Terpene Synthase (TPS) Family Member Identification and Expression Pattern Analysis in Flowers of Dendrobium chrysotoxum
Abstract
1. Introduction
2. Methods
2.1. Identification of TPS Genes in D. chrysotoxum
2.2. Characteristics of TPS Genes
2.3. Phylogenetic, Conserved Motifs and Gene Structure Analysis
2.4. Cis-Elements in the Promoter Regions Analysis
2.5. Protein–Protein Interaction (PPI) Network Prediction Analysis
2.6. GO Classification and Enrichment Analysis
2.7. Cultivation of D. chrysotoxum
2.8. RNA Extraction and qRT-PCR Expression Analysis
2.9. Transient Expression of DcTPS Genes in Nicotiana benthamiana
2.10. GC–MS Analysis
3. Results
3.1. Identification and Characterization of TPS Gene Family in D. chrysotoxum Genome Resources
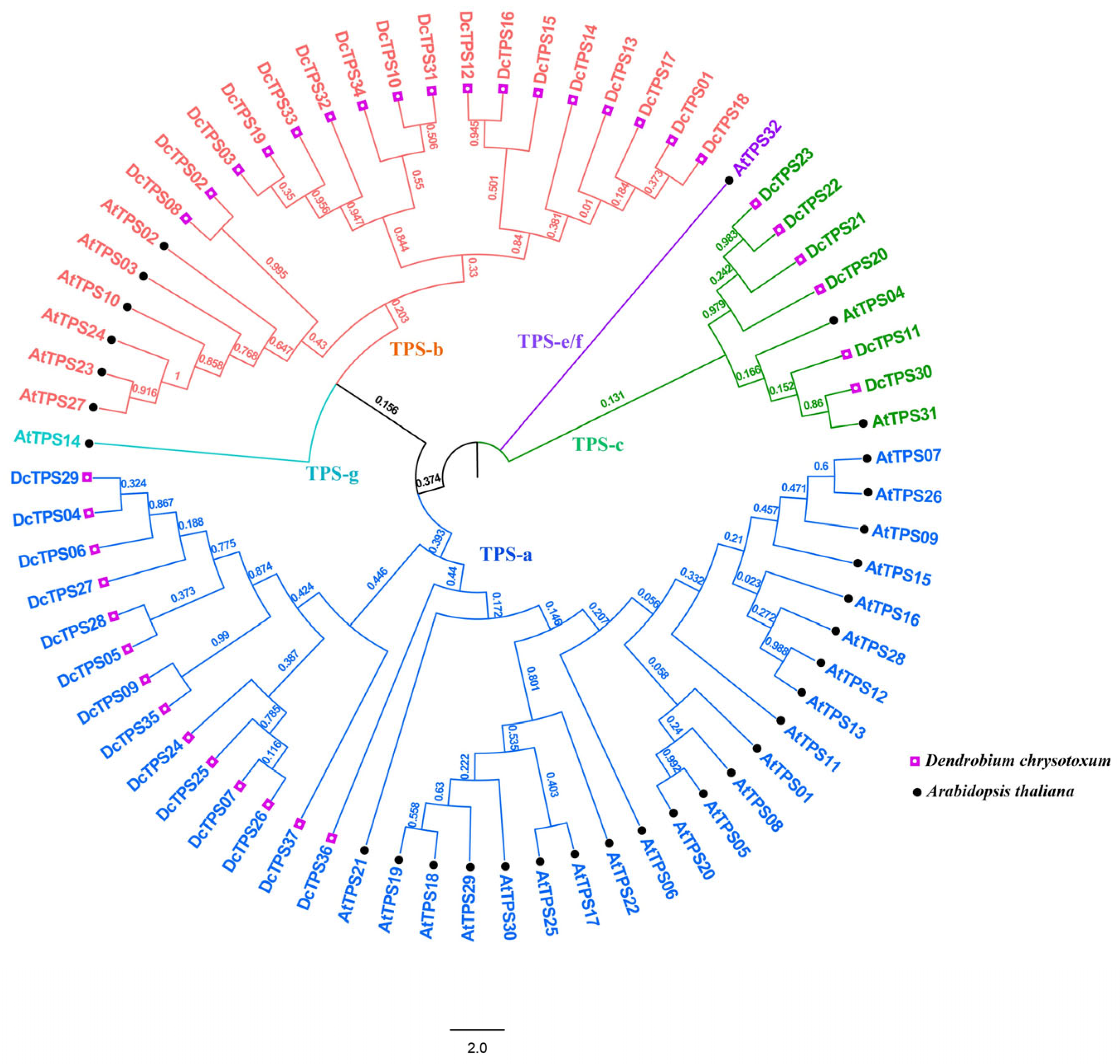
3.2. Phylogenetic Relationship of DcTPS Family
3.3. Conserved Motifs and Gene Structure Analysis
3.4. Promoters Cis-Elements Analysis
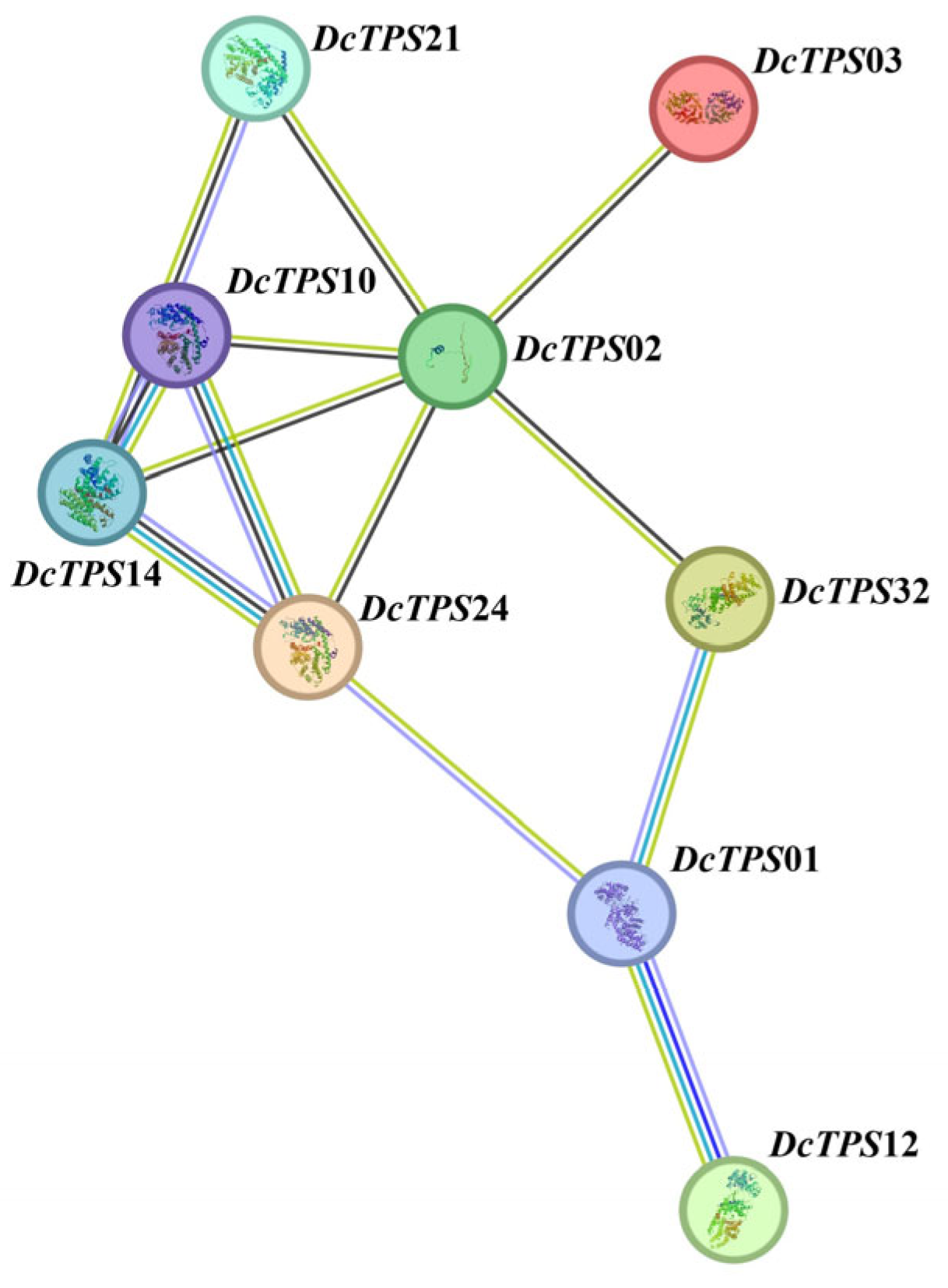
3.5. Protein–Protein Interaction (PPI) Network Analysis
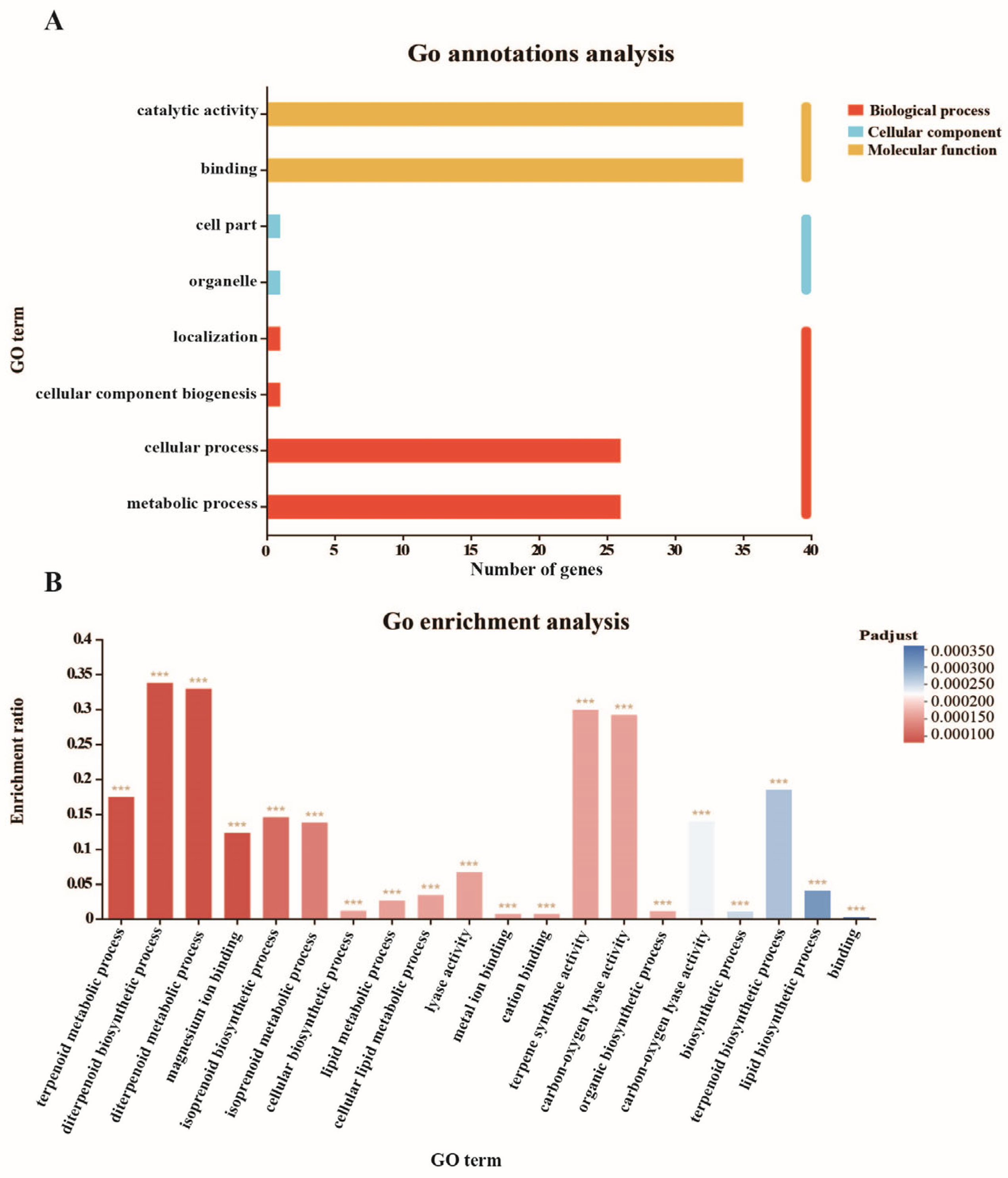
