Genome Wide Identification of Terpenoid Metabolism Pathway Genes in Chili and Screening of Key Regulatory Genes for Fruit Terpenoid Aroma Components
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Treatment
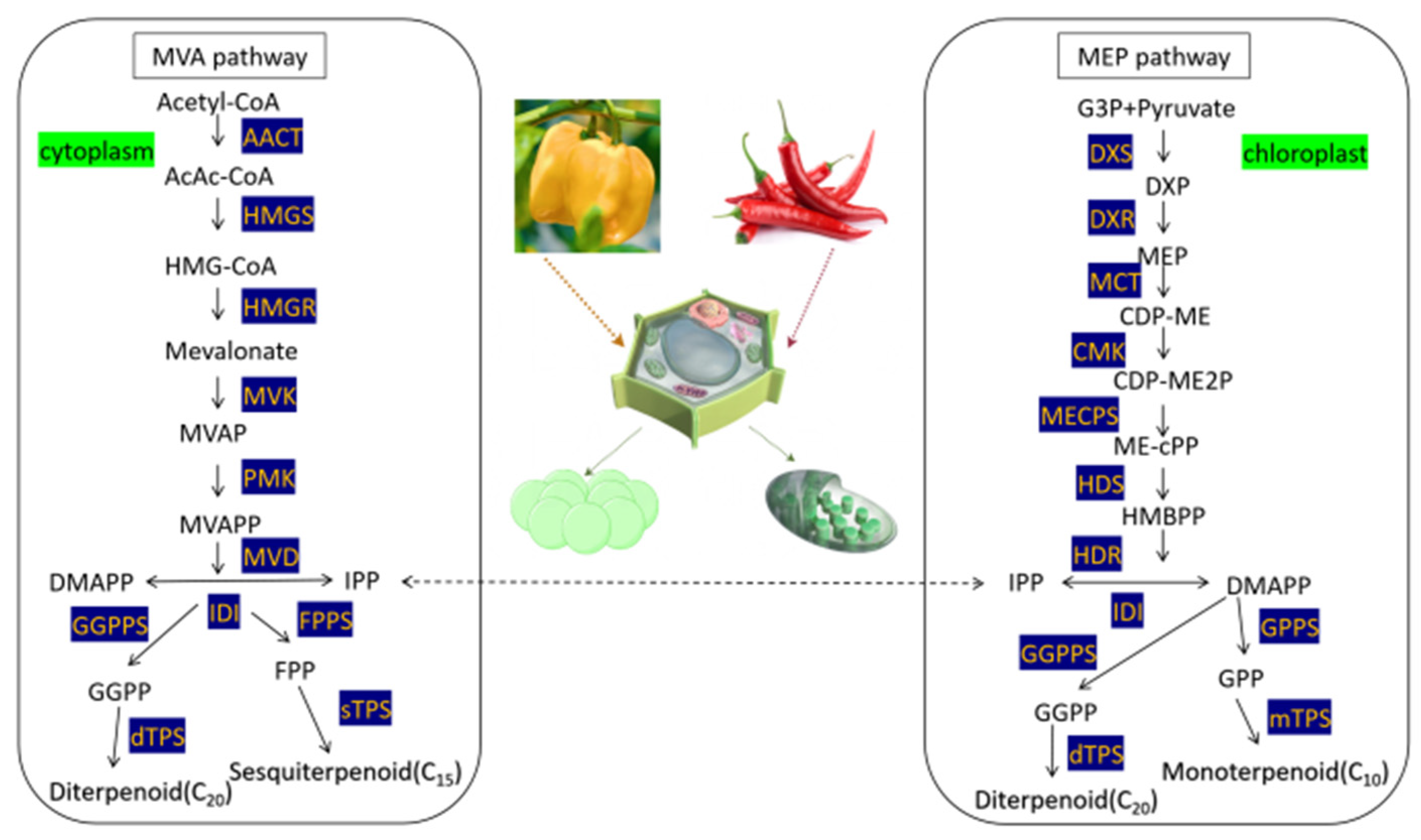
2.2. Selection of Genes Related to Biosynthesis of Terpene Metabolic Pathway
2.3. Sequence Analysis of Genes Related to Terpene Metabolic Pathway Biosynthesis
2.4. Chromosome Mapping, Phylogenetic Tree Analysis, Intron–Exon Analysis, and Promoter Analysis of Related Genes
2.5. GC-MS Experiments and Transcriptome Experiments
2.5.1. Sample Preparation and Extraction
2.5.2. GC-MS Analysis of VACs
2.5.3. Screening of Differential VACs and Marker Aroma Compounds
2.5.4. RNA Sequencing Analysis
2.6. RNA Extraction and qRT-PCR Analysis
2.7. Screening of Enzyme Genes Related to Terpenoid Metabolic Pathway in C. annuum
2.8. Statistical Analysis
3. Results
3.1. Physicochemical Properties of Terpene Metabolic Pathway Gene Proteins
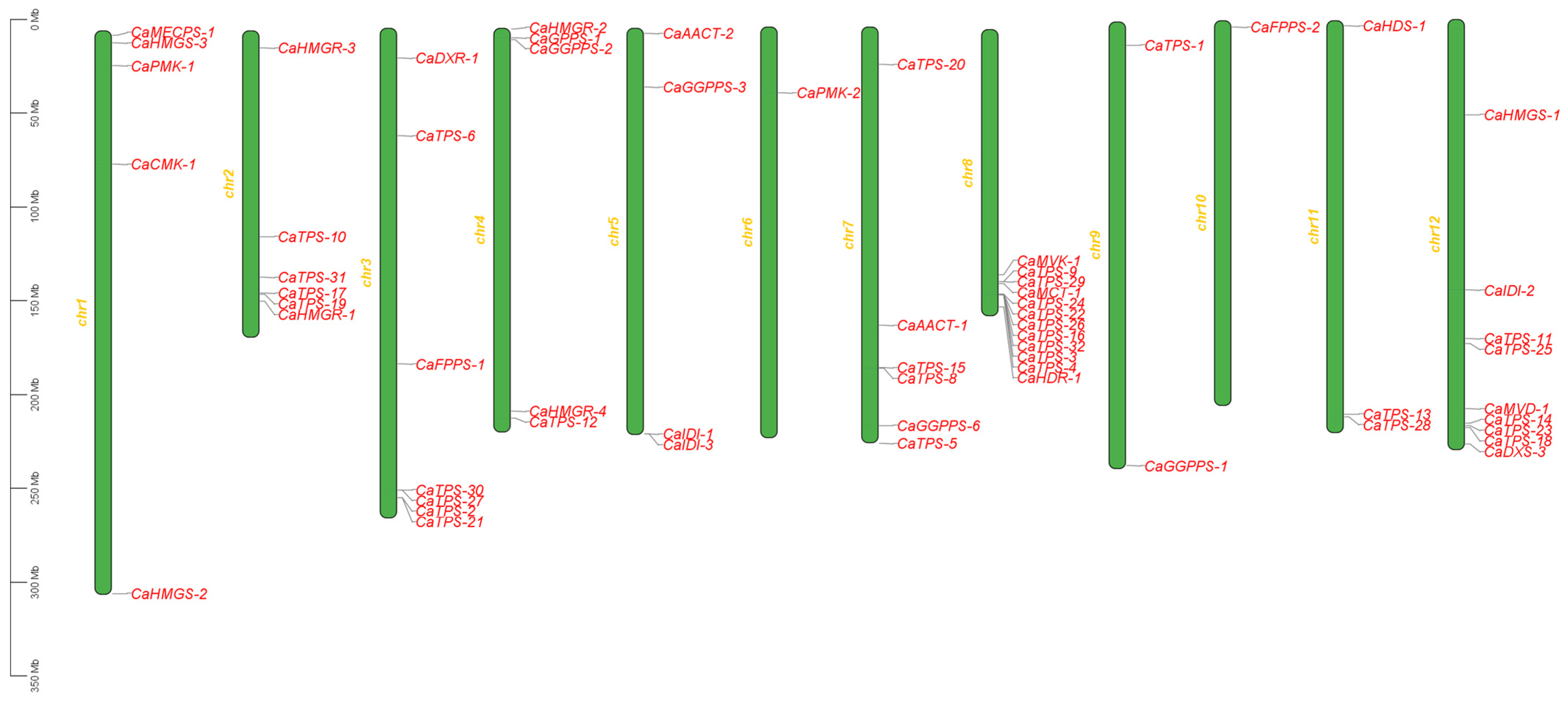
3.2. Chromosomal Localization Analysis of Terpene Metabolic Pathway Genes
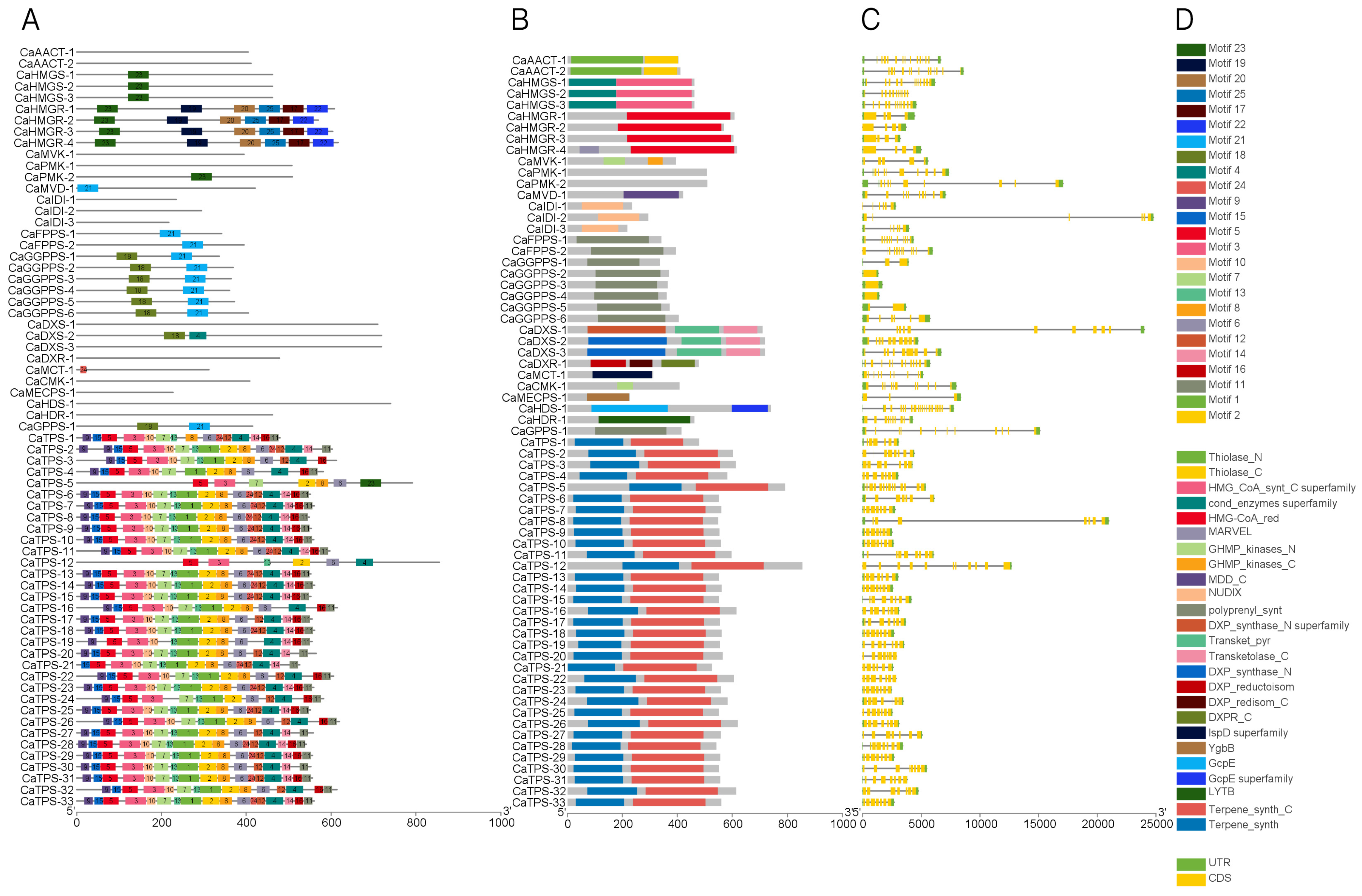
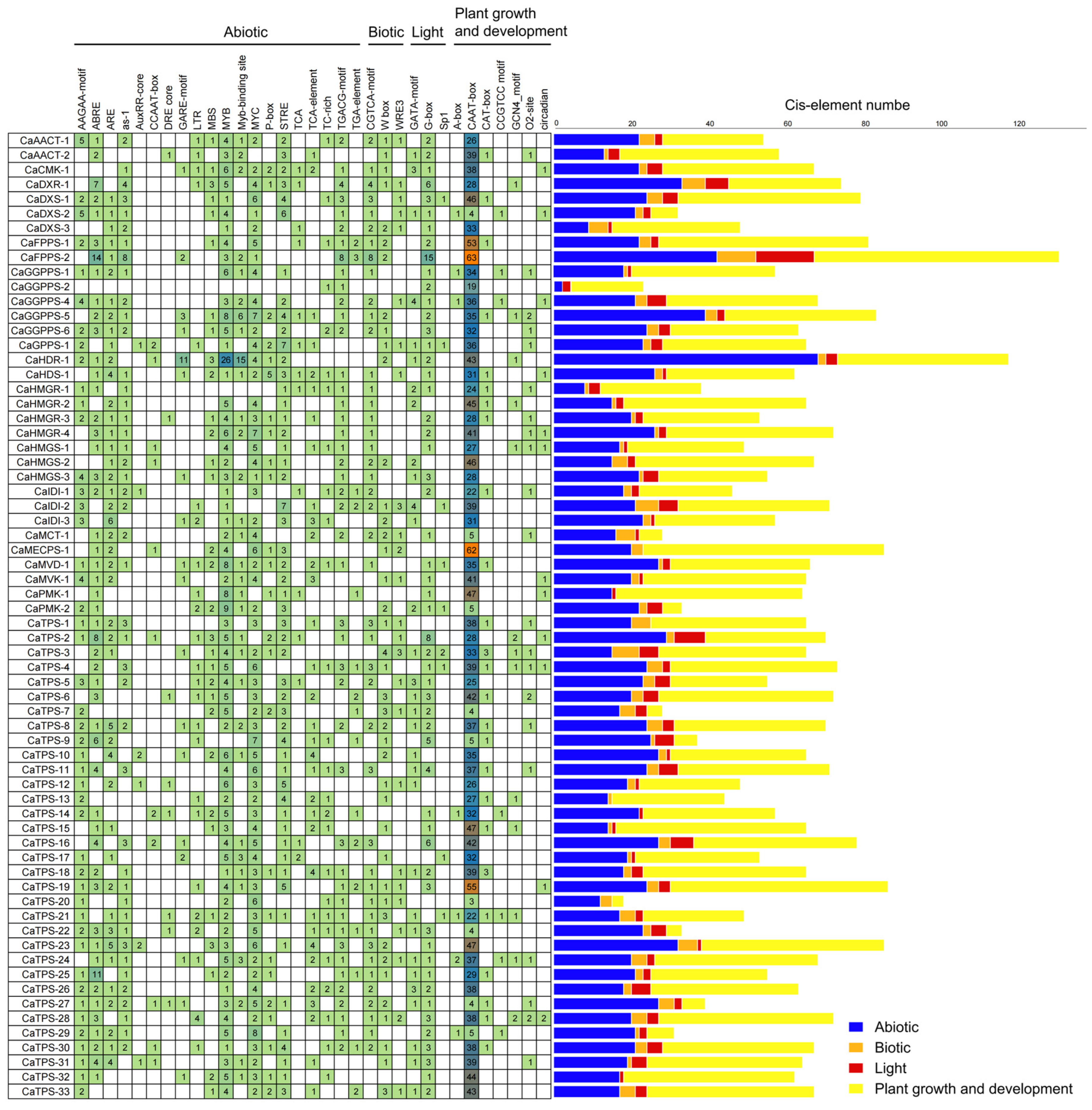
3.3. Phylogenetic, Gene Structure, and Promoter Analysis of Terpene Metabolism Genes
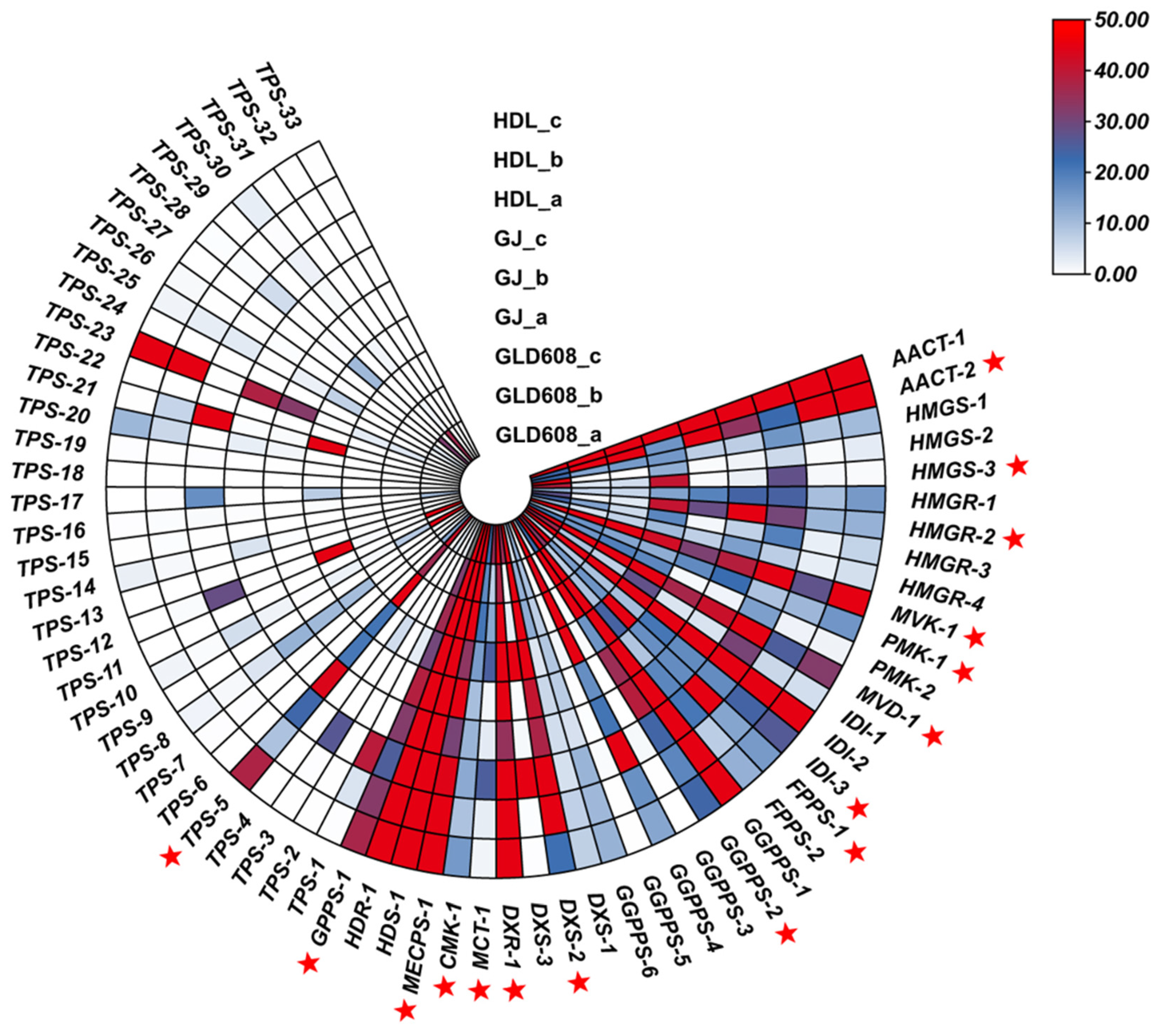
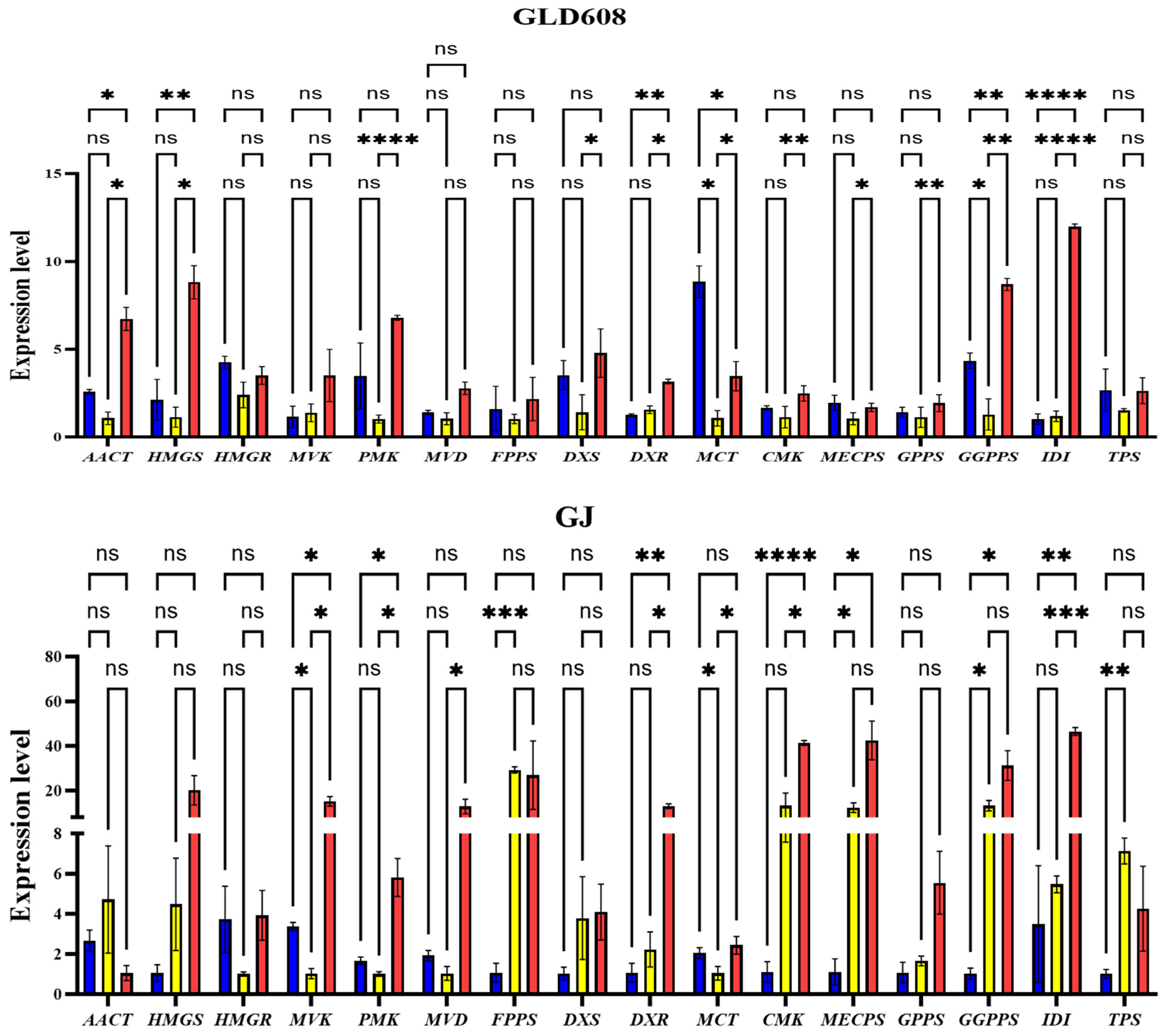
3.4. Expression Patterns of Enzyme Genes for Terpene Metabolism and Synthesis During Chili Fruit Development
3.5. Changes in Expression of Enzymes and Correlation with Content of Corresponding Metabolites
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zou, X.X.; Ma, Y.Q.; Li, X.F.; Yang, S. The spread and industrial development of peppers in China. Acta Hortic. 2020, 9, 1715–1726. [Google Scholar] [CrossRef]
- Hu, F.X.; Chu, Q.; Bai, R.X.; Zheng, Z.J.; Wang, Z.S. Research progress on the activity and utilization improvement technology of pepper alkaloids. Sci. Technol. Food Ind. 2021, 42, 412–419. [Google Scholar]
- Guo, T.R.; Zhang, L.; Wu, W.L.; Ke, Y.; Peng, H.C.; Pei, B.W. Analysis of characteristic volatile aroma components in puff pastry dried peppers. Chin. Condiments 2021, 4, 148–152. [Google Scholar]
