × 4.5 cm); after that, the R. delavayi seedlings were continuously watered until the water level in the trays reached 4 cm. In the waterlogging treatment groups, daily irrigation maintained a waterlogging depth of 4 cm, with stress durations of 0 (W0), 10 (W10), 20 (W20), and 30 (W30) days. After the waterlogging stress period, water in the trays was removed, and the seedlings were allowed to recover for durations corresponding to their respective waterlogging times: 10 days of waterlogging followed by 10 days of recovery (WR10), 20 days of waterlogging followed by 20 days of recovery (WR20), and 30 days of waterlogging followed by 30 days of recovery (WR30). The soil was normally irrigated in the later part of the restoration process. Three biological replicates were included for each treatment.
4.2. Analysis of Root Metabolites by UPLC–MS/MS
Using a mixer mill (MM 400, Retsch, Haan, Germany), the freeze-dried root sample was crushed with a zirconia bead for 1.5 min at 30 Hz. The 100 mg powder sample was then weighed and dissolved in 1.2 mL of 70% methanol solution, followed by vortexing for 30 s every 30 min for 6 times in total. Subsequently, the samples were placed overnight at a temperature of 4 °C. Before UPLC–MS/MS analysis, the sample extracts were filtered (SCAA–104, 0.22 μm pore size, ANPEL, Shanghai, China) after centrifugation at a speed of 12,000× g rpm for a duration of 10 min. The sample extracts were analyzed using a UPLC–ESI–MS/MS system (UPLC, SHIMADZU Nexera X2, Shimadzu, Japan; MS, Applied Biosystems 4500 Q TRAP, Foster City, CA, USA). The UPLC conditions were as follows: Column, Agilent SB–C18 (1.8 µm, 2.1 mm × 100 mm). Mobile Phase, solvent A: pure water with 0.1% formic acid; solvent B: acetonitrile with 0.1% formic acid. Sample Measurements: The starting conditions were 95% A and 5% B. A linear gradient to 5% A and 95% B was applied over 9 min, and 5% A and 95% B were kept for 1 min. Subsequently, 95% A and 5.0% B were adjusted within 1.10 min and kept for 2.9 min. Flow velocity: 0.35 mL per minute. Column Oven, 40 °C. Injection Volume, 4 μL. The effluent was alternatively connected to an ESI–triple quadrupole–linear ion trap (QTRAP)–MS. The ESI source was operated in turbo spray mode with the following parameters: source temperature 550 °C, ion spray voltage ±5500 V (positive/negative modes), gas flows (GS1 50 psi, GS2 60 psi, curtain gas 25 psi), and high collision–activated dissociation. System calibration used 10 μmol/L (QQQ mode) and 100 μmol/L (LIT mode) polypropylene glycol solutions. MRM acquisitions employed nitrogen collision gas at medium pressure. Qualitative analysis of the secondary mass spectrometry data was performed using a self–built database, MWDB (v2.0), and the quantitative analysis of the metabolites was performed using the multiple reaction monitoring mode. After operation in positive– and negative–ion modes, the raw data were controlled and processed by Analyst 1.6.3 software (AB Sciex, Framingham, MA, USA) [55]. Chromatographic peak areas (Area) reflect the relative abundance of corresponding metabolites. To enable cross–sample comparison of metabolite abundance while ensuring accurate annotation and quantification, we implemented retention time–based alignment and chromatographic peak shape validation across all detected features.
4.3. Microbial DNA Extraction and 16S rRNA Gene V4 Region Sequencing
5 g root samples were first vortexed with 5 mL of phosphate–buffered saline (PBS) solution (pH 7.2) and then centrifuged (5 min at 3000× g) to collect all the sediment from the root surfaces, which was treated as the rhizosphere microbial samples. Total genome DNA from the samples was extracted using the MOBIO PowerSoil ® DNA Isolation Kit (12888–50, Carlsbad, CA, USA). A sample was extracted three times and mixed before being dissolved in 60 μL TE buffer. Then we quantified the DNA ND1000 and stored it at –80 °C. The V4 region of the 16S rRNA gene was amplified using the 20~30 ng DNA from each sample and then sequenced using the Illumina NovaSeq platform. The PCR primers were 515F′–‘GTGCCAGCMGCCGCGGTAA,’ and 806R′–‘GGACTACHVGGGTWTCTAAT’. PCR reactions were carried out with 15 µL of Phusion® High–Fidelity PCR Master Mix (New England Biolabs), 2 µM of forward and reverse primers, and about 10 ng template DNA. Sequencing libraries were generated using the TruSeq® DNA PCR–Free Sample Preparation Kit (Illumina, San Diego, CA, USA) following the manufacturer’s recommendations, and index codes were added.