3.6. GO Classification and Enrichment of DcTPS Genes
3.7. Different Tissues Expression Analysis of DcTPS Genes
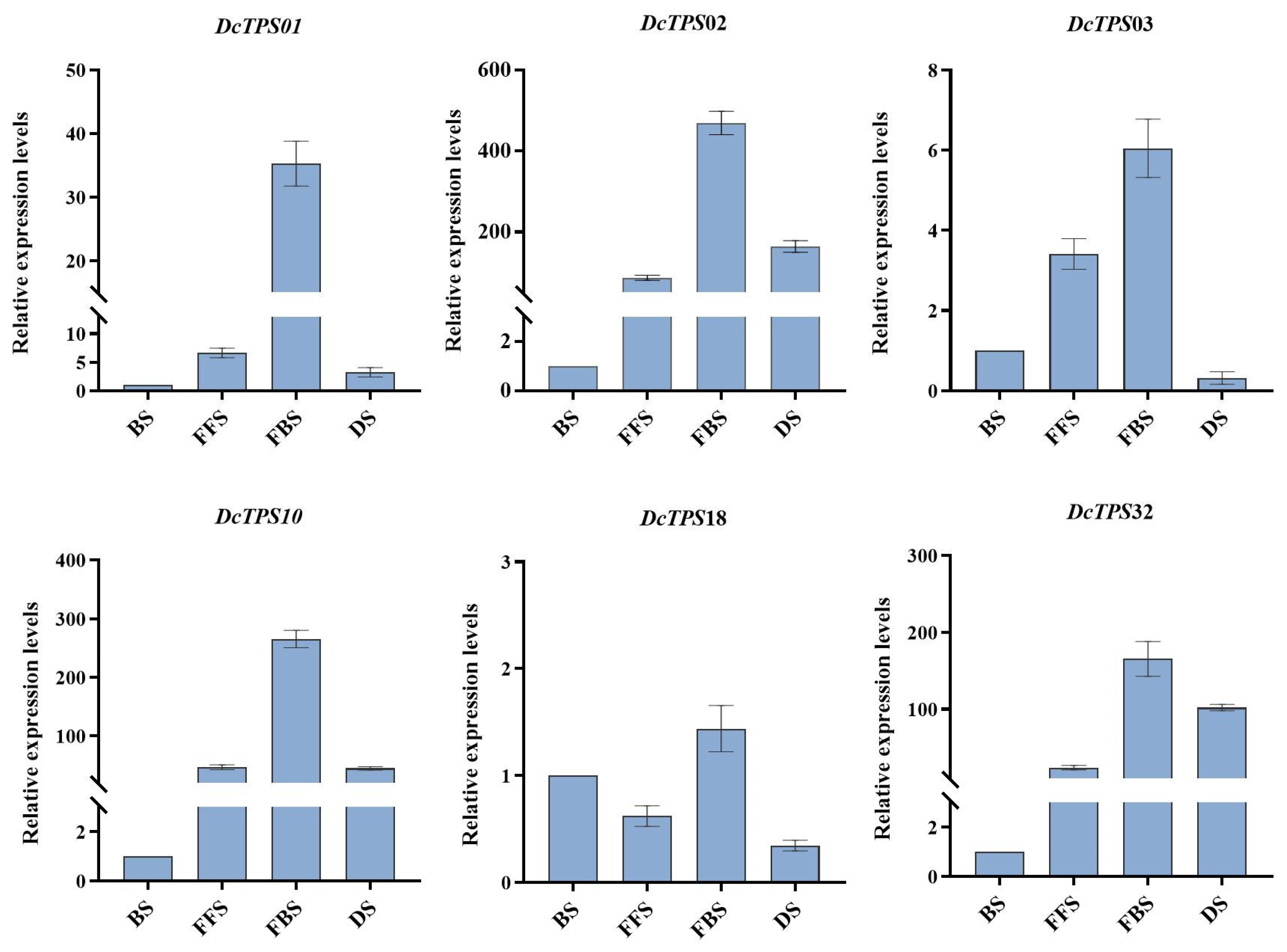
3.8. Different Fluorescence Stages’ Expression Analysis of DcTPS Genes
3.9. Functional Identification of DcTPS Genes in Plants
4. Discussion
4.1. Comprehensively Investigated TPS Gene Family in D. chrysotoxum
4.2. TPS-b Was a Dominant Subfamily in D. chrysotoxum
4.3. TPS-b Subfamily Genes Have Conserved Motifs Encoding Monoterpenes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| aa | amino acid |
References
- Sun, Z.; Song, Y.; Zeng, R. Advances in studies on intraspecific and interspecific relationships mediated by plant volatiles. J. S. China Agric. Univ. 2019, 40, 166–174. [Google Scholar] [CrossRef]
- Wu, J.N.; Liu, Z.Y.; Wu, M.Q. Analysis of Volatile Components in the Flowers of Seven Species of Dendrobium. Mol. Plant Breed. 2023, 3, 1–16. Available online: http://kns.cnki.net/kcms/detail/46.1068.S.20230314.0905.004.html (accessed on 8 January 2022).