- Qu, J.; Liu, L.; Guo, Z.; Li, X.; Pan, F.; Sun, D.; Yin, L. The ubiquitous position effect, synergistic effect of recent generated tandem duplicated genes in grapevine, and their co-response and over activity to biotic stress. Fruit Res. 2023, 16, 16. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, H.; Zong, Y.; Tu, Z.; Li, H. Isolation, expression, and functional analysis of the geranylgeranyl pyrophosphate synthase (GGPPS) gene from Liriodendron tulipifera. Plant Physiol. Biochem. 2021, 166, 700–711. [Google Scholar] [CrossRef]
- Fang, X.; Shi, L.; Ren, A.; Jiang, A.-L.; Wu, F.-L.; Zhao, M.-W. The cloning, characterization and functional analysis of a gene encoding an acetyl-CoA acetyltransferase involved in triterpene biosynthesis in Ganoderma lucidum. Int. J. Med. Mushrooms 2013, 54, 100–105. [Google Scholar] [CrossRef]
- Rodrı’guez-Concepcion, M.; Boronat, A. Breaking new ground in the regulation of the early steps of plant isoprenoid biosynthesis. Curr. Opin. Plant Biol. 2015, 25, 17–22. [Google Scholar] [CrossRef]
- Lluch, M.A.; Masferrer, A.; Arró, M.; Boronat, A.; Ferrer, A. Molecular cloning and expression analysis of the mevalonate kinase gene from Arabidopsis thaliana. Plant Mol. Biol. 2000, 42, 365–376. [Google Scholar] [CrossRef]
- Liu, C.; Meng, Y.L.; Hou, S.S.; Chen, X. Cloning and sequencing of a cDNA encoding farnesyl pyrophosphate synthase from Gossypium arboreum and its expression pattern in the developing seeds of Gossypium hirsutum cv. “Sumian-6”. Acta Bot. Sin. 1998, 40, 703–710. Available online: https://api.semanticscholar.org/CorpusID:87622179 (accessed on 16 August 2024).
- Chen, D.; Ye, H.; Li, G. Expression of a chimeric farnesyl diphosphate synthase gene in Artemisia annua L. transgenic plants via Agrobacterium tumefaciens-mediated transformation. Plant Sci. 2000, 155, 179–185. [Google Scholar] [CrossRef]
- Kim, O.T.; Kim, S.H.; Ohyama, K.; Muranaka, T.; Choi, Y.E.; Lee, H.Y.; Kim, M.Y.; Hwang, B. Upregulation of phytosterol and triterpene biosynthesis in Centella asiatica hairy roots overexpressed ginseng farnesyl diphosphate synthase. Plant Cell Rep. 2010, 29, 403–411. [Google Scholar] [CrossRef]