4.4. Sequencing Data Analysis
Quality filtering on the raw tags was performed to obtain the high–quality clean tags according to QIIME. Quality filtering parameters included maximum expected errors per read (2.0), removing low–quality regions (≤19), and removing the chimeric sequences. Using the VSERCH algorithm to detect chimera sequences, the Effective Tags were finally obtained. Tags with ≥97% similarity were assigned to the same OTUs using the UPARSE method. In total, 1,386,614 high–quality 16S rRNA V4 amplicons for 21 rhizosphere samples were obtained and analyzed (Supplementary Table S1). Then, using the Silva Database based on the USEARCH algorithm, OTUs were annotated with taxonomic information [56]. OTU abundance information was normalized using a standard of sequence number corresponding to the sample with the least sequences.
4.5. Network Construction and Characterization
The OTUs were screened for significant clusters using STEM. OTUs that could not be annotated to a genus or had an abundance of less than 10 were excluded [57]. A total of 634 OTUs were screened. Based on OTU abundance, a microbial co–occurrence network was established using Pearson co–occurrence coefficients (r) exceeding 0.7 or falling below −0.7, in conjunction with false discovery rate-adjusted p-values of less than 0.05 [58]. The co-occurrence networks were visualized with the interactive R package v4.2.2 ggClusterNet [59]. To estimate the complexity of networks, we calculated the characterization of the networks by R package igraph, including total number of nodes, number of links, network diameter, relative modularity (RM), average clustering coefficient (Average CC), and average degree [60]. Robustness is defined as the proportion of the remaining species in this network after random removal, in which 50% of the nodes were randomly removed. Cohesion can be used as an indicator to measure microbial interactions [44]. The robustness, cohesion, and negative co–occurrence ratio were calculated and visualized by the R package ggClusterNet. In this study, two different methods were used to jointly define core microbes. First, potential specialist microbiotas were identified by SPEC–OCCU plots [61]. Second, according to within–module connectivity (Zi) and among–module connectivity (Pi), hub microbiotas were identified [32]. Cytoscape 3.7.0 was utilized to visualize and clarify the network of root metabolites and microbiotas, which was established based on Spearman’s co–occurrence coefficients (r > 0.7 or r < −0.7, p < 0.05).
4.6. Analysis of the Microbial Community Assembly Processes and the Effect of Root Metabolites on Community Assembly
By calculating the βNTI among samples, the phylogenetic pattern of the rhizosphere microbial community is assessed by employing the null model approach [62]. Sample pairs with |βNTI| > 2 were considered to result from a deterministic process, while |βNTI| < 2 indicated that the selective pressure was relatively weak, suggesting that community assembly may be under the control of a stochastic process [63]. The values obtained from the βNTI analysis reflected the driving force of the relevant factors. For the computation of RCbray, the number of simulated communities with a Bray–Curtis dissimilarity exceeding the observed value was aggregated with half of the number of simulated communities with dissimilarity equal to the observed value, and the sum was subsequently divided by the total number of simulations (999). By combining the results of βNTI and RCbray, the relative contributions of deterministic and stochastic processes to overall community assembly within each sample were determined. All analyses of microbial community assembly processes and the effect of root metabolites on community assembly were carried out within the R “stats”, “minpack.lm”, “Hmisc”, and “picante” packages [52].
4.7. Statistical Analyses
DAMs were identified using OPLS–DA in the R package MetaboAnalystR (VIP > 1, |log2Fold change| ≥ 1) [64], with permutation testing (200 iterations) to validate model robustness. Enriched metabolic pathways were derived via metabolite set enrichment analysis using hypergeometric tests (p < 0.05). Root metabolite structures were visualized by PCA of Bray–Curtis distances (vegan package). For rhizosphere microbiota, α– and β–diversity indices were computed in QIIME and visualized in R. Community structure differences were assessed via Non–Metric Multidimensional Scaling (NMDS; Bray–Curtis) and permutational multivariate analysis of variance (PERMANOVA) by the vegan package. Taxonomic differentials between groups were evaluated using likelihood ratio tests (p < 0.05). The co–occurrence between microbial community and root metabolites was tested via Mantel tests (vegan; Bray–Curtis vs. Euclidean matrices, 999 permutations). Analyses were conducted in R version v4.2.2 with vegan v2.6-6.