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant3volatile organic compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef]
- Abbas, F.; Yu, Y.; Yue, R.; Yue, Y.; Amanullah, S.; Jahangir, M.M.; Fan, Y. Volatile terpenoids: Multiple functions, biosynthesis, modulation and manipulation by genetic engineering. Planta Int. J. Plant Biol. 2017, 246, 115–129. [Google Scholar] [CrossRef]
- Yu, F.; Utsumi, R. Diversity, regulation, and genetic manipulation of plant mono- and sesquiterpenoid biosynthesis. Cell. Mol. Life Sci. 2009, 66, 3043–3052. [Google Scholar] [CrossRef] [PubMed]
- Baek, Y.S.; Ramya, M.; An, H.R.; Park, P.M.; Lee, S.Y.; Baek, N.I.; Park, P.H. Volatiles Profile of the Floral Organs of a New Hybrid Cymbidium, ‘Sunny Bell’ Using Headspace Solid-Phase Microextraction Gas Chromatography-Mass Spectrometry Analysis. Plants 2019, 8, 251. [Google Scholar] [CrossRef]
- Bohlmann, J.; Meyer-Gauen, G.; Croteau, R. Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA 1998, 95, 4126–4133. [Google Scholar] [CrossRef]
- Huang, L.-M.; Huang, H.; Chuang, Y.-C.; Chen, W.-H.; Wang, C.-N.; Chen, H.-H. Evolution of Terpene Synthases in Orchidaceae. Int. J. Mol. Sci. 2021, 22, 6947. [Google Scholar] [CrossRef]
- Flath, R.A.; Ohinata, K. Volatile Components of the Orchid Dendrobium superbum Rchb. f. J. Agric. Food Chem. 1982, 30, 841–842. [Google Scholar] [CrossRef]
- Vranová, E.; Coman, D.; Gruissem, W. Network Analysis of the MVA and MEP Pathways for Isoprenoid Synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef]
- Pulido, P.; Perello, C.; Rodriguez-Concepcion, M. New Insights into Plant Isoprenoid Metabolism. Mol. Plant 2012, 5, 964–967. [Google Scholar] [CrossRef] [PubMed]
- Simkin, A.J.; Guirimand, G.; Papon, N.; Courdavault, V.; Thabet, I.; Ginis, O.; Bouzid, S.; Giglioli-Guivarc’h, N.; Clastre, M. Peroxisomal localisation of the final steps of the mevalonic acid pathway in planta. Plant Signal. Behav. 2011, 234, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Sandra, I.; Krause, S.T.; Grit, K.; Jonathan, G.; Jörg, D.; Köllner, T.G. The organ-specific expression of terpene synthase genes contributes to the terpene hydrocarbon composition of chamomile essential oils. Bmc Plant Biol. 2012, 12, 84–97. [Google Scholar] [CrossRef]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef]
- Degenhardt, J.; Kllner, T.G.; Gershenzon, J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 2009, 69, 1621–1637. [Google Scholar] [CrossRef]
- Gao, F.; Liu, B.; Li, M.; Gao, X.; Fang, Q.; Liu, C.; Ding, H.; Wang, L.; Gao, X. Identification and Characterization of Terpene Synthase Genes Accounting for the Volatile Terpene Emissions in Flowers of Freesia × hybrida. J. Exp. Bot. 2018, 69, 4249–4265. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L.L.; et al. The Pfam Protein Families Database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef]
- Jiang, S.Y.; Jin, J.J.; Sarojam, R.; Ramachandran, S. A Comprehensive Survey on the Terpene Synthase Gene Family Provides New Insight into Its Evolutionary Patterns. Genome Biol. Evol. 2019, 11, 2078–2098. [Google Scholar] [CrossRef]
- Trapp, S.C.; Croteau, R.B. Genomic Organization of Plant Terpene Synthases and Molecular Evolutionary Implications. Genetics 2001, 158, 811–832. [Google Scholar] [CrossRef]
- Tholl, D.; Lee, S. Terpene Specialized Metabolism in Arabidopsis thaliana. Arab. Book 2011, 9, e0143. [Google Scholar] [CrossRef]
- Falara, V.; Akhtar, T.A.; Nguyen, T.T.H.; Spyropoulou, E.A.; Bleeker, P.M. The tomato (Solanum lycopersicum) terpene synthase gene family. Plant Physiol. 2011, 157, 770–789. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.M.; Aubourg, S.; Schouwey, M.B.; Daviet, L.; Schalk, M.; Toub, O.; Lund, S.T.; Bohlmann, J. Functional Annotation, Genome Organization and Phylogeny of the Grapevine (Vitis vinifera) Terpene Synthase Gene Family Based on Genome Assembly, FLcDNA Cloning, and Enzyme Assays. BMC Plant Biol. 2010, 10, 226. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.Q.; Liu, K.W.; Li, Z.; Lohaus, R.; Hsiao, Y.Y.; Niu, S.C.; Wang, J.Y.; Lin, Y.C.; Xu, Q.; Chen, L.J.; et al. The Apostasia genome and the evolution of orchids. Nature 2017, 549, 379–383. [Google Scholar] [CrossRef]