- Nieuwenhuizen, N.J.; Chen, X.; Wang, M.Y.; Matich, A.J.; Perez, R.L.; Allan, A.C.; Green, S.A.; Atkinson, R.G. Natural variation in monoterpene synthesis in kiwifruit: Transcriptional regulation of terpene synthases by NAC and ETHYLENE-INSENSITIVE3-like transcription factors. Plant Physiol. 2015, 167, 1243–1258. [Google Scholar] [CrossRef]
- Ruiz-Sola, M.A.; Barja, M.V.; Manzano, D.; Llorente, B.; Schipper, B.; Beekwilder, J.; Rodriguez-Concepcion, M. A single Arabidopsis gene encodes two differentially targeted geranylgeranyl diphosphate synthase isoforms. Plant Physiol. 2016, 172, 1393–1402. [Google Scholar] [CrossRef]
- Hu, Z.H.; Tang, B.; Wu, Q.; Zheng, J.; Leng, P.S.; Zhang, K.Z. Transcriptome sequencing analysis reveals a difference in monoterpene biosynthesis between scented Lilium ‘Siberia’ and unscented Lilium ‘Novano’. Front. Plant Sci. 2017, 8, 1351. [Google Scholar] [CrossRef]
- Tian, J.P.; Ma, Z.Y.; Zhao, K.G.; Zhang, J.; Xiang, L.; Chen, L.Q. Transcriptomic and proteomic approaches to explore the differences in monoterpene and benzenoid biosynthesis between scented and unscented genotypes of wintersweet. Physiol. Plant. 2019, 166, 478–493. [Google Scholar] [CrossRef]
- Gao, F.Z.; Liu, B.F.; Li, M.; Gao, X.Y.; Fang, Q.; Liu, C.; Ding, H.; Wang, L.; Gao, X. Identification and characterization of terpene synthase genes accounting for volatile terpene emissions in flowers of Freesia x hybrida. J. Exp. Bot. 2018, 69, 4249–4265. [Google Scholar] [CrossRef]
- Liu, S.; Shan, B.; Zhou, X.; Gao, W.; Liu, Y.; Zhu, B.; Sun, L. Transcriptome and Metabolomics Integrated Analysis Reveals Terpene Synthesis Genes Controlling Linalool Synthesis in Grape Berries. J. Agric. Food Chem. 2022, 70, 9084–9094. [Google Scholar] [CrossRef]
- Jiang, F.; Liu, D.; Dai, J.; Yang, T.; Zhang, J.; Che, D. Cloning and Functional Characterization of 2-C-methyl-d-erythritol-4-phosphate cytidylyltransferase (LiMCT) Gene in Oriental Lily (Lilium ‘Sorbonne’). Mol. Biotechnol. 2024, 66, 56–67. [Google Scholar] [CrossRef]