5. Conclusions
This study provides an analysis of root metabolites and rhizosphere microbiota in the rhizosphere soil of R. delavayi under waterlogging stress and post–waterlogging recovery. Our findings revealed significant alterations in the root metabolites, rhizosphere microbiota, and microbial co–occurrence network during waterlogging stress, with these changes persisting throughout the recovery period. Moreover, we found that the root metabolites are closely related to the composition and assembly process of rhizosphere microorganisms. Notably, certain root metabolites were found to enrich potentially beneficial rhizosphere microbiota, which collectively played a role in mitigating waterlogging stress. For example, the addition of L-homoserine and the beneficial bacteria Sphingomonas sp. may synthesize highly advantageous plant hormones to promote plant growth and resist waterlogging stress. However, further detailed time–series studies are needed to better understand the dynamic interactions between root metabolites, rhizosphere microbiota, and the duration of waterlogging stress and recovery. Such studies would provide critical insights into the intricate relationships between the root metabolome and rhizosphere microbiome. This knowledge is essential for developing future strategies that integrate “metabolites + rhizosphere microbiota” to enhance the waterlogging resistance of R. delavayi.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae11070770/s1.
Author Contributions
J.T.: Writing—review and editing, Writing—original draft, Visualization, Methodology, Data curation, Conceptualization, Funding acquisition. Q.H.: Writing—original draft, Investigation, Visualization, Methodology, Data curation. Q.W.: Data curation, Investigation, Software. F.S.: Investigation. S.W.: Methodology and execution of experiments. X.Z.: Funding acquisition, Supervision. M.T.: Investigation, Project administration; Y.Y.: Writing—review and editing, Supervision, Project administration, Funding acquisition, Conceptualization. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Guizhou Provincial Natural Science Foundation Project (QKHJC–ZD [2025]046), Science and Technology Support Project of Guizhou Province (QKHZC [2021]YB459), the Natural Science Foundation of China(NSFC) (32260393); Key Laboratory of Environment Friendly Management on Alpine Rhododendron Diseases and Pests of Institutions of Higher Learning in Guizhou Province ([2022]044) and Karst Mountain Ecological Security Engineering Re–search Center, grant number [2021]007.
Data Availability Statement
The datasets presented in this study can be found in NCBI online repositories. The names of the repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/sra, (12-Nov-2024) PRJNA1184993.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflicts of interest.
References
- Ma, H.; Liu, Y.B.; Liu, D.T.; Sun, W.B.; Liu, X.F.; Wan, Y.M.; Zhang, X.J.; Zhang, R.G.; Yun, Q.Z.; Wang, J.H.; et al. Chromosomelevel genome assembly and population genetic analysis of a critically endangered rhododendron provide insights into its conservation. Plant J. 2021, 107, 1533–1545. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.M.; Duan, S.G.; Xia, Y.; Li, J.T.; Liu, L.X.; Tang, M.; Tang, J.; Sun, W.; Yi, Y. Transcriptomic, Physiological, and Metabolomic Response of an Alpine Plant, Rhododendron delavayi, to Waterlogging Stress and PostWaterlogging Recovery. Int. J. Mol. Sci. 2023, 24, 10509. [Google Scholar] [CrossRef]