- Kaundal, R.; Zhao, S.P.X. Combining Machine Learning and Homology-Based Approaches to Accurately Predict Subcellular Localization in Arabidopsis. Plant Physiol. 2010, 154, 36–54. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Ojeda, D.I.; Santosguerra, A.; Olivatejera, F.; Valido, A.; Xue, X.; Marrero, A.; Caujapé-Castells, J.; Cronk, Q. Bird-pollinated Macaronesian Lotus (Leguminosae) evolved within a group of entomophilous ancestors with post-anthesis flower color change. Perspect. Plant Ecol. Evol. Syst. 2013, 15, 193–204. [Google Scholar] [CrossRef]
- Byers, K.J.R.P.; Vela, J.P.; Peng, F.; Riffell, J.A.; Bradshaw, H.D. Floral volatile alleles can contribute to pollinator-mediated reproductive isolation in monkeyflowers (Mimulus). Plant J. Cell Mol. Biol. 2015, 80, 1031–1042. [Google Scholar] [CrossRef]
- Knudsen, J.T.; Eriksson, R.; Gershenzon, J.; Ståhl, B. Diversity and distribution of floral scent. Bot. Rev. 2006, 72, 1. [Google Scholar] [CrossRef]
- Ramsey, J.; Bradshaw, H.D.; Schemske, D.W. Components of reproductive isolation between the monkeyflowers mimulus lewisii and m. cardinalis (phrymaceae). Evolution 2003, 57, 1520–1534. [Google Scholar] [CrossRef]
- Dudareva, N.; Murfitt, L.M.; Mann, C.J.; Gorenstein, N.; Kolosova, N.; Kish, C.M.; Bonham, C.; Wood, K. Developmental Regulation of Methyl Benzoate Biosynthesis and Emission in Snapdragon Flowers. Plant Cell 2000, 12, 949–961. [Google Scholar] [CrossRef]
- Yang, Y.; Xia, K.; Wu, Q.; Lu, X.; Lu, S.; Zhao, Z.; Qiu, S. Combined Analysis of Volatile Compounds and Extraction of Floral Fragrance Genes in Two Dendrobium Species. Horticulturae 2023, 9, 745. [Google Scholar] [CrossRef]
- Burkhardt, D.; Mosandl, A. Biogenetic Studies in Syringa vulgaris L.: Bioconversion of 18O(2H)-Labeled Precursors into Lilac Aldehydes and Lilac Alcohols. J. Agric. Food Chem. 2003, 51, 7391–7395. [Google Scholar] [CrossRef] [PubMed]
- Shalit, M.; Guterman, I.; Volpin, H.; Bar, E.; Tamari, T.; Menda, N.; Adam, Z.; Zamir, D.; Vainstein, A.; Weiss, D.; et al. Ester Formation in Roses. Identification of an Acetyl-Coenzyme A. Geraniol/Citronellol Acetyltransferase in Developing Rose Petals. Plant Physiol. 2003, 131, 1868–1876. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.M.; Zhao, C.H.; Zhang, G.H.; Silva, J.A.T.; Duan, J. Genome-Wide Identification and Expression Profile of TPS Gene Family in Dendrobium officinale and the Role of DoTPS10 in Linalool Biosynthesis. Int. J. Mol. Sci. 2020, 21, 5419. [Google Scholar] [CrossRef]
- Wang, Q.Q.; Zhu, M.J.; Yu, X.; Bi, Y.Y.; Zhou, Z.; Chen, M.K.; Chen, J.; Zhang, D.; Ai, Y.; Liu, Z.J.; et al. Genome-Wide Identification and Expression Analysis of Terpene Synthase Genes in Cymbidium faberi. Front. Plant Sci. 2021, 12, 751853. [Google Scholar] [CrossRef]
- Chuang, Y.C.; Hung, Y.C.; Tsai, W.C.; Chen, H.H. PbbHLH4 regulates floral monoterpene biosynthesis in Phalaenopsis orchids. J. Exp. Bot. 2018, 69, 4363–4377. [Google Scholar] [CrossRef]
- Green, S.A.; Chen, X.; Nieuwenhuizen, N.J.; Matich, A.J.; Wang, M.Y.; Bunn, B.J.; Yauk, Y.K.; Atkinson, R.G. Identification, functional characterization, and regulation of the enzyme responsible for floral (E)-nerolidol biosynthesis in kiwifruit (Actinidia chinensis). J. Exp. Bot. 2011, 63, 1951–1967. [Google Scholar] [CrossRef]
- Nieuwenhuizen, N.J.; Wang, M.Y.; Matich, A.J.; Green, S.A.; Chen, X.; Yauk, Y.K.; Beuning, L.L.; Nagegowda, D.A.; Dudareva, N.; Atkinson, R.G. Two terpene synthases are responsible for the major sesquiterpenes emitted from the flowers of kiwifruit (Actinidia deliciosa). J. Exp. Bot. 2009, 60, 3203–3219. [Google Scholar] [CrossRef]
- Feng, L.G.; Chen, C.; Sheng, L.X.; Liu, P.; Tao, J.; Su, J.L.; Zhao, L.Y. Comparative analysis of headspace volatiles of Chinese Rosa rugosa. Molecules 2010, 15, 8390–8399. [Google Scholar] [CrossRef]
- Heller, S.R.; Milne, G.W. EPA/NIH Mass Spectral Data Base; U.S. Governwent Printing Office: Washington, DC, USA, 1980.