- Zhang, X.H.; Lian, M.Y.; Li, H.; Tao, G.; Xinyu, D.; Jiayue, X.; Lingui, Z.; Zhijian, S.; Xiaojun, L.; Liang, Z. Quality Analysis of Key Nodes of Natural Drying Chilis in Xinjiang Region. Sci. Technol. Food Ind. 2023, 44, 119–125. [Google Scholar] [CrossRef]
- Huang, C.; Sun, P.; Yu, S.; Fu, G.; Deng, Q.; Wang, Z.; Cheng, S. Analysis of volatile aroma components and regulatory genes in different kinds and development stages of pepper fruits based on non-Targeted metabolome combined with transcriptome. Int. J. Mol. Sci. 2023, 24, 7901. [Google Scholar] [CrossRef]
- Lange, B.M.; Ghassemian, M. Genome organization in Arabidopsis thaliana: A survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol. Biol. 2003, 51, 925–948. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Peng, M.; Fang, H.; Wang, Z.; Zhou, S.; Jing, X.; Zhang, M.; Yang, C.; Guo, H.; Li, Y.; et al. An Oryza-specific hydroxycinnamoyl tyramine gene cluster contributes to enhanced disease resistance. Sci. Bull. 2021, 66, 2369–2380. [Google Scholar] [CrossRef] [PubMed]
- Birchler, J.A.; Yang, H. The multiple fates of gene duplications: Deletion, hypofunctionalization, subfunctionalization, neofunctionalization, dosage balance constraints, and neutral variation. Plant Cell 2022, 34, 2466–2474. [Google Scholar] [CrossRef] [PubMed]
- Lü, H.; Liu, W.; He, L.; Xu, Z.; Luo, H. Advances on the study of gene clusters involved in plant secondary metabolism. Plant Sci. J. 2017, 35, 609–621. [Google Scholar] [CrossRef]
- Fang, H.; Shen, S.; Wang, D.; Zhang, F.; Zhang, C.; Wang, Z.; Zhou, Q.; Wang, R.; Tao, H.; He, F.; et al. A monocot-specific hydroxycinnamoyl putrescine gene cluster contributes to immunity and cell death in rice. Sci. Bull. 2021, 66, 2381–2393. [Google Scholar] [CrossRef]
- Frey, M.; Chomet, P.; Glawischnig, E.; Stettner, C.; Grün, S.; Winklmair, A.; Eisenreich, W.; Bacher, A.; Meeley, R.B.; Briggs, S.P.; et al. Analysis of a chemical plant defense mechanism in grasses. Science 1997, 277, 696–699. [Google Scholar] [CrossRef]
- Qi, X.; Bakht, S.; Leggett, M.; Maxwell, C.; Melton, R.; Osbourn, A. A gene cluster for secondary metabolism in oat: Implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. USA 2004, 101, 8233–8238. [Google Scholar] [CrossRef] [PubMed]