- Barbara, R.H.; Wiebke, B.; Claudia, S.B.; Mugdha, S.; Thomas, H. Roots Shaping Their Microbiome: Global Hotspots for Microbial Activity. Annu. Rev. Phytopathol. 2015, 53, 403–424. [Google Scholar] [CrossRef]
- Zhou, S.Y.D.; Lie, Z.Y.; Liu, X.J.; Zhu, Y.G.; Peñuelas, J.; Neilson, R.; Su, X.X.; Liu, Z.F.; Chu, G.W.; Meng, Z.; et al. Distinct patterns of soil bacterial and fungal community assemblages in subtropical forest ecosystems under warming. Glob. Change Biol. 2023, 29, 1501–1513. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wang, M.H.; Cui, R.F.; Li, B.C.; Wu, T.; Liu, Y.L.; Geng, G.; Xu, Y.; Wang, Y.G. Waterlogging stress alters the structure of sugar beet rhizosphere microbial community structure and recruiting potentially beneficial bacterial. Ecotoxicol. Environ. Saf. 2023, 262, 115172. [Google Scholar] [CrossRef]
- Francioli, D.; Cid, G.; Kanukollu, S.; Ulrich, A.; Hajirezaei, M.R.; Kolb, S. Waterlogging Causes Dramatic Compositional Shifts and Depletion of Putative Beneficial Bacteria on the Spring Wheat Microbiota. Front. Microbiol. 2021, 12, 773116. [Google Scholar] [CrossRef]
- Yu, T.; Cheng, L.; Liu, Q.; Wang, S.; Zhou, Y.; Zhong, H.; Tang, M.; Nian, H.; Lian, T. Effects of Waterlogging on Soybean Rhizosphere Bacterial Community Using V4, LoopSeq, and PacBio 16S rRNA Sequence. Microbiol. Spectr. 2022, 10, e0201121. [Google Scholar] [CrossRef]
- Martínez-Arias, C.; Witzell, J.; Solla, A.; Martin, J.A.; Rodríguez-Calcerrada, J. Beneficial and pathogenic plant–microbe interactions during waterlogging stress. Plant Cell Environ. 2022, 45, 2875–2897. [Google Scholar] [CrossRef]
- Moche, M.; Gutknecht, J.; Schulz, E.; Langer, U.; Rinklebe, J. Monthly dynamics of microbial community structure and their controlling factors in three floodplain soils. Soil Biol. Biochem. 2015, 90, 169–178. [Google Scholar] [CrossRef]
- Bhaskar, J.P.; Indrani, S.; Niraj, A. Root exudation drives abiotic stress tolerance in plants by recruiting beneficial microbes. Appl. Soil Ecol. 2024, 198, 105351. [Google Scholar] [CrossRef]
- Bouwmeester, H.J.; Fonne-Pfister, R.; Screpanti, C.; De Mesmaeker, A. Strigolactones: Plant Hormones with Promising Features. Angew. Chem. Int. Ed. 2019, 58, 12778–12786. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Lyu, D.G.; Jia, L.T.; He, J.L.; Qin, S.J. Physiological and de novo transcriptome analysis of the fermentation mechanism of Cerasus sachalinensis roots in response to short-term waterlogging. BMC Genom. 2017, 18, 649. [Google Scholar] [CrossRef]
- Song, Y.; Wilson, A.J.; Zhang, X.C.; Thoms, D.; Sohrabi, R.; Song, S.Y.; Geissmann, Q.; Liu, Y.; Walgren, L.; He, S.Y.; et al. FERONIA restricts Pseudomonas in the rhizosphere microbiome via regulation of reactive oxygen species. Nat. Plants 2021, 7, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Vives-Peris, V.; de Ollas, C.; Gómez-Cadenas, A.; Pérez-Clemente, R.M. Root exudates: From plant to rhizosphere and beyond. Plant Cell Rep. 2020, 39, 3–17. [Google Scholar] [CrossRef]
- Savchenko, T.; Rolletschek, H.; Heinzel, N.; Tikhonov, K.; Dehesh, K. Waterlogging tolerance rendered by oxylipin-mediated metabolic reprogramming in Arabidopsis. J. Exp. Bot. 2019, 70, 2919–2932. [Google Scholar] [CrossRef]
- Yeung, E.; Bailey-Serres, J.L.; Sasidharan, R. After The Deluge: Plant Revival Post-Waterlogging. Trends Plant Sci. 2019, 24, 443–454. [Google Scholar] [CrossRef]
- Herren, C.M.; McMahon, K.D. Cohesion: A method for quantifying the connectivity of microbial communities. ISME J. 2017, 11, 2426–2438. [Google Scholar] [CrossRef]
- Coyte, K.Z.; Schluter, J.; Foster, K.R. The ecology of the microbiome: Networks, competition, and stability. Science 2015, 350, 663–666. [Google Scholar] [CrossRef]