- Zhang, H.; Chen, M.; Wang, X.; Dai, J.; Zhang, X.; Zhang, Z.; Zhang, X.; Tang, M.; Tang, J.; Gong, J.; et al. Transcriptome Analysis of Rhododendron liliiflorum H. Lév. Flower Colour Differences. Horticulturae 2023, 9, 82. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. Cold Spring Harb. Lab. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Hsiao, Y.Y.; Pan, Z.J.; Hsu, C.C.; Yang, Y.P.; Hsu, Y.C.; Chuang, Y.C.; Shih, H.H.; Chen, W.H.; Tsai, W.C.; Chen, H.H. Research on Orchid Biology and Biotechnology. Plant Cell Physiol. 2011, 52, 1467–1486. [Google Scholar] [CrossRef] [PubMed]
- Aubourg, S.; Lecharny, A.; Bohlmann, J. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genom. 2002, 267, 730–745. [Google Scholar] [CrossRef] [PubMed]
- Li, G.L.; Köllner, T.G.; Yin, Y.B.; Jiang, Y.F.; Chen, H.; Xu, Y.; Gershenzon, J.; Pichersky, E.; Chen, F. Nonseed plant Selaginella moellendorfii has both seed plant and microbial types of terpene synthases. Proc. Natl. Acad. Sci. USA 2012, 109, 14711–14715. [Google Scholar] [CrossRef]
- Cao, Z.; Ma, Q.; Weng, Y.; Shi, J.; Chen, J.; Hao, Z. Genome-Wide Identification and Expression Analysis of TPS Gene Family in Liriodendron chinense. Genes 2023, 14, 770. [Google Scholar] [CrossRef]
- Li, N.; Dong, Y.; Lv, M.; Qian, L.; Sun, X.; Liu, L.; Cai, Y.; Fan, H. Combined Analysis of Volatile Terpenoid Metabolism and Transcriptome Reveals Transcription Factors Related to Terpene Synthase in Two Cultivars of Dendrobium officinale Flowers. Front. Genet. 2021, 12, 661296. [Google Scholar] [CrossRef]
- Zeng, X.; Liu, C.; Zheng, R.; Cai, X.; Luo, J.; Zou, J.; Wang, C. Emission and Accumulation of Monoterpene and the Key Terpene Synthase (TPS) Associated with Monoterpene Biosynthesis in Osmanthus fragrans Lour. Front. Plant Sci. 2016, 12, 1232. [Google Scholar] [CrossRef]






| DcTPS s Name | ProteinLength (a.a) | Molecular Weight Mw/kDa | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity |
|---|---|---|---|---|---|---|
| DcTPS 1 | 510 | 58.84 | 8.16 | 40.98 | 88.14 | −0.174 |
| DcTPS 2 | 427 | 48.79 | 5.71 | 43.00 | 93.70 | −0.085 |
| DcTPS 3 | 1286 | 150.38 | 5.99 | 45.44 | 92.86 | −0.242 |
| DcTPS 4 | 670 | 77.97 | 6.56 | 45.94 | 96.06 | −0.195 |
| DcTPS 5 | 530 | 62.76 | 6.94 | 47.18 | 94.38 | −0.206 |
| DcTPS 6 | 512 | 60.52 | 5.58 | 46.53 | 96.93 | −0.215 |
| DcTPS 7 | 553 | 64.91 | 5.55 | 40.07 | 97.32 | −0.263 |
| DcTPS 8 | 510 | 58.45 | 6.00 | 39.76 | 97.18 | −0.105 |
| DcTPS 9 | 438 | 51.35 | 5.30 | 45.07 | 97.72 | −0.205 |
| DcTPS 10 | 598 | 69.48 | 5.21 | 47.26 | 92.81 | −0.236 |
| DcTPS 11 | 764 | 88.92 | 7.07 | 39.42 | 93.69 | −0.280 |
| DcTPS 12 | 458 | 53.88 | 6.07 | 44.15 | 93.89 | −0.191 |