- Wilderman, P.R.; Xu, M.; Jin, Y.; Coates, R.M.; Peters, R.J. Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 2004, 135, 2098–2105. [Google Scholar] [CrossRef]
- Pütter, K.M.; Van Deenen, N.; Unland, K.; Prüfer, D.; Schulze Gronover, C. Isoprenoid biosynthesis in dandelion latex is enhanced by the overexpression of three key enzymes involved in the mevalonate pathway. BMC Plant Biol. 2017, 17, 88. [Google Scholar] [CrossRef]
- Ezquerro, M.; Li, C.; Pérez-Pérez, J.; Burbano-Erazo, E.; Barja, M.V.; Wang, Y.; Dong, L.; Lisón, P.; López-Gresa, M.P.; Bouwmeester, H.J.; et al. Tomato geranylgeranyl diphosphate synthase isoform1 is involved in the stress-triggered production of diterpenes in leaves and strigolactones in roots. New Phytol. 2023, 239, 2292–2306. [Google Scholar] [CrossRef]
- Sakamoto, T.; Miura, K.; Itoh, H.; Tatsumi, T.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Agrawal, G.K.; Takeda, S.; Abe, K.; et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004, 134, 1642–1653. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Cui, M.; Liu, Z.; Shi, X.; Sun, R.; Zhang, S.; Du, G. Bioinformatics and expression analysis of the intron less genes of the grape genome. J. Nanjing Agric. Univ. 2018, 41, 655–661. [Google Scholar] [CrossRef]
- Xie, X.; Wu, N. Introns in higher plant genes. Chin. Sci. Bull. 2002, 47, 1409–1415. [Google Scholar] [CrossRef]
- Wittkopp, P.; Kalay, G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2012, 13, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhou, X.; Fan, H.; Gao, X.; Yang, L. Comparative analysis on volatile terpenoids in nine aromatic plants of Lamiaceae. Bull. Bot. Res. 2020, 40, 696–705. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, L.; Cai, X.; Li, X.; Bian, L.; Luo, Z.; Li, Z.; Chen, Z.; Xin, Z. (E)-Nerolidol is a volatile signal that induces defenses against insects and pathogens in tea plants. Hortic. Res. 2020, 37, 1741–1752. [Google Scholar] [CrossRef]