- Zhou, Y.Y.; Liu, D.H.; Li, F.Q.; Dong, Y.H.; Jin, Z.L.; Liao, Y.W.K.; Li, X.H.; Peng, S.G.; Delgado-Baquerizo, M.; Li, X.G. Superiority of native soil core microbiomes in supporting plant growth. Nat. Commun 2024, 15, 6599. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.C.; Jiang, T.; Liu, Y.X.; Bai, Y.C.; Reed, J.; Qu, B.Y.; Goossens, A.; Nützmann, H.W.; Bai, Y.; Osbourn, A. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 2019, 364, eaau6389. [Google Scholar] [CrossRef]
- Wang, L.X.; Chen, M.X.; Lam, P.Y.; Dini-Andreote, F.; Dai, L.; Wei, Z. Multifaceted roles of flavonoids mediating plant-microbe interactions. Microbiome 2022, 10, 233. [Google Scholar] [CrossRef]
- Yuan, L.B.; Chen, M.X.; Wang, L.N.; Sasidharan, R.; Voesenek, L.A.C.J.; Xiao, S. Multi-stress resilience in plants recovering from submergence. Plant Biotechnol. J. 2023, 21, 466–481. [Google Scholar] [CrossRef] [PubMed]
- Yeung, E.; van Veen, H.; Vashisht, D.; Sobral Paiva, A.L.; Hummel, M.; Rankenberg, T.; Steffens, B.; Steffen-Heins, A.; Sauter, M.; de Vries, M.; et al. A stress recovery signaling network for enhanced waterlogging tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2018, 115, E6085–E6094. [Google Scholar] [CrossRef] [PubMed]
- Shikov, A.E.; Chirkova, T.V.; Yemelyanov, V.V. Post-Anoxia in Plants: Reasons, Consequences, and Possible Mechanisms. Russ. J. Plant Physiol. 2020, 67, 45–59. [Google Scholar] [CrossRef]
- Bai, H.; He, S.; Qin, T.L.; Yan, D.H.; Weng, B.S.; Zhao, X.X.; Li, X.N.; Bai, Y.J.; Ma, J. Influences of irrigation amount on the rhizospheric microorganism composition and carbon dioxide flux of maize crops. Geoderma 2019, 343, 1–9. [Google Scholar] [CrossRef]
- Etesami, H.; Maheshwari, D.K. Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: Action mechanisms and future prospects. Ecotoxicol. Environ. Saf. 2018, 156, 225–246. [Google Scholar] [CrossRef]
- Yin, J.; Guo, H.; Fry, E.L.; De Long, J.R.; Tang, S.; Yuan, T.; Ren, W. Plant roots send metabolic signals to microbes in response to long-term overgrazing. Sci. Total Environ. 2022, 842, 156241. [Google Scholar] [CrossRef]
- Li, P.F.; Tedersoo, L.; Crowther, T.W.; Dumbrell, A.J.; Dini-Andreote, F.; Bahram, M.; Kuang, L.; Li, T.; Wu, M.; Jiang, Y.J.; et al. Fossil-fuel-dependent scenarios could lead to a significant decline of global plant-beneficial bacteria abundance in soils by 2100. Nat. Food 2023, 4, 996–1006. [Google Scholar] [CrossRef]
- Compant, S.; Cassan, F.; Kostić, T.; Johnson, L.; Brader, G.; Trognitz, F.; Sessitsch, A. Harnessing the plant microbiome for sustainable crop production. Nat. Rev. Microbiol. 2024, 23, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.Z.; Wang, T.T.; Huang, Q.W.; Guo, H.Y.; Zhang, H.; Xu, Q.C.; Shen, Q.R.; Ling, N. Core species impact plant health by enhancing soil microbial cooperation and network complexity during community coalescence. Soil Biol. Biochem. 2024, 188, 109231. [Google Scholar] [CrossRef]
- Trivedi, K.; Kumar, R.; Vijay Anand, K.G.; Bhojani, G.; Kubavat, D.; Ghosh, A. Structural and functional changes in soil bacterial communities by drifting spray application of a commercial red seaweed extract as revealed by metagenomics. Arch. Microbiol. 2021, 204, 72. [Google Scholar] [CrossRef] [PubMed]
- Ajilogba, C.F.; Olanrewaju, O.S.; Babalola, O.O. Plant Growth Stage Drives the Temporal and Spatial Dynamics of the Bacterial Microbiome in the Rhizosphere of Vigna subterranea. Front. Microbiol. 2022, 13, 825377. [Google Scholar] [CrossRef]