| DcTPS 13 | 606 | 71.03 | 6.08 | 45.46 | 88.18 | −0.314 |
| DcTPS 14 | 648 | 75.92 | 5.48 | 41.55 | 93.49 | −0.249 |
| DcTPS 15 | 940 | 109.39 | 5.69 | 40.37 | 93.46 | −0.160 |
| DcTPS 16 | 627 | 73.15 | 5.61 | 46.33 | 97.99 | −0.134 |
| DcTPS 17 | 657 | 76.62 | 5.23 | 42.66 | 97.20 | −0.157 |
| DcTPS 18 | 395 | 46.25 | 5.49 | 36.83 | 97.44 | −0.137 |
| DcTPS 19 | 619 | 72.61 | 6.41 | 41.03 | 93.47 | −0.262 |
| DcTPS 20 | 834 | 97.32 | 7.12 | 47.90 | 86.24 | −0.359 |
| DcTPS 21 | 1305 | 152.19 | 8.46 | 43.40 | 86.08 | −0.349 |
| DcTPS 22 | 608 | 71.10 | 7.88 | 41.18 | 85.84 | −0.375 |
| DcTPS 23 | 563 | 65.79 | 5.68 | 49.30 | 84.01 | −0.334 |
| DcTPS 24 | 842 | 98.76 | 5.37 | 40.22 | 94.87 | −0.113 |
| DcTPS 25 | 504 | 58.97 | 5.29 | 39.83 | 93.27 | −0.221 |
| DcTPS 26 | 486 | 56.69 | 5.19 | 42.22 | 97.08 | −0.163 |
| DcTPS 27 | 550 | 64.78 | 5.65 | 49.26 | 95.20 | −0.216 |
| DcTPS 28 | 446 | 52.78 | 5.54 | 46.71 | 94.01 | −0.238 |
| DcTPS 29 | 499 | 58.93 | 5.70 | 44.76 | 98.48 | −0.156 |
| DcTPS 30 | 923 | 105.17 | 6.13 | 45.04 | 93.69 | −0.084 |
| DcTPS 31 | 360 | 41.82 | 9.79 | 57.04 | 83.17 | −0.480 |
| DcTPS 32 | 778 | 91.89 | 5.98 | 44.67 | 89.29 | −0.355 |
| DcTPS 33 | 598 | 70.24 | 5.91 | 42.60 | 91.84 | −0.270 |
| DcTPS 34 | 617 | 72.14 | 5.86 | 51.28 | 93.42 | −0.243 |
| DcTPS 35 | 539 | 63.33 | 5.51 | 46.90 | 99.83 | −0.194 |
| DcTPS 36 | 549 | 64.36 | 5.69 | 38.12 | 92.66 | −0.397 |
| DcTPS 37 | 509 | 59.57 | 5.54 | 39.41 | 86.07 | −0.300 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Gong, J.; Nong, R.; Liu, Q.; Xia, K.; Qiu, S.; Wang, Z. Terpene Synthase (TPS) Family Member Identification and Expression Pattern Analysis in Flowers of Dendrobium chrysotoxum. Horticulturae 2025, 11, 566. https://doi.org/10.3390/horticulturae11060566
Yang Y, Gong J, Nong R, Liu Q, Xia K, Qiu S, Wang Z. Terpene Synthase (TPS) Family Member Identification and Expression Pattern Analysis in Flowers of Dendrobium chrysotoxum. Horticulturae. 2025; 11(6):566. https://doi.org/10.3390/horticulturae11060566
Chicago/Turabian StyleYang, Yanni, Jianying Gong, Rongrong Nong, Qiao Liu, Ke Xia, Shuo Qiu, and Zaihua Wang. 2025. "Terpene Synthase (TPS) Family Member Identification and Expression Pattern Analysis in Flowers of Dendrobium chrysotoxum" Horticulturae 11, no. 6: 566. https://doi.org/10.3390/horticulturae11060566
APA StyleYang, Y., Gong, J., Nong, R., Liu, Q., Xia, K., Qiu, S., & Wang, Z. (2025). Terpene Synthase (TPS) Family Member Identification and Expression Pattern Analysis in Flowers of Dendrobium chrysotoxum. Horticulturae, 11(6), 566. https://doi.org/10.3390/horticulturae11060566