- Dai, H.; Liu, B.; Yang, L.; Yao, Y.; Liu, M.; Xiao, W.; Li, S.; Ji, R.; Sun, Y. Investigating the regulatory mechanism of the sesquiterpenes nerolidol from a plant on juvenile hormone-related genes in the insect Spodoptera exiguity. Int. J. Mol. Sci. 2023, 24, 13330. [Google Scholar] [CrossRef]
- Mączka, W.; Duda-Madej, A.; Grabarczyk, M.; Wińska, K. Natural compounds in the battle against microorganisms-linalool. Molecule 2022, 27, 6928. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Du, Y.; Xu, N.; Yue, C.; Ye, L. Improved linalool production in Saccharomyces cerevisiae by combining directed evolution of linalool synthase and overexpression of the complete mevalonate pathway. Biochem. Eng. J. 2020, 161, 107655. [Google Scholar] [CrossRef]
- Li, Y.; Ren, F.; Chen, D.; Chen, H.; Chen, W. Antibacterial mechanism of linalool against Pseudomonas fragi: A transcriptomic study. Foods 2022, 11, 2058. [Google Scholar] [CrossRef]
- Moreno, E.; Fita, A.M.; González-Mas, M.D.; Rodríguez-Burruezo, A. HS-SPME study of the volatile fraction of Capsicum accessions and hybrids in different parts of the fruit. Sci. Hortic. 2012, 135, 87–97. [Google Scholar] [CrossRef]
- Xiong, X.; Xia, Y.; Zhang, X.; Deng, H. GC-MS analysis of volatile components of different varieties of paprika. Sci. Technol. Food Ind. 2012, 33, 4. [Google Scholar]
- Liu, H. Functional study of HMGR and DXR genes in the biosynthesis of terpenoids in tobacco in Amomum villosum L. Guangzhou Univ. Tradit. Chin. Med. 2012, 24, 12532. [Google Scholar]

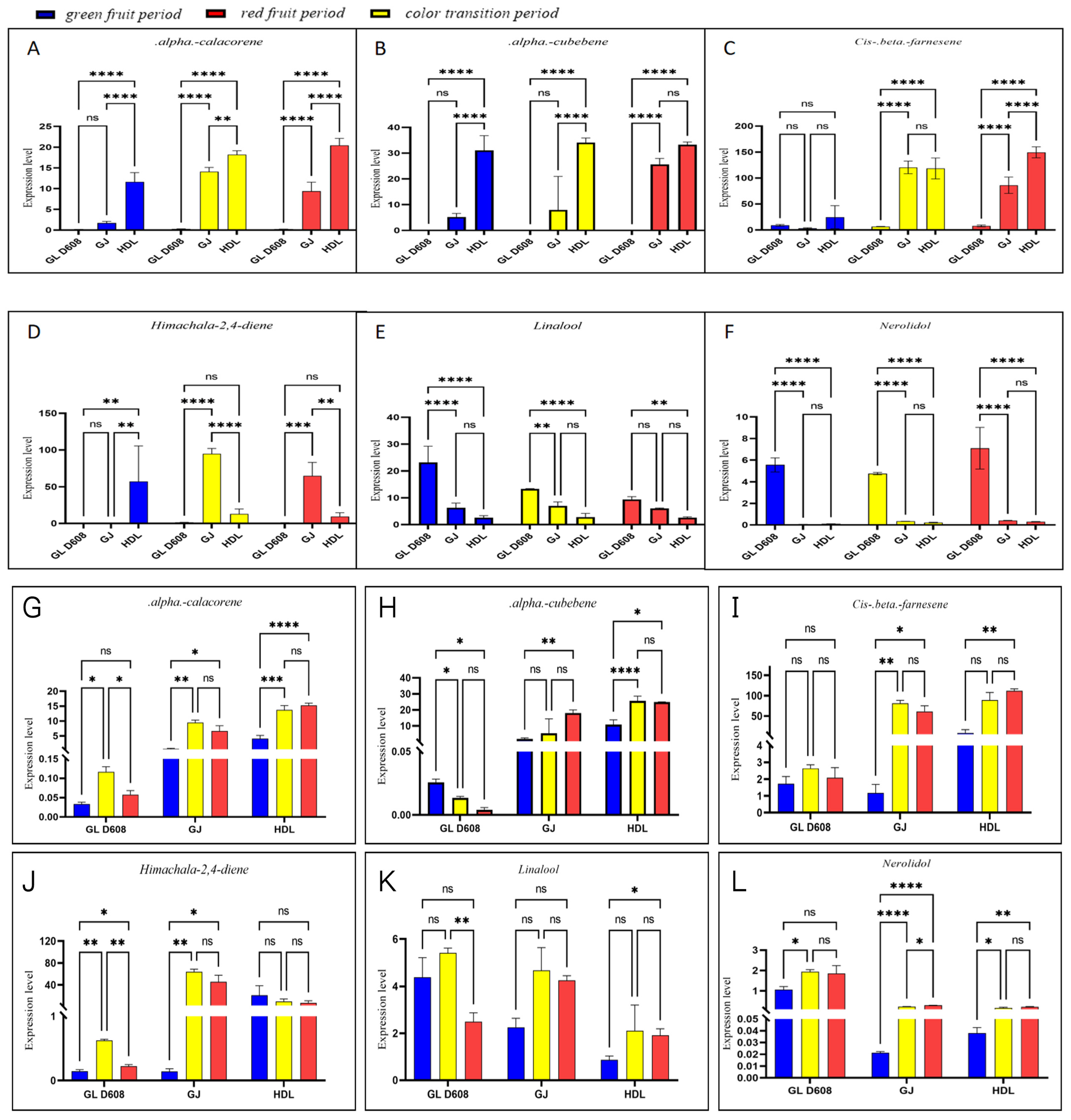

| Retention Indices | Retention Time | Formula | Stage | Green Stage | ||||||||
| Aroma Volatile Profile (μg/kg) | GLD608 | GJ | HDL | |||||||||
| 1540.658602 | 29.957 | C15H20 | α-calacorene | 0.189501185 | 0.169392939 | 0.161516859 | 2.026104808 | 1.83741121 | 1.256568456 | 9.08122777 | 12.42851114 | 13.34401757 |
| 1346.721064 | 23.854 | C15H24 | α-cubebene | 0.189501185 | 0.169392939 | 0.161516859 | 6.750840996 | 4.859400783 | 4.038638958 | 24.42060494 | 34.12703405 | 34.63315802 |
| 1450.990415 | 27.213 | C15H24 | cis-β-farnesene | 9.914039453 | 6.998453542 | 9.900740257 | 4.282773783 | 3.054284601 | 2.100333178 | 29.31950673 | 44.11500031 | 0.566948561 |