- Das, V.A.; Gautam, B.; Yadav, P.K.; Singh, S. Identification of Conserved Pathways in Bacillus Strains Known for Plant Growth-Promoting Behavior Using a Multifaceted Computational Approach. Agriculture 2024, 14, 838. [Google Scholar] [CrossRef]
- Zarraonaindia, I.; Martínez-Goñi, X.S.; Liñero, O.; Muñoz-Colmenero, M.; Aguirre, M.; Abad, D.; Baroja-Careaga, I.; de Diego, A.; Gilbert, J.A.; Estonba, A. Response of Horticultural Soil Microbiota to Different Fertilization Practices. Plants 2020, 9, 1051. [Google Scholar] [CrossRef]
- Li, D.; Zhou, C.R.; Wu, Y.L.; An, Q.S.; Zhang, J.B.; Fang, Y.; Li, J.Q.; Pan, C.P. Nanoselenium integrates soil-pepper plant homeostasis by recruiting rhizosphere-beneficial microbiomes and allocating signaling molecule levels under Cd stress. J. Hazard. Mater. 2022, 432, 128763. [Google Scholar] [CrossRef]
- Chen, J.Y.; Liu, S.; Deng, W.K.; Niu, S.H.; Liao, X.D.; Xiang, L.; Xing, S.C. The effect of manure-borne doxycycline combined with different types of oversized microplastic contamination layers on carbon and nitrogen metabolism in sandy loam. J. Hazard. Mater. 2023, 456, 131612. [Google Scholar] [CrossRef]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef]
- Zheng, B.X.; Bi, Q.F.; Hao, X.L.; Zhou, G.W.; Yang, X.R. Massilia phosphatilytica sp. nov., a phosphate solubilizing bacteria isolated from a long-term fertilized soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 2514–2519. [Google Scholar] [CrossRef]
- Wang, D.N.; He, X.M.; Baer, M.; Lami, K.; Yu, B.G.; Tassinari, A.; Salvi, S.; Schaaf, G.; Hochholdinger, F.; Yu, P. Lateral root enriched Massilia associated with plant flowering in maize. Microbiome 2024, 12, 124. [Google Scholar] [CrossRef]
- Steenhoudt, O.; Vanderleyden, J. Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: Genetic, biochemical and ecological aspects. FEMS Microbiol. Rev. 2000, 24, 487–506. [Google Scholar] [CrossRef] [PubMed]
- Pereyra, M.A.; Zalazar, C.A.; Barassi, C.A. Root phospholipids in Azospirillum-inoculated wheat seedlings exposed to water stress. Plant Physiol. Biochem. 2006, 44, 873–879. [Google Scholar] [CrossRef]
- Wu, H.; Yang, J.J.; Fu, W.; Rillig, M.C.; Cao, Z.J.; Zhao, A.; Hao, Z.P.; Zhang, X.; Chen, B.D.; Han, X.G. Identifying thresholds of nitrogen enrichment for substantial shifts in arbuscular mycorrhizal fungal community metrics in a temperate grassland of northern China. New Phytol. 2023, 237, 279–294. [Google Scholar] [CrossRef]
- Aurilia, V.; Martin, J.C.; McCrae, S.I.; Scott, K.P.; Rincon, M.T.; Flint, H.J. Three multidomain esterases from the cellulolytic rumen anaerobe Ruminococcus flavefaciens 17 that carry divergent dockerin sequences. Microbiology 2000, 146, 1391–1397. [Google Scholar] [CrossRef]
- Tanaka, K.; Stackebrandt, E.; Tohyama, S.; Eguchi, T. Desulfovirga adipica gen. nov., sp. nov., an adipate-degrading, gram-negative, sulfate-reducing bacterium. Int. J. Syst. Evol. Microbiol. 2000, 50, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Morotomi, M.; Nagai, F.; Watanabe, Y. Parasutterella secunda sp. nov., isolated from human faeces and proposal of Sutterellaceae fam. nov. in the order Burkholderiales. Int. J. Syst. Evol. Microbiol. 2011, 61, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Lee, K.; Hsu, M.; Nau, G.; Mylonakis, E.; Ramratnam, B. Lactobacillus-derived extracellular vesicles enhance host immune responses against vancomycin-resistant enterococci. BMC Microbiol. 2017, 17, 66. [Google Scholar] [CrossRef] [PubMed]