| 1481.916933 | 28.181 | C15H24 | himachala-2,4-diene | 0.672396897 | 0.728896278 | 0.890786308 | 0.46487621 | 0.375202389 | 0.295370111 | 1.781478646 | 81.0652433 | 89.05190136 |
| 1560.719086 | 30.554 | C15H26O | nerolidol | 5.100512268 | 5.275688126 | 6.298854821 | 0.052066552 | 0.059313761 | 0.064776714 | 0.105807802 | 0.108882511 | 0.114321495 |
| 1098.606177 | 15.15 | C10H28O | linalool | 17.72278692 | 29.74326537 | 21.83802079 | 5.307310451 | 5.187160583 | 8.333778717 | 3.449577756 | 2.020037205 | 2.212748445 |
| Retention Indices | Retention Time | Formula | Stage | Transition Stage | ||||||||
| Aroma Volatile Profile (μg/kg) | GLD608 | GJ | HDL | |||||||||
| 1540.658602 | 29.957 | C15H20 | α-calacorene | 0.290480591 | 0.307122624 | 0.261394513 | 14.57378854 | 14.76637357 | 12.91119967 | 17.3293049 | 19.20997281 | 18.01882563 |
| 1346.721064 | 23.854 | C15H24 | α-cubebene | 0.032606452 | 0.03005341 | 0.037267364 | 22.99147365 | 0.257943059 | 0.588673996 | 32.22838886 | 35.82612365 | 34.24461297 |
| 1450.990415 | 27.213 | C15H24 | cis-β-farnesene | 6.203308506 | 6.999444129 | 6.297113557 | 111.2381479 | 134.2963163 | 115.9745627 | 118.6495515 | 98.17047579 | 138.5707145 |
| 1481.916933 | 28.181 | C15H24 | himachala-2,4-diene | 1.519322631 | 1.519458133 | 1.559081741 | 92.49001943 | 102.9068201 | 88.94440934 | 19.41318347 | 5.577432573 | 13.49877299 |
| 1560.719086 | 30.554 | C15H26O | nerolidol | 4.725287206 | 4.891903877 | 4.684003949 | 0.343238587 | 0.336734435 | 0.336889952 | 0.195508473 | 0.192212047 | 0.257280851 |
| 1098.606177 | 15.15 | C10H28O | linalool | 13.28365092 | 13.26640641 | 13.47729543 | 5.776609437 | 8.67423761 | 6.456407814 | 2.099548198 | 4.399658607 | 1.901405828 |
| Retention Indices | Retention Time | Formula | Stage | Red stage | ||||||||
| Aroma Volatile Profile (μg/kg) | GLD608 | GJ | HDL | |||||||||
| 1540.658602 | 29.957 | C15H20 | α-calacorene | 0.196212603 | 0.250821045 | 0.204236846 | 11.19527999 | 9.958240804 | 6.920142137 | 18.75339771 | 20.40489899 | 22.11276696 |
| 1346.721064 | 23.854 | C15H24 | α-cubebene | 0.007219878 | 0.018750759 | 0.020920522 | 26.13886164 | 27.64447898 | 23.07029552 | 32.35776509 | 33.17996989 | 34.38264749 |
| 1450.990415 | 27.213 | C15H24 | cis-β-farnesene | 7.369725556 | 9.761146872 | 6.212745679 | 102.0934501 | 85.58305552 | 70.58480417 | 137.6160577 | 152.7423527 | 158.0275569 |
| 1481.916933 | 28.181 | C15H24 | himachala-2,4-diene | 0.704335791 | 0.82826117 | 1.036842206 | 60.51347448 | 84.83262687 | 49.36515377 | 14.03306576 | 3.924964995 | 10.53879978 |
| 1560.719086 | 30.554 | C15H26O | nerolidol | 5.001703282 | 7.459903041 | 8.821545182 | 0.386440769 | 0.409054335 | 0.423033271 | 0.262315722 | 0.3180631 | 0.297885325 |
| 1098.606177 | 15.15 | C10H28O | linalool | 10.41722888 | 8.321212038 | 9.494170242 | 5.968380794 | 6.227085797 | 5.889582503 | 2.839207021 | 2.164262988 | 2.674034191 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Wu, K.; Fu, G.; Yu, S.; Huang, R.; Wang, Z.; Lu, X.; Fu, H.; Deng, Q.; Cheng, S. Genome Wide Identification of Terpenoid Metabolism Pathway Genes in Chili and Screening of Key Regulatory Genes for Fruit Terpenoid Aroma Components. Horticulturae 2025, 11, 586. https://doi.org/10.3390/horticulturae11060586
Yang M, Wu K, Fu G, Yu S, Huang R, Wang Z, Lu X, Fu H, Deng Q, Cheng S. Genome Wide Identification of Terpenoid Metabolism Pathway Genes in Chili and Screening of Key Regulatory Genes for Fruit Terpenoid Aroma Components. Horticulturae. 2025; 11(6):586. https://doi.org/10.3390/horticulturae11060586
Chicago/Turabian StyleYang, Mengxian, Kun Wu, Genying Fu, Shuang Yu, Renquan Huang, Zhiwei Wang, Xu Lu, Huizhen Fu, Qin Deng, and Shanhan Cheng. 2025. "Genome Wide Identification of Terpenoid Metabolism Pathway Genes in Chili and Screening of Key Regulatory Genes for Fruit Terpenoid Aroma Components" Horticulturae 11, no. 6: 586. https://doi.org/10.3390/horticulturae11060586
APA StyleYang, M., Wu, K., Fu, G., Yu, S., Huang, R., Wang, Z., Lu, X., Fu, H., Deng, Q., & Cheng, S. (2025). Genome Wide Identification of Terpenoid Metabolism Pathway Genes in Chili and Screening of Key Regulatory Genes for Fruit Terpenoid Aroma Components. Horticulturae, 11(6), 586. https://doi.org/10.3390/horticulturae11060586