- de la Maza, L.M.; Pezzlo, M.T.; Bittencourt, C.E.; Peterson, E.M. Bacteroides, Porphyromonas, Prevotella, Fusobacterium, and Other Anaerobic Gram-Negative Bacteria. In Color Atlas of Medical Bacteriology; American Society for Microbiology: Washington, DC, USA, 2020; pp. 252–260. [Google Scholar] [CrossRef]
- Northen, T.R.; Kleiner, M.; Torres, M.; Kovács, Á.T.; Nicolaisen, M.H.; Krzyżanowska, D.M.; Sharma, S.; Lund, G.; Jelsbak, L.; Baars, O.; et al. Community standards and future opportunities for synthetic communities in plant-microbiota research. Nat. Microbiol. 2024, 9, 2774–2784. [Google Scholar] [CrossRef]
- Vorholt, J.A.; Vogel, C.; Carlström, C.I.; Müller, D.B. Establishing Causality: Opportunities of Synthetic Communities for Plant Microbiome Research. Cell Host Microbe 2017, 22, 142–155. [Google Scholar] [CrossRef]
- Wen, T.; Xie, P.; Penton, C.R.; Hale, L.; Thomashow, L.S.; Yang, S.; Ding, Z.; Su, Y.; Shen, Q.R.; Yuan, J. Specific metabolites drive the deterministic assembly of diseased rhizosphere microbiome through weakening microbial degradation of autotoxin. Microbiome 2022, 10, 177. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.R.; Zhalnina, K.; Yuan, M.T.; Herman, D.; Ceja-Navarro, J.A.; Sasse, J.; Jordan, J.S.; Bowen, B.P.; Wu, L.Y.; Fossum, C.; et al. Nutrient and moisture limitations reveal keystone metabolites linking rhizosphere metabolomes and microbiomes. Proc. Natl. Acad. Sci. USA 2024, 121, e2303439121. [Google Scholar] [CrossRef] [PubMed]
- Rizaludin, M.S.; Stopnisek, N.; Raaijmakers, J.M.; Garbeva, P. The Chemistry of Stress: Understanding the ‘Cry for Help’ of Plant Roots. Metabolites 2021, 11, 357. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.L.; Wang, W.S.; Zhang, H.Y.; Liu, X.Q.; Yu, S.B.; Xiong, L.Z.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.X.; Chen, L.; Ma, T.F.; Li, X.F.; Zheng, M.S.; Zhou, X.; Chen, L.; Qian, X.B.; Xi, J.; Lu, H.Y.; et al. EasyAmplicon: An easy-to-use, open-source, reproducible, and community-based pipeline for amplicon data analysis in microbiome research. IMeta 2023, 2, e83. [Google Scholar] [CrossRef]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef]
- Jiao, S.; Qi, J.J.; Jin, C.J.; Liu, Y.; Wang, Y.; Pan, H.B.; Chen, S.; Liang, C.L.; Peng, Z.H.; Chen, B.B.; et al. Core phylotypes enhance the resistance of soil microbiome to environmental changes to maintain multifunctionality in agricultural ecosystems. Glob. Change Biol. 2022, 28, 6653–6664. [Google Scholar] [CrossRef]
- Wen, T.; Xie, P.H.; Yang, S.D.; Niu, G.Q.; Liu, X.Y.; Ding, Z.X.; Xue, C.; Liu, Y.X.; Shen, Q.; Yuan, J. ggClusterNet: An R package for microbiome network analysis and modularity-based multiple network layouts. IMeta 2022, 1, e32. [Google Scholar] [CrossRef]
- Yuan, M.M.; Guo, X.; Wu, L.W.; Zhang, Y.; Xiao, N.J.; Ning, D.L.; Shi, Z.; Zhou, X.S.; Wu, L.Y.; Yang, Y.F.; et al. Climate warming enhances microbial network complexity and stability. Nat. Clim. Change 2021, 11, 343–348. [Google Scholar] [CrossRef]
- Gweon, H.S.; Bowes, M.J.; Moorhouse, H.L.; Oliver, A.E.; Bailey, M.J.; Acreman, M.C.; Read, D.S. Contrasting community assembly processes structure lotic bacteria metacommunities along the river continuum. Environ. Microbiol. 2021, 23, 484–498. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.J.; Konopka, A.E.; Fredrickson, J.K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012, 6, 1653–1664. [Google Scholar] [CrossRef] [PubMed]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Xia, J.G. MetaboAnalystR: An R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 2018, 34, 4313–4314. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).